Comparative Genomics and Pathogenicity Analysis of Two Bacterial Symbionts of Entomopathogenic Nematodes: The Role of the GroEL Protein in Virulence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Maintenance of Insects and Nematodes
2.2. Bacterial Growth
2.3. Genome Collection
2.4. Genome Sequencing and Annotation
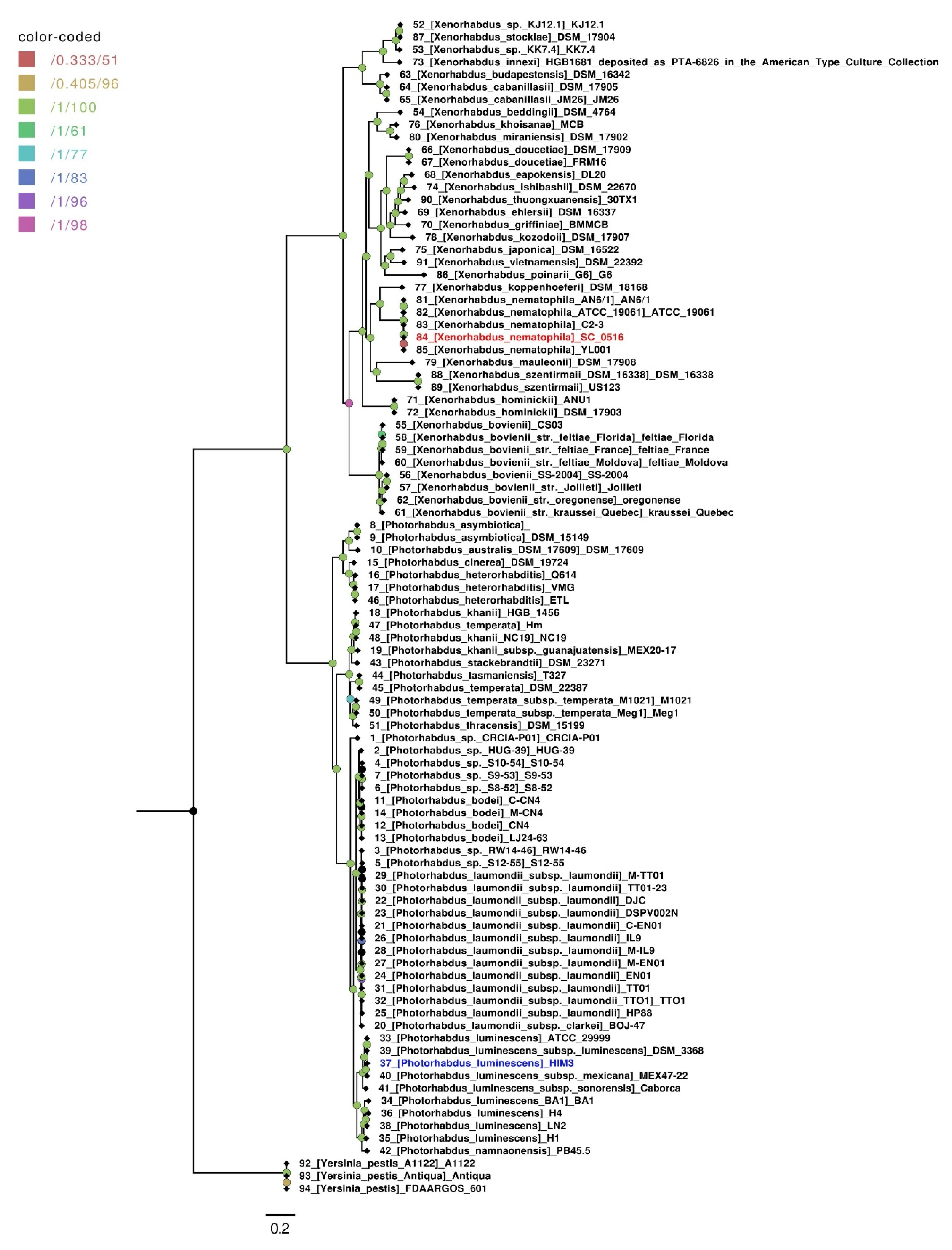
2.5. Phylogenomic Analyses
2.6. Pathogenicity Assays in Galleria mellonella
2.7. Cloning, Expression and Purification of GroEL Proteins
2.8. Evaluation of the Insecticidal Activity of GroEL Proteins
2.9. Sequence and Structure Analysis
2.10. Statistical Analysis
3. Results
3.1. Genomic Features and Assembly Metrics of the SC 0516 Strain
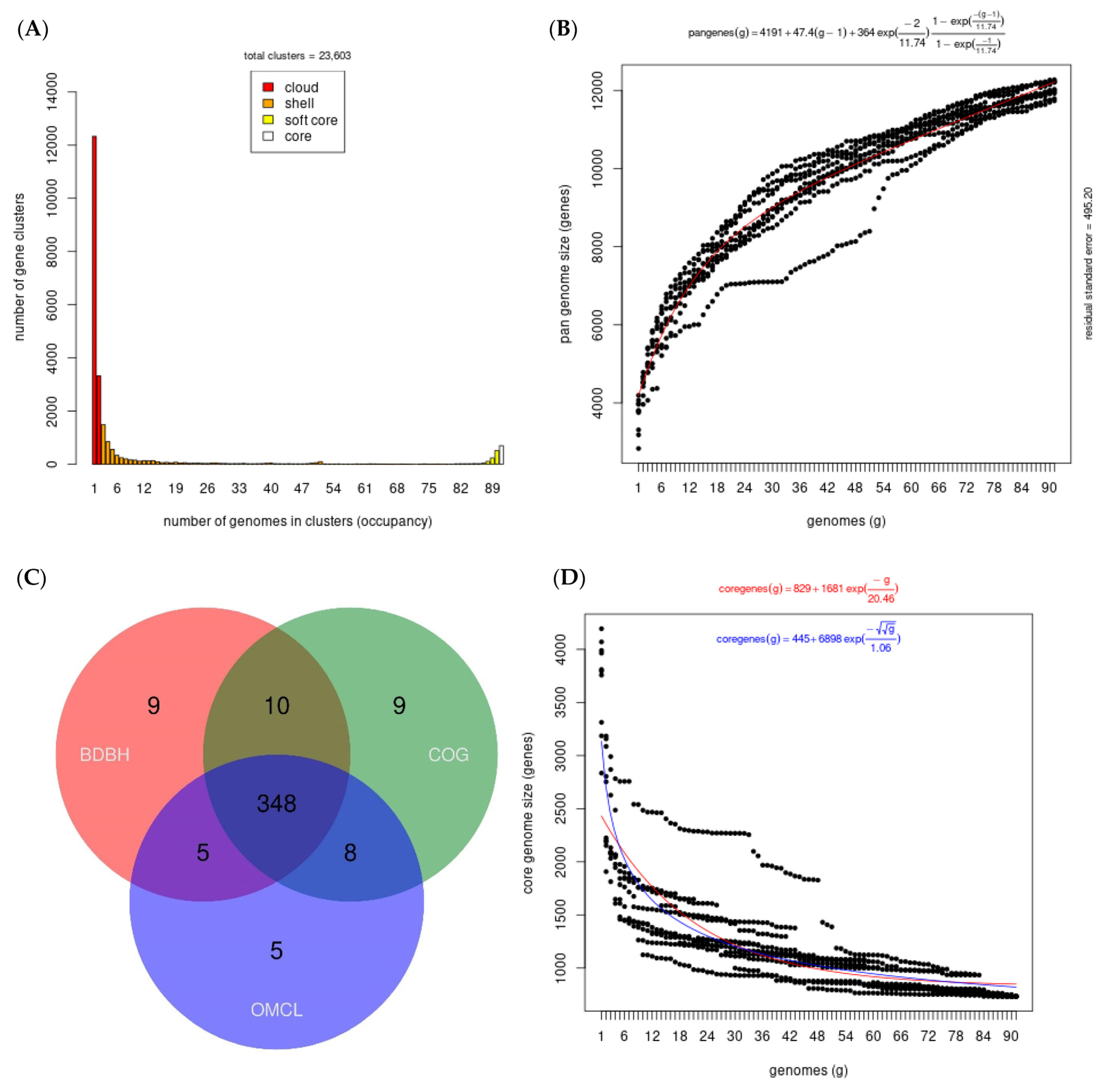
3.2. Pangenome and Phylogenetic Analysis of Xenorhabdus nematophila SC 0516
3.3. Evaluation of Pathogenicity in Galleria mellonella
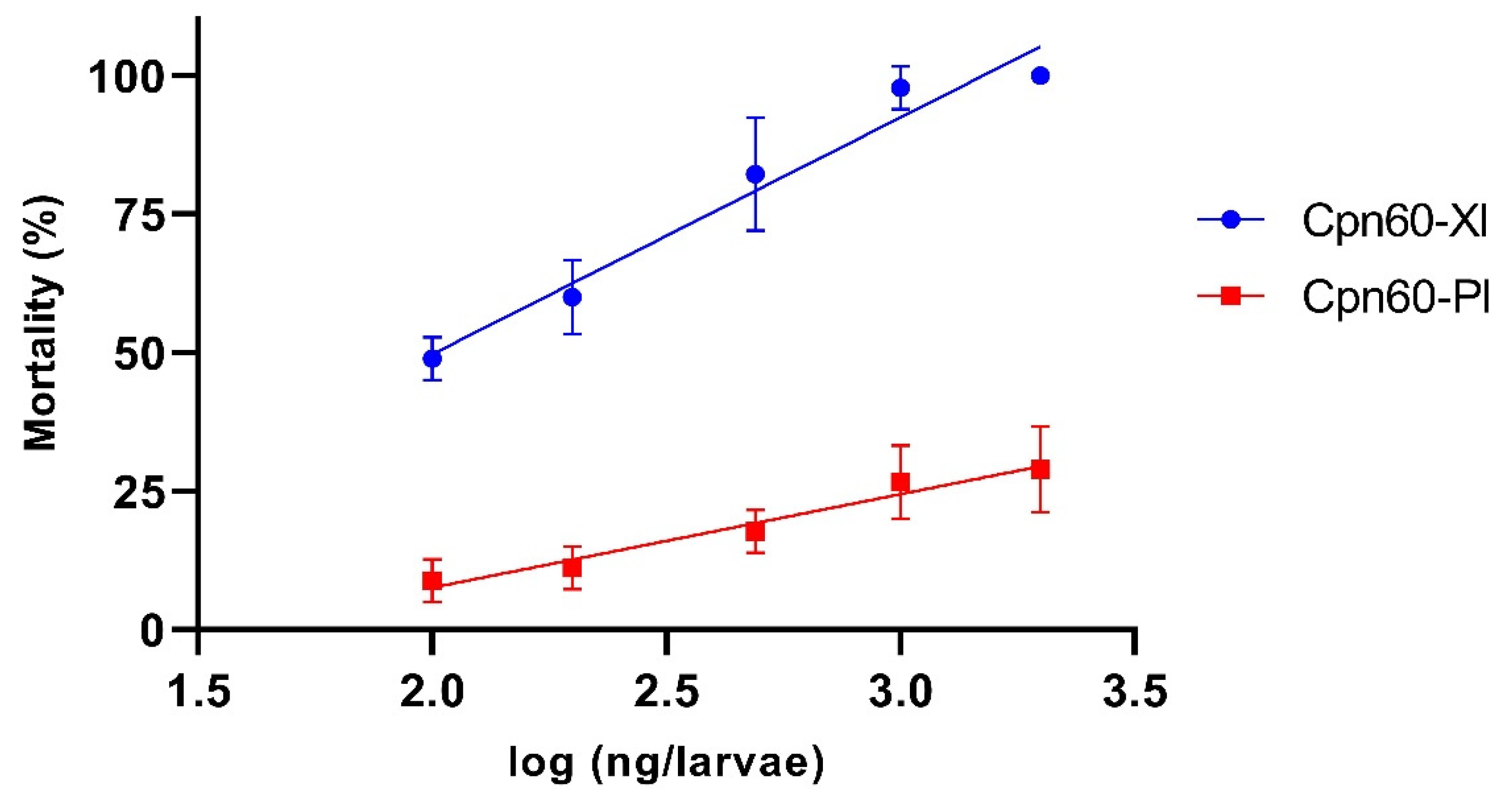
3.4. Evaluation of the Insecticidal Activity of GroEL Proteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Akhurst, R.J.; Dunphy, G.B. Tripartite interactions between symbiotically associated entomopathogenic bacteria, nematodes, and their insect hosts. In Parasites and Pathogens of Insects; Academic Press: Cambridge, MA, USA, 1993; Volume 2, pp. 1–23. [Google Scholar]
- Akhurst, R.J. Bacterial symbionts of entomopathogenic nematodes: The power behind the throne. In Nematodes and The Biological Control of Insect Pests; CSIRO: East Melbourne, Australia, 1993; pp. 127–135. [Google Scholar]
- Forst, S.; Dowds, B.; Boemare, N.; Stackebrandt, E. Xenorhabdus and Photorhabdus spp.: Bugs That Kill Bugs. Annu. Rev. Microbiol. 1997, 51, 47–72. [Google Scholar] [CrossRef] [PubMed]
