Transcriptomic Stress Response in Streptococcus mutans following Treatment with a Sublethal Concentration of Chlorhexidine Digluconate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chlorhexidine Digluconate
2.2. Bacterial Culture and Treatment
2.3. RNA Extraction, Library Preparation and RNA Sequencing
2.4. RNA Sequencing Data Analysis
2.5. Primer Design and qRT-PCR
3. Results
3.1. Sublethal Concentration of CHX
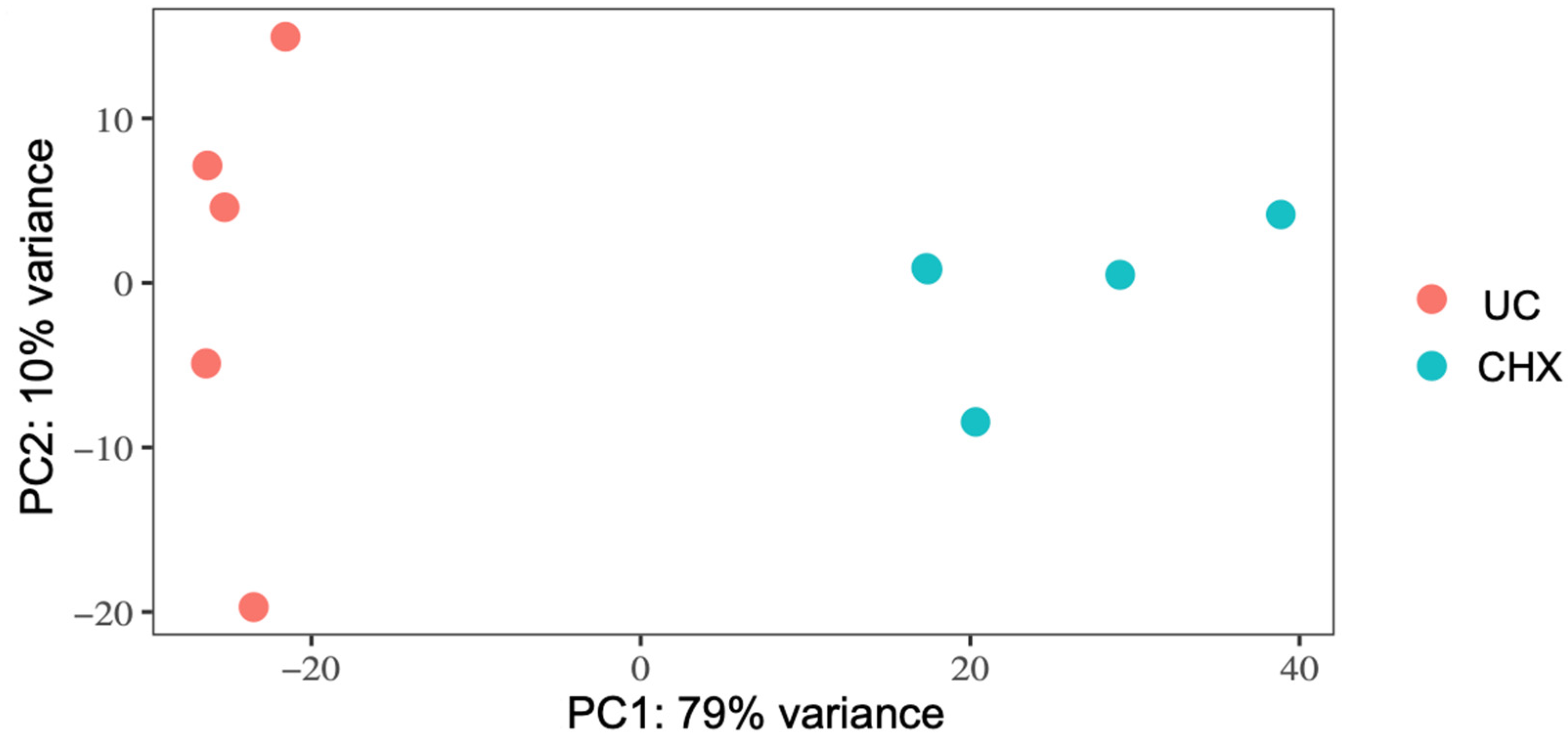
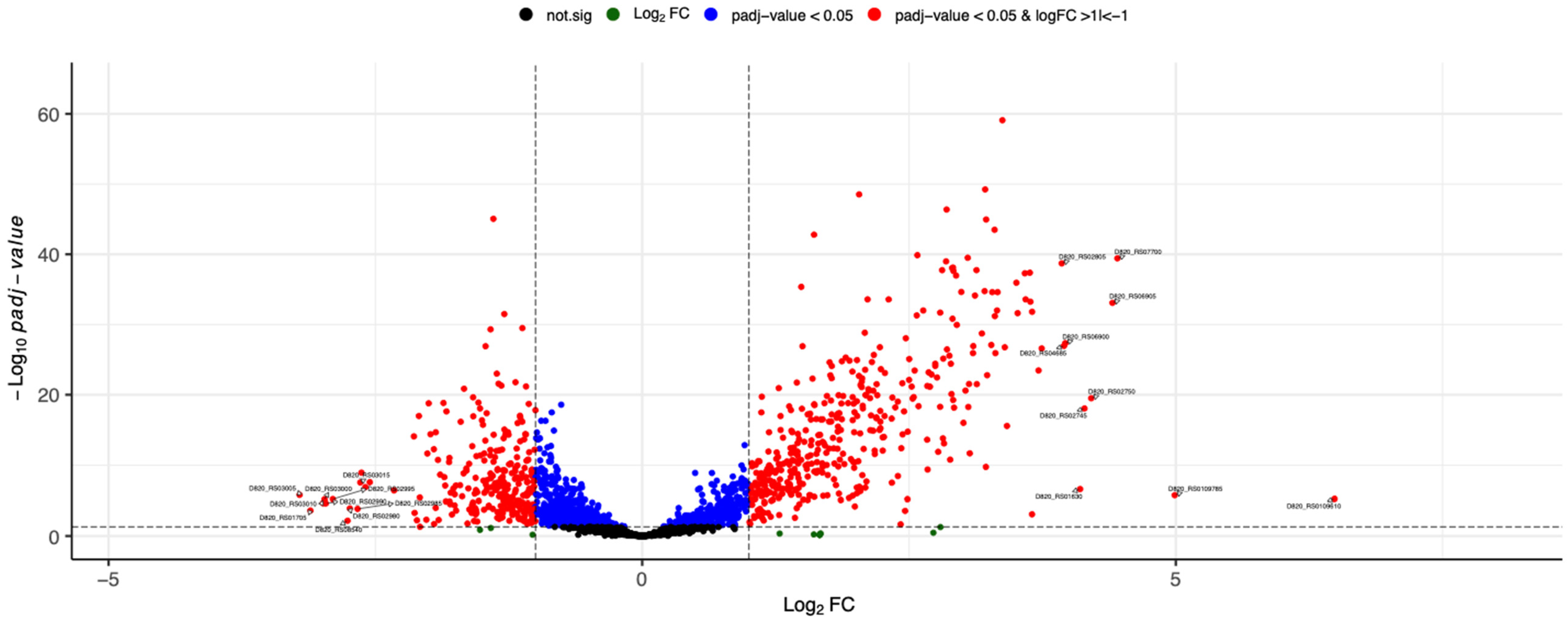
3.2. RNA-Seq and Analysis of Differentially Expressed Genes
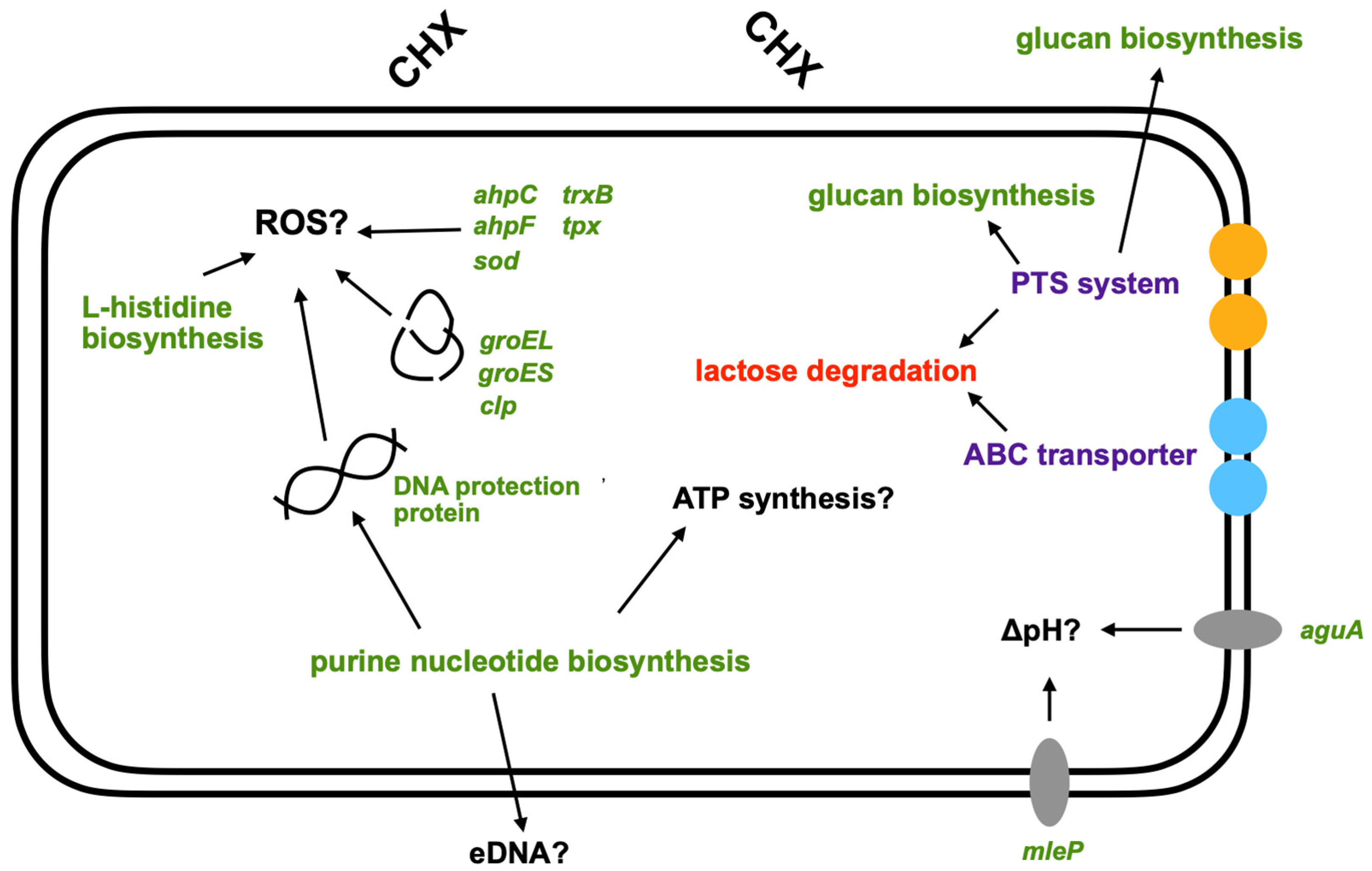
3.3. Pathway Enrichment Analysis with Differentially Expressed Genes
3.4. Validation of RNA-Seq Data Using qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tacconelli, E.; Pezzani, M.D. Public health burden of antimicrobial resistance in Europe. Lancet Infect. Dis. 2019, 19, 4–6. [Google Scholar] [CrossRef] [Green Version]
- Knight, G.M.; Glover, R.E.; McQuaid, C.F.; Olaru, I.D.; Gallandat, K.; Leclerc, O.J.; Fuller, N.M.; Willcocks, S.J.; Hasan, R.; van Kleef, E.; et al. Antimicrobial resistance and COVID-19: Intersections and implications. Elife 2021, 10, e64139. [Google Scholar] [CrossRef] [PubMed]
- OECD. Stemming the Superbug Tide; OECD: Paris, France, 2018. [Google Scholar]
- Cieplik, F.; Jakubovics, N.S.; Buchalla, W.; Maisch, T.; Hellwig, E.; Al-Ahmad, A. Resistance Toward Chlorhexidine in Oral Bacteria-Is There Cause for Concern? Front. Microbiol. 2019, 10, 587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, X.; Auer, D.L.; Buchalla, W.; Hiller, K.A.; Maisch, T.; Hellwig, E.; Al-Ahmad, A.; Cieplik, F. Cetylpyridinium Chloride: Mechanism of Action, Antimicrobial Efficacy in Biofilms, and Potential Risks of Resistance. Antimicrob. Agents Chemother. 2020, 64, e00576-20. [Google Scholar] [CrossRef]
- Kampf, G. Acquired resistance to chlorhexidine-is it time to establish an ‘antiseptic stewardship’ initiative? J. Hosp. Infect. 2016, 94, 213–227. [Google Scholar] [CrossRef]
- Venter, H.; Henningsen, M.L.; Begg, S.L. Antimicrobial resistance in healthcare, agriculture and the environment: The biochemistry behind the headlines. Essays Biochem. 2017, 61, 1–10. [Google Scholar] [CrossRef]
- Davies, G.E.; Francis, J.; Martin, A.R.; Rose, F.L.; Swain, G. 1:6-Di-4′-chlorophenyldiguanidohexane (hibitane); laboratory investigation of a new antibacterial agent of high potency. Br. J. Pharmacol. Chemother. 1954, 9, 192–196. [Google Scholar] [CrossRef]
- Gjermo, P. Chlorhexidine in dental practice. J. Clin. Periodontol. 1974, 1, 143–152. [Google Scholar] [CrossRef]
- Jones, C.G. Chlorhexidine: Is it still the gold standard? Periodontol 2000 1997, 15, 55–62. [Google Scholar] [CrossRef]
- Van der Weijden, F.A.; Van der Sluijs, E.; Ciancio, S.G.; Slot, D.E. Can Chemical Mouthwash Agents Achieve Plaque/Gingivitis Control? Dent. Clin. N. Am. 2015, 59, 799–829. [Google Scholar] [CrossRef] [Green Version]
- Pithon, M.M.; Sant’Anna, L.I.; Baião, F.C.; dos Santos, R.L.; Coqueiro Rda, S.; Maia, L.C. Assessment of the effectiveness of mouthwashes in reducing cariogenic biofilm in orthodontic patients: A systematic review. J. Dent. 2015, 43, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Solderer, A.; Kaufmann, M.; Hofer, D.; Wiedemeier, D.; Attin, T.; Schmidlin, P.R. Efficacy of chlorhexidine rinses after periodontal or implant surgery: A systematic review. Clin. Oral Investig. 2019, 23, 21–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanz, M.; Serrano, J.; Iniesta, M.; Santa Cruz, I.; Herrera, D. Antiplaque and antigingivitis toothpastes. Monogr. Oral Sci. 2013, 23, 27–44. [Google Scholar]