- Owuama, C.I. Entomopathogenic Symbiotic Bacteria, Xenorhabdus and Photorhabdus of Nematodes. World J. Microbiol. Biotechnol. 2001, 17, 505–515. [Google Scholar] [CrossRef]
- Adams, B.J.; Fodor, A.; Koppenhöfer, H.S.; Stackebrandt, E.; Patricia Stock, S.; Klein, M.G. Reprint of “Biodiversity and Systematics of Nematode–Bacterium Entomopathogens”. Biol. Control. 2006, 38, 4–21. [Google Scholar] [CrossRef]
- Gulcu, B.; Cimen, H.; Raja, R.K.; Hazir, S. Entomopathogenic Nematodes and Their Mutualistic Bacteria: Their Ecology and Application as Microbial Control Agents. Biopestic. Int. 2017, 13, 79–112. [Google Scholar]
- Sajnaga, E.; Kazimierczak, W. Evolution and Taxonomy of Nematode-Associated Entomopathogenic Bacteria of the Genera Xenorhabdus and Photorhabdus: An Overview. Symbiosis 2020, 80, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Hinchliffe, S.J. Insecticidal Toxins from the Photorhabdus and Xenorhabdus Bacteria. TOTNJ 2013, 3, 101–118. [Google Scholar] [CrossRef]
- Poinar, G. Nematode biopesticides Fundam. Appl. Nematol. 1998, 21, 733–737. [Google Scholar]
- Goodrich-Blair, H.; Clarke, D.J. Mutualism and Pathogenesis in Xenorhabdus and Photorhabdus: Two Roads to the Same Destination. Mol. Microbiol. 2007, 64, 260–268. [Google Scholar] [CrossRef]
- Chaston, J.M.; Suen, G.; Tucker, S.L. The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: Convergent lifestyles from divergent genomes. PLoS ONE 2011, 6, e27909. [Google Scholar] [CrossRef]
- Lu, Q.-F.; Cao, D.-M.; Su, L.-L.; Li, S.-B.; Ye, G.-B.; Zhu, X.-Y.; Wang, J.-P. Genus-Wide Comparative Genomics Analysis of Neisseria to Identify New Genes Associated with Pathogenicity and Niche Adaptation of Neisseria Pathogens. Int. J. Genom. 2019, 2019, e6015730. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Gu, C.; Kim, H.U.; Lee, S.Y. Current Status of Pan-Genome Analysis for Pathogenic Bacteria. Curr. Opin. Biotechnol. 2020, 63, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J. Genome Analysis of Multiple Pathogenic Isolates of Streptococcus agalactiae: Implications for the Microbial “Pan-Genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef] [Green Version]
- Tettelin, H.; Riley, D.; Cattuto, C.; Medini, D. Comparative Genomics: The Bacterial Pan-Genome. Curr. Opin. Microbiol. 2008, 11, 472–477. [Google Scholar] [CrossRef]
- McInerney, J.O.; McNally, A.; O’Connell, M.J. Why Prokaryotes Have Pangenomes. Nat. Microbiol. 2017, 2, 17040. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.W.; Andrews, S. FastQ Screen: A Tool for Multi-Genome Mapping and Quality Control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. SolexaQA: At-a-Glance Quality Assessment of Illumina Second-Generation Sequencing Data. BMC Bioinform. 2010, 11, 485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. Spades: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Bosi, E.; Donati, B.; Galardini, M.; Brunetti, S.; Sagot, M.-F.; Lió, P.; Crescenzi, P.; Fani, R.; Fondi, M. MeDuSa: A Multi-Draft Based Scaffolder. Bioinformatics 2015, 31, 2443–2451. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and Rapid Annotation of Ribosomal RNA Genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Moreira, B.; Vinuesa, P. GET_HOMOLOGUES, a Versatile Software Package for Scalable and Robust Microbial Pangenome Analysis. Appl. Environ. Microbiol. 