- Cieplik, F.; Kara, E.; Muehler, D.; Enax, J.; Hiller, K.A.; Maisch, T.; Buchalla, W. Antimicrobial efficacy of alternative compounds for use in oral care toward biofilms from caries-associated bacteria in vitro. Microbiologyopen 2019, 8, e00695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marui, V.C.; Souto, M.L.S.; Rovai, E.S.; Romito, G.A.; Chambrone, L.; Pannuti, C.M. Efficacy of preprocedural mouthrinses in the reduction of microorganisms in aerosol: A systematic review. J. Am. Dent. Assoc. 2019, 150, 1015–1026.e1. [Google Scholar] [CrossRef] [PubMed]
- Arweiler, N.B.; Boehnke, N.; Sculean, A.; Hellwig, E.; Auschill, T.M. Differences in efficacy of two commercial 0.2% chlorhexidine mouthrinse solutions: A 4-day plaque re-growth study. J. Clin. Periodontol. 2006, 33, 334–339. [Google Scholar] [CrossRef]
- Auschill, T.M.; Hein, N.; Hellwig, E.; Follo, M.; Sculean, A.; Arweiler, N.B. Effect of two antimicrobial agents on early in situ biofilm formation. J. Clin. Periodontol. 2005, 32, 147–152. [Google Scholar] [CrossRef]
- Zaura-Arite, E.; van Marle, J.; ten Cate, J.M. Confocal Microscopy Study of Undisturbed and Chlorhexidine-treated Dental Biofilm. J. Dent. Res. 2001, 80, 1436–1440. [Google Scholar] [CrossRef]
- Schwarz, S.R.; Hirsch, S.; Hiergeist, A.; Kirschneck, C.; Muehler, D.; Hiller, K.A.; Maisch, T.; Al-Ahmad, A.; Gessner, A.; Buchalla, W.; et al. Limited antimicrobial efficacy of oral care antiseptics in microcosm biofilms and phenotypic adaptation of bacteria upon repeated exposure. Clin. Oral Investig. 2021, 25, 2939–2950. [Google Scholar] [CrossRef]
- Jakubovics, N.S.; Goodman, S.D.; Mashburn-Warren, L.; Stafford, G.P.; Cieplik, F. The dental plaque biofilm matrix. Periodontol. 2000 2021, 86, 32–56. [Google Scholar] [CrossRef]