2013, 79, 7696–7701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinuesa, P.; Ochoa-Sánchez, L.E.; Contreras-Moreira, B. GET_PHYLOMARKERS, a Software Package to Select Optimal Orthologous Clusters for Phylogenomics and Inferring Pan-Genome Phylogenies, Used for a Critical Geno-Taxonomic Revision of the Genus Stenotrophomonas. Front. Microbiol. 2018, 9, 771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinuesa, P.; Contreras-Moreira, B. Robust Identification of Orthologues and Paralogues for Microbial Pan-Genomics Using GET_HOMOLOGUES: A Case Study of PIncA/C Plasmids. In Bacterial Pangenomics: Methods and Protocols; Mengoni, A., Galardini, M., Fondi, M., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2015; pp. 203–232. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree v1.4.4; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2010. [Google Scholar]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, Y. I-TASSER Server: New Development for Protein Structure and Function Predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Roberts, E.; Eargle, J.; Wright, D.; Luthey-Schulten, Z. MultiSeq: Unifying Sequence and Structure Data for Evolutionary Analysis. BMC Bioinform. 2006, 7, 382. [Google Scholar] [CrossRef] [Green Version]
- Treangen, T.J.; Rocha, E.P.C. Horizontal Transfer, Not Duplication, Drives the Expansion of Protein Families in Prokaryotes. PLoS Genet. 2011, 7, e1001284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brockhurst, M.A.; Harrison, E.; Hall, J.P.J.; Richards, T.; McNally, A.; MacLean, C. The Ecology and Evolution of Pangenomes. Curr. Biol. 2019, 29, R1094–R1103. [Google Scholar] [CrossRef] [PubMed]
- Brugirard-Ricaud, K.; Givaudan, A.; Parkhill, J.; Boemare, N.; Kunst, F.; Zumbihl, R.; Duchaud, E. Variation in the Effectors of the Type III Secretion System among Photorhabdus Species as Revealed by Genomic Analysis. J. Bacteriol. 2004, 186, 4376–4381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brugirard-Ricaud, K.; Duchaud, E.; Givaudan, A.; Girard, P.A.; Kunst, F.; Boemare, N.; Brehélin, M.; Zumbihl, R. Site-Specific Antiphagocytic Function of the Photorhabdus luminescens Type III Secretion System during Insect Colonization. Cell 2005, 7, 363–371. [Google Scholar] [CrossRef]
- Salgado-Morales, R.; Martínez-Ocampo, F.; Obregón-Barboza, V.; Vilchis-Martínez, K.; Jiménez-Pérez, A.; Dantán-González, E. Assessing the Pathogenicity of Two Bacteria Isolated from the Entomopathogenic Nematode Heterorhabditis indica against Galleria mellonella and Some Pest Insects. Insects 2019, 10, 83. [Google Scholar] [CrossRef] [Green Version]
- An, R.; Sreevatsan, S.; Grewal, P.S. Comparative in Vivo Gene Expression of the Closely Related Bacteria Photorhabdus temperata and Xenorhabdus koppenhoeferi upon Infection of the Same Insect Host, Rhizotrogus majalis. BMC Genom. 2009, 10, 433. [Google Scholar] [CrossRef] [Green Version]
- Cowles, K.N.; Goodrich-Blair, H. Expression and Activity of a Xenorhabdus nematophila Haemolysin Required for Full Virulence towards Manduca sexta Insects. Cell. Microbiol. 2005, 7, 209–219. [Google Scholar] [CrossRef]
- Darsouei, R.; Karimi, J.; Dunphy, G.B. Functional Characterization of Outer Membrane Proteins (OMPs) in Xenorhabdus nematophila and Photorhabdus luminescens through Insect Immune Defense Reactions. Insects 2019, 10, 352. [Google Scholar] [CrossRef] [Green Version]
- Schwechheimer, C.; Kuehn, M.J. Outer-Membrane Vesicles from Gram-Negative Bacteria: Biogenesis and Functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef] [Green Version]