- Kitagawa, H.; Izutani, N.; Kitagawa, R.; Maezono, H.; Yamaguchi, M.; Imazato, S. Evolution of resistance to cationic biocides in Streptococcus mutans and Enterococcus faecalis. J. Dent. 2016, 47, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Wand, M.E.; Bock, L.J.; Bonney, L.C.; Sutton, J.M. Mechanisms of Increased Resistance to Chlorhexidine and Cross-Resistance to Colistin following Exposure of Klebsiella pneumoniae Clinical Isolates to Chlorhexidine. Antimicrob. Agents Chemother. 2017, 61, e01162-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verspecht, T.; Rodriguez Herrero, E.; Khodaparast, L.; Khodaparast, L.; Boon, N.; Bernaerts, K.; Quirynen, M.; Teughels, W. Development of antiseptic adaptation and cross-adapatation in selected oral pathogens in vitro. Sci. Rep. 2019, 9, 8326. [Google Scholar] [CrossRef]
- Laumen, J.G.E.; Van Dijck, C.; Manoharan-Basil, S.S.; Abdellati, S.; De Baetselier, I.; Cuylaerts, V.; De Block, T.; Van den Bossche, D.; Xavier, B.B.; Malhotra-Kumar, S.; et al. Sub-Inhibitory Concentrations of Chlorhexidine Induce Resistance to Chlorhexidine and Decrease Antibiotic Susceptibility in Neisseria gonorrhoeae. Front. Microbiol. 2021, 12, 776909. [Google Scholar] [CrossRef] [PubMed]
- Lemos, J.A.; Palmer, S.R.; Zeng, L.; Wen, Z.T.; Kajfasz, J.K.; Freires, I.A.; Abranches, J.; Brady, L.J. The Biology of Streptococcus mutans. Microbiol. Spectr. 2019, 7, GPP3-0051-2018. [Google Scholar] [CrossRef] [PubMed]
- Lemos, J.A.; Burne, R.A. A model of efficiency: Stress tolerance by Streptococcus mutans. Microbiology 2008, 154 Pt 11, 3247–3255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miles, A.A.; Misra, S.S.; Irwin, J.O. The estimation of the bactericidal power of the blood. J. Hyg. 1938, 38, 732–749. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC. A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 22 January 2022).
- Krueger, F. Trim Galore: A Wrapper Tool around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files, with Some Extra Functionality for MspI-Digested RRBS-Type (Reduced Representation Bisufite-Seq) Libraries. 2012. Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 22 January 2022).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2012, 29, 15–21. [Google Scholar] [CrossRef]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q.; et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief Bioinform. 2019, 20, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Pedley, A.M.; Benkovic, S.J. A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem. Sci. 2017, 42, 141–154. [Google Scholar] [CrossRef] [Green Version]
- Thomas, V.C.; Thurlow, L.R.; Boyle, D.; Hancock, L.E. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 2008, 190, 5690–5698. [Google Scholar] [CrossRef] [Green Version]
- Dengler, V.; Foulston, L.; DeFrancesco, A.S.; Losick, R. An electrostatic net model for the role of extracellular DNA in biofilm formation by Staphylococcus aureus. J. Bacteriol. 2015, 197, 3779–3787. [Google Scholar] [CrossRef] [Green Version]
- Fahmi, T.; Faozia, S.; Port, G.C.; Cho, K.H. The second messenger c-di-AMP regulates diverse cellular pathways involved in stress response, biofilm formation, cell wall homeostasis, SpeB expression, and virulence in Streptococcus pyogenes. Infect. Immun. 2019, 87, e00147-19. [Google Scholar] [CrossRef] [Green Version]
- Gélinas, M.; Museau, L.; Milot, A.; Beauregard, P.B. Cellular adaptation and the importance of the purine biosynthesis pathway during biofilm formation in Gram-positive pathogens. bioRxiv 2020, 2020. [Google Scholar] [CrossRef]
- Mongodin, E.; Finan, J.; Climo, M.W.; Rosato, A.; Gill, S.; Archer, G.L. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 2003, 185, 4638–4643. [Google Scholar] [CrossRef] [Green Version]
- Yee, R.; Cui, P.; Shi, W.; Feng, J.; Zhang, Y. Genetic Screen Reveals the Role of Purine Metabolism in Staphylococcus aureus Persistence to Rifampicin. Antibiotics 2015, 4, 627–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, A.L.; Dassa, E.; Orelle, C.; Chen, J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 2008, 72, 317–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraburtty, K. Translational regulation by ABC systems. Res. Microbiol. 2001, 152, 391–399. [Google Scholar] [CrossRef]
- Goosen, N.; Moolenaar, G.F. Role of ATP hydrolysis by UvrA and UvrB during nucleotide excision repair. Res. Microbiol. 2001, 152, 401–409. [Google Scholar] [CrossRef]
- Hassan, K.A.; Jackson, S.M.; Penesyan, A.; Patching, S.G.; Tetu, S.G.; Eijkelkamp, B.A.; Brown, M.H.; Henderson, P.J.F.; Paulsen, I.T. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 20254–20259. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Zhang, J.; Guo, L.; Zhao, W.; Hu, X.; Wei, X. Inactivation of a putative efflux pump (LmrB) in Streptococcus mutans results in altered biofilm structure and increased exopolysaccharide synthesis: Implications for biofilm resistance. Biofouling 2017, 33, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Hassan, K.A.; Liu, Q.; Henderson, P.J.F.; Paulsen, I.T. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. mBio 2015, 6, e01982-14. [Google Scholar] [CrossRef] [Green Version]