- Avila-Calderón, E.D.; Ruiz-Palma, M.d.S.; Aguilera-Arreola, M.G.; Velázquez-Guadarrama, N.; Ruiz, E.A.; Gomez-Lunar, Z.; Witonsky, S.; Contreras-Rodríguez, A. Outer Membrane Vesicles of Gram-Negative Bacteria: An Outlook on Biogenesis. Front. Microbiol. 2021, 12, 557902. [Google Scholar] [CrossRef]
- Khandelwal, P.; Banerjee-Bhatnagar, N. Insecticidal Activity Associated with the Outer Membrane Vesicles of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 2003, 69, 2032–2037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khandelwal, P.; Bhatnagar, R.; Choudhury, D.; Banerjee, N. Characterization of a Cytotoxic Pilin Subunit of Xenorhabdus nematophila. Biochem. Biophys. Res. Commun. 2004, 314, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, N.; Oeda, K.; Watanabe, E.; Mikami, T.; Fukita, Y.; Nishimura, K.; Komai, K.; Matsuda, K. Chaperonin Turned Insect Toxin. Nature 2001, 411, 44. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.C.; Sharma, A.; Kant, S.; Birah, A.; Gupta, G.P.; Khan, S.R.; Bhatnagar, R.; Banerjee, N. An Insecticidal GroEL Protein with Chitin Binding Activity from Xenorhabdus nematophila. J. Biol. Chem. 2008, 283, 28287–28296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Zeng, H.-M.; Lin, H.-F.; Yang, X.-F.; Liu, Z.; Guo, L.-H.; Yuan, J.-J.; Qiu, D.-W. An Insecticidal Protein from Xenorhabdus budapestensis That Results in Prophenoloxidase Activation in the Wax Moth, Galleria mellonella. J. Invertebr. Pathol. 2012, 110, 60–67. [Google Scholar] [CrossRef]
- Shi, H.; Zeng, H.; Yang, X.; Zhao, J.; Chen, M.; Qiu, D. An Insecticidal Protein from Xenorhabdus ehlersii Triggers Prophenoloxidase Activation and Hemocyte Decrease in Galleria mellonella. Curr. Microbiol. 2012, 64, 604–610. [Google Scholar] [CrossRef]