- Mombeshora, M.; Mukanganyama, S. Development of an accumulation assay and evaluation of the effects of efflux pump inhibitors on the retention of chlorhexidine digluconate in Pseudomonas aeruginosa and Staphylococcus aureus. BMC Res. Notes 2017, 10, 328. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Guo, L.; Lui, J.; Zhang, J.; Zeng, H.; Ning, Y.; Wei, Y. Identification of an Efflux Transporter LmrB Regulating Stress Response and Extracellular Polysaccharide Synthesis in Streptococcus mutans. Front. Microbiol. 2017, 8, 962. [Google Scholar] [CrossRef] [Green Version]
- Vadeboncoeur, C.; Pelletier, M. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol. Rev. 1997, 19, 187–207. [Google Scholar] [CrossRef]
- Deutscher, J.; Francke, C.; Postma, P.W. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 2006, 70, 939–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nobre dos Santos, M.; Melo dos Santos, L.; Francisco, S.B.; Cury, J.A. Relationship among dental plaque composition, daily sugar exposure and caries in the primary dentition. Caries Res. 2002, 36, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Niu, Y.; Zhou, X.; Zheng, X.; Wang, S.; Guo, Q.; Li, Y.; Li, M.; Li, J.; Yang, Y.; et al. Streptococcus mutans copes with heat stress by multiple transcriptional regulons modulating virulence and energy metabolism. Sci. Rep. 2015, 5, 12929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busuioc, M.; Mackiewicz, K.; Buttaro, B.A.; Piggot, P.J. Role of intracellular polysaccharide in persistence of Streptococcus mutans. J. Bacteriol. 2009, 191, 7315–7322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colby, S.M.; Russell, R.R. Sugar metabolism by mutans streptococci. Soc. Appl. Bacteriol. Symp. Ser. 1997, 26, 80s–88s. [Google Scholar] [CrossRef] [PubMed]
- Rainey, K.; Michalek, S.M.; Wen, Z.T.; Wu, H. Glycosyltransferase-Mediated Biofilm Matrix Dynamics and Virulence of Streptococcus mutans. Appl. Environ. Microbiol. 2019, 85, e02247-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, L.; Tong, Z.; Linghu, D.; Lin, Y.; Tao, R.; Liu, J.; Tian, Y.; Ni, L. Effects of sub-minimum inhibitory concentrations of antimicrobial agents on Streptococcus mutans biofilm formation. Int. J. Antimicrob. Agents 2012, 39, 390–395. [Google Scholar] [CrossRef]
- Kajfasz, J.K.; Ganguly, T.; Hardin, E.L.; Abranches, J.; Lemos, J.A. Transcriptome responses of Streptococcus mutans to peroxide stress: Identification of novel antioxidant pathways regulated by Spx. Sci. Rep. 2017, 7, 16018. [Google Scholar] [CrossRef]
- Cabiscol, E.; Tamarit, J.; Ros, J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 2000, 3, 3–8. [Google Scholar]
- Imlay, J.A. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef] [Green Version]
- Lemos, J.A.; Burne, R.A. Regulation and Physiological Significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 2002, 184, 6357–6366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, X.H.; Zhang, J.Q.; Song, X.Y.; Ma, X.B.; Zhang, S.Y. Contribution of ClpP to stress tolerance and virulence properties of Streptococcus mutans. J. Basic Microbiol. 2014, 54, 1222–1232. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.M.; Ten Cate, J.M.; Crielaard, W. The adaptive response of Streptococcus mutans towards oral care products: Involvement of the ClpP serine protease. Eur. J. Oral Sci. 2007, 115, 363–370. [Google Scholar] [CrossRef]
- Svensäter, G.; Larsson, U.B.; Greif, E.C.; Cvitkovitch, D.G.; Hamilton, I.R. Acid tolerance response and survival by oral bacteria. Oral Microbiol. Immunol. 1997, 12, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Burne, R.A.; Marquis, R.E. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol. Lett. 2000, 193, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Baldeck, J.D.; Nguyen, P.T.M.; Quivey, R.G., Jr.; Marquis, R.E. Alkali production associated with malolactic fermentation by oral streptococci and protection against acid, oxidative, or starvation damage. Can. J. Microbiol. 2010, 56, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Marquis, R.E. Malolactic fermentation by Streptococcus mutans. FEMS Microbiol. Lett. 2007, 272, 196–201. [Google Scholar] [CrossRef] [Green Version]
- Simon, J.P.; Stalon, V. Enzymes of agmatine degradation and the control of their synthesis in Streptococcus faecalis. J. Bacteriol. 1982, 152, 676–681. [Google Scholar] [CrossRef]
- Griswold, A.R.; Jameson-Lee, M.; Burne, R.A. Regulation and physiologic significance of the agmatine deiminase system of Streptococcus mutans UA159. J. Bacteriol. 2006, 188, 834–841. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
| Target Gene | Sequence 5′-3′ (F = Forward; R = Reverse) | Product Size |
|---|---|---|
| gyrB | F: GCACAAGAGTACGATGCCAGT | 119 bp |
| R: TCCCAAACAAGGTGATGCAGC | ||
| D820_RS03005 | F: CGTGGTTATCAAGTATCGTGTGA | 148 bp |
| R: AAAGAATTGGTCCTGCATCCA | ||
| D820_RS09005 | F: CAGTAGGTGCCGCTCAAACT | 128 bp |
| R: AAGTCCGCCGCCAAACATAT | ||
| glgA | F: GTGCCTTGCCCAAATCCCTT | 145 bp |
| R: ACATATTCACGACGCCAGCC | ||
| ssrS | F: CGGAAGCAACTAAAGTCAGAGCG | 80 bp |
| R: TGGCACCGATGATTCACGTT |
| Gene ID | Gene Name | Product | Log2 FC | padj |
|---|---|---|---|---|
| D820_RS03005 | NA | PTS fructose transporter subunit IIB | −3.2 | 1.60 × 10−6 |
| D820_RS01705 | NA | tRNA-Thr | −3.1 | 2.46 × 10−4 |
| D820_RS03010 | NA | PTS mannitol/fructose, IIC component | −3.0 | 2.26 × 10−5 |
| D820_RS03000 | NA | PTS mannitol transporter subunit IIB | −3.0 | 6.57 × 10−6 |
| D820_RS02990 | lacC | Tagatose-6-phosphate kinase | −3.0 | 2.40 × 10−5 |
| D820_RS02995 | lacD | Tagatose-bisphosphate aldolase | −2.9 | 6.31 × 10−6 |
| D820_RS08540 | NA | tRNA-Ser | −2.8 | 6.90 × 10−3 |
| D820_RS02980 | lacA | Galactose-6-phosphate isomerase | −2.7 | 1.24 × 10−4 |
| D820_RS02985 | lacB | Galactose-6-phosphate isomerase | −2.7 | 1.43 × 10−4 |
| D820_RS03015 | NA | Lactose-specific phosphotransferase enzyme IIA component | −2.6 | 2.85 × 10−8 |
| D820_RS02805 | glgA | Glycogen synthase | 3.9 | 0.00 |
| D820_RS04685 | NA | ABC transporter (ATP-binding protein) | 4.0 | 0.00 |
| D820_RS06900 | NA | Hypothetical protein | 4.0 | 0.00 |
| D820_RS01630 | NA | 16S ribosomal RNA | 4.1 | 2.44 × 10−7 |
| D820_RS02745 | NA | ABC transporter (ATP-binding protein) | 4.1 | 0.00 |
| D820_RS02750 | NA | ABC transporter permease | 4.2 | 0.00 |
| D820_RS06905 | lrgB | Antiholin | 4.4 | 0.00 |
| D820_RS07700 | pflB | Formate C-acetyltransferase | 4.5 | 0.00 |
| D820_RS0109785 | NA | 23S ribosomal RNA | 5.0 | 1.79 × 10−6 |
| D820_RS0109610 | NA | 23S ribosomal RNA | 6.5 | 5.73 × 10−6 |
| Gene ID | Gene Name | Product | Log2 FC | padj |
|---|---|---|---|---|
| D820_RS06120 | ahpC | Peroxiredoxin | 1.1 | 1.50 × 10−6 |
| D820_RS06115 | ahpF | Alkyl hydroperoxide reductase subunit F | 1.8 | 6.00 × 10−17 |
| D820_RS06685 | sod | Superoxide dismutase | 1.6 | 1.71 × 10−6 |
| D820_RS07435 | trxB | Thioredoxin disulfide reductase | 1.1 | 5.15 × 10−4 |
| D820_RS05415 | tpx | 2-Cys-peroxiredoxine | 1.0 | 1.02 × 10−2 |
| D820_RS07050 | NA | DNA protection protein | 1.9 | 2.41 × 10−12 |
| D820_RS00990 | groES | Co-Chaperone GroES | 1.1 | 8.15 × 10−6 |
| D820_RS00995 | groEL | Chaperone GroEL | 1.5 | 0.00 |
| D820_RS05270 | NA | ATP-dependent Clp protease ATP-binding subunit | 1.1 | 1.04 × 10−7 |