- Guo, L.; Fatig, R.O.; Orr, G.L.; Schafer, B.W.; Strickland, J.A.; Sukhapinda, K.; Woodsworth, A.T.; Petell, J.K. Photorhabdus luminescens W-14 Insecticidal Activity Consists of at Least Two Similar but Distinct Proteins: Purification and characterization of toxin a and toxin b. J. Biol. Chem. 1999, 274, 9836–9842. [Google Scholar] [CrossRef] [Green Version]

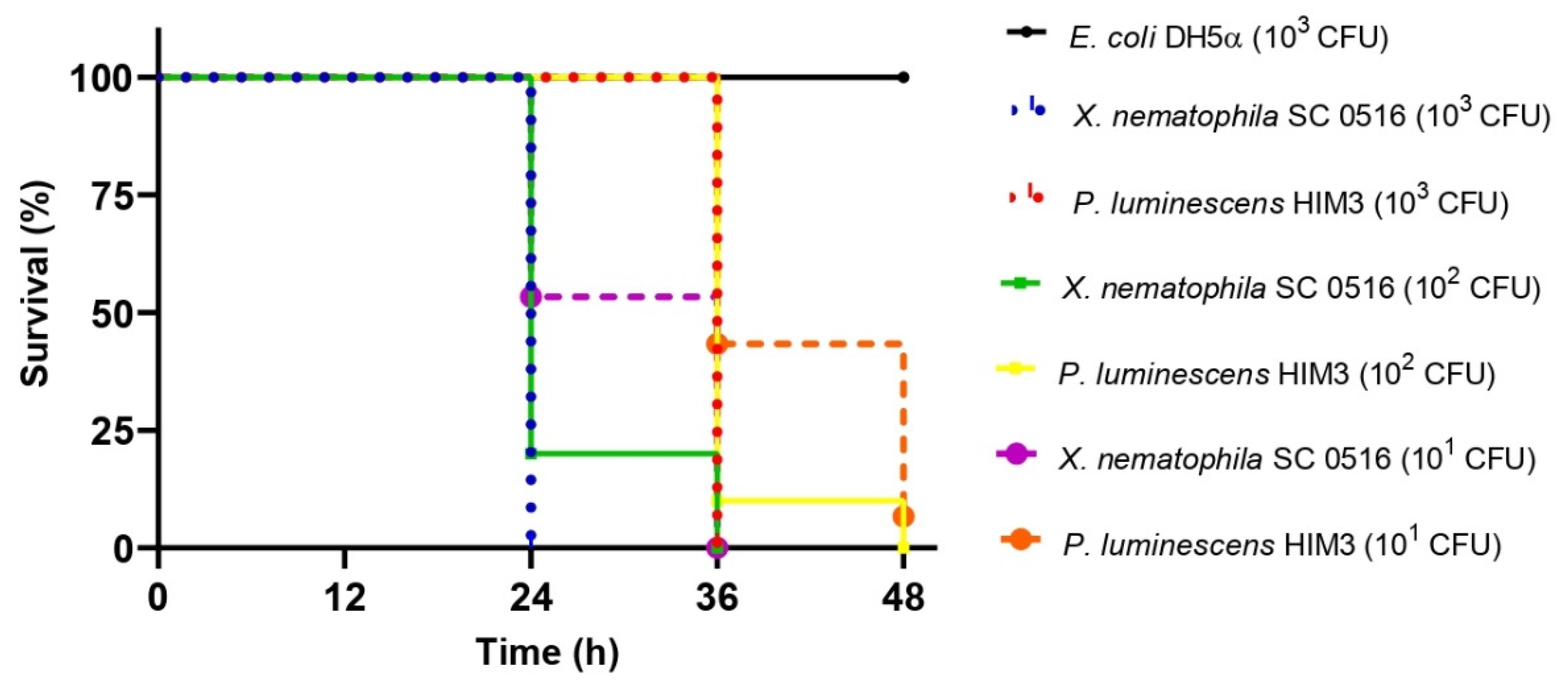

| Strain | 101 CFU | 102 CFU | 103 CFU |
|---|---|---|---|
| E. coli DH5α | - | 48 A | |
| P. luminescens HIM3 | 41 ± 0.771 A | 37.2 ± 0.469 A | 36 B |
| X. nematophila SC 0516 | 30. 4 ± 0.779 B | 26.4 ± 0.625 B | 24 C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rivera-Ramírez, A.; Salgado-Morales, R.; Jiménez-Pérez, A.; Pérez-Martínez, R.; García-Gómez, B.I.; Dantán-González, E. Comparative Genomics and Pathogenicity Analysis of Two Bacterial Symbionts of Entomopathogenic Nematodes: The Role of the GroEL Protein in Virulence. Microorganisms 2022, 10, 486. https://doi.org/10.3390/microorganisms10030486
Rivera-Ramírez A, Salgado-Morales R, Jiménez-Pérez A, Pérez-Martínez R, García-Gómez BI, Dantán-González E. Comparative Genomics and Pathogenicity Analysis of Two Bacterial Symbionts of Entomopathogenic Nematodes: The Role of the GroEL Protein in Virulence. Microorganisms. 2022; 10(3):486. https://doi.org/10.3390/microorganisms10030486
Chicago/Turabian StyleRivera-Ramírez, Abraham, Rosalba Salgado-Morales, Alfredo Jiménez-Pérez, Rebeca Pérez-Martínez, Blanca Inés García-Gómez, and Edgar Dantán-González. 2022. "Comparative Genomics and Pathogenicity Analysis of Two Bacterial Symbionts of Entomopathogenic Nematodes: The Role of the GroEL Protein in Virulence" Microorganisms 10, no. 3: 486. https://doi.org/10.3390/microorganisms10030486