| D820_RS03310 | clpB | Clp proteinase ATP-binding subunit ClpB | 2.4 | 4.29 × 10−15 |
| D820_RS06955 | NA | ATP-dependent Clp protease ATP-binding subunit | 1.4 | 3.43 × 10−10 |
| D820_RS08310 | aguA | Agmatine deiminase | 1.3 | 1.27 × 10−14 |
| D820_RS08960 | mleP | Malate permease | 3.7 | 0.00 |
| Pathway | Genes Involved in Pathway (Ensemble IDs) | p-Value | |
|---|---|---|---|
| Upregulated | |||
| Carboxylate degradation | D820_RS00885, D820_RS07700, D820_RS07305, D820_RS08215, D820_RS08265, D820_RS08275, D820_RS08270, D820_RS04975, D820_RS04980, D820_RS08965 | 5.00 × 10−5 | |
| 5-Aminoimidazole ribonucleotide biosynthesis | D820_RS09340, D820_RS09355, D820_RS09335, D820_RS09300, D820_RS09345 | 2.00 × 10−3 | |
| Glycan pathway | D820_RS07040, D820_RS02810, D820_RS02685, D820_RS02800, D820_RS02795, D820_RS02790, D820_RS02805 | 3.00 × 10−3 | |
| L-ascorbate degradation | D820_RS08215, D820_RS08265, D820_RS08275, D820_RS08270 | 6.00 × 10−3 | |
| L-histidine biosynthesis | D820_RS03965, D820_RS03970, D820_RS03955, D820_RS03950, D820_RS03960, D820_RS03940, D820_RS03930 | 7.00 × 10−3 | |
| Purine nucleotide biosynthesis | D820_RS04105, D820_RS04110, D820_RS04100, D820_RS09285, D820_RS09290, D820_RS09325, D820_RS09250, D820_RS08295, D820_RS09340, D820_RS09355, D820_RS09335, D820_RS09300, D820_RS09345 | 1.00 × 10−2 | |
| Inosine-5’-phosphate biosynthesis | D820_RS09285, D820_RS09290, D820_RS09325, D820_RS09250 | 2.00 × 10−2 | |
| Downregulated | |||
| Lactose degradation | D820_RS02995, D820_RS02980, D820_RS02985, D820_RS03025, D820_RS02990 | 4.00 × 10−6 | |
| Galactose degradation | D820_RS02995, D820_RS02980, D820_RS02985, D820_RS03025, D820_RS02990 | 4.00 × 10−6 | |
| Protein modification | D820_RS05715, D820_RS02255, D820_RS08010 | 2.00 × 10−2 | |
| L-phenylalanine biosynthesis | D820_RS03740, D820_RS07095 | 2.00 × 10−2 |
| Gene | RNA-Seq | qRT-PCR |
|---|---|---|
| D820_RS03005 | −3.2 | −2.4 |
| D820_RS09005 | 3.3 | 3.4 |
| ssrS | 3.4 | 2.5 |
| glgA | 3.9 | 4.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muehler, D.; Mao, X.; Czemmel, S.; Geißert, J.; Engesser, C.; Hiller, K.-A.; Widbiller, M.; Maisch, T.; Buchalla, W.; Al-Ahmad, A.; et al. Transcriptomic Stress Response in Streptococcus mutans following Treatment with a Sublethal Concentration of Chlorhexidine Digluconate. Microorganisms 2022, 10, 561. https://doi.org/10.3390/microorganisms10030561
Muehler D, Mao X, Czemmel S, Geißert J, Engesser C, Hiller K-A, Widbiller M, Maisch T, Buchalla W, Al-Ahmad A, et al. Transcriptomic Stress Response in Streptococcus mutans following Treatment with a Sublethal Concentration of Chlorhexidine Digluconate. Microorganisms. 2022; 10(3):561. https://doi.org/10.3390/microorganisms10030561
Chicago/Turabian StyleMuehler, Denise, Xiaojun Mao, Stefan Czemmel, Janina Geißert, Christina Engesser, Karl-Anton Hiller, Matthias Widbiller, Tim Maisch, Wolfgang Buchalla, Ali Al-Ahmad, and et al. 2022. "Transcriptomic Stress Response in Streptococcus mutans following Treatment with a Sublethal Concentration of Chlorhexidine Digluconate" Microorganisms 10, no. 3: 561. https://doi.org/10.3390/microorganisms10030561
APA StyleMuehler, D., Mao, X., Czemmel, S., Geißert, J., Engesser, C., Hiller, K.-A., Widbiller, M., Maisch, T., Buchalla, W., Al-Ahmad, A., & Cieplik, F. (2022). Transcriptomic Stress Response in Streptococcus mutans following Treatment with a Sublethal Concentration of Chlorhexidine Digluconate. Microorganisms, 10(3), 561. https://doi.org/10.3390/microorganisms10030561







