Lactiplantibacillus plantarum-Derived Biosurfactant Attenuates Quorum Sensing-Mediated Virulence and Biofilm Formation in Pseudomonas aeruginosa and Chromobacterium violaceum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of Lactic Acid Bacteria
2.2. Production and Extraction of Biosurfactant
2.3. Biosurfactant Assays
2.3.1. Bacterial Strains and Growth Conditions
2.3.2. Determination of Minimum Inhibitory Concentration (MIC)
2.4. Quorum Sensing Inhibitory Activity in C. violaceum
2.4.1. Evaluation of Anti–Quorum Sensing Activity
2.4.2. Violacein Inhibition Assay
2.4.3. Quantification of Acyl Homoserine Lactones (AHLs)
2.5. Quorum Sensing Inhibitory Activity in P. aeruginosa
2.5.1. Quantitative Analysis of Pyocyanin Production in P. aeruginosa
2.5.2. LasA Staphylolytic Assay
2.5.3. LasB Elastase Assay
2.5.4. Azocasein Assay for Proteolytic Activity
2.6. Swarming Motility Assay
2.7. Extraction and Estimation of Exopolysaccharides (EPS)
2.8. Antibiofilm Assay
2.9. In Situ Visualization of Biofilms
2.10. Gas Chromatography–Mass Spectrometry (GC–MS) Analysis
2.11. Molecular Docking Analysis
2.12. Statistical Analysis
3. Results
3.1. Identification and Screening of Biosurfactant Production by Isolated LAB
3.2. Antibacterial Activity
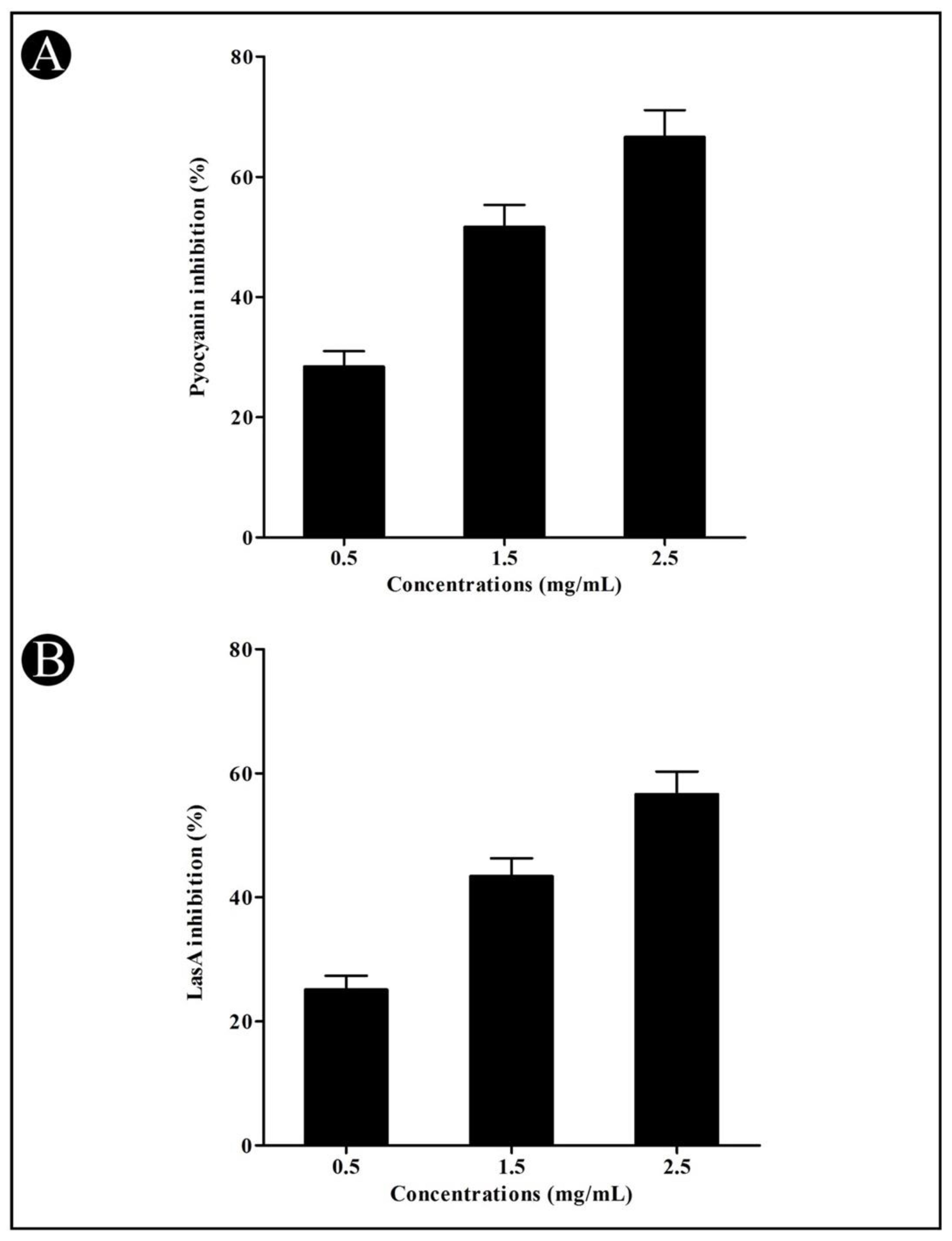
3.3. Effect of Crude L. plantarum Biosurfactant on QS-Regulated Virulence Factors
3.4. Antibiofilm Potential of Crude L. plantarum Biosurfactant
3.5. Identification of Chemical Constituents by GC–MS
3.6. Docking Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abdula, N.; Macharia, J.; Motsoaledi, A.; Swaminathan, S.; VijayRaghavan, K. National action for global gains in antimicrobial resistance. Lancet 2016, 387, e3–e5. [Google Scholar] [CrossRef]
- Adnan, M.; Alshammari, E. Significance and potential of marine microbial natural bioactive compounds against biofilms/biofouling: Necessity for green chemistry. PeerJ 2018, 6, e5049. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Ashraf, M.S.; Siddiqui, A.J.; Ashraf, S.A.; Sachidanandan, M.; Snoussi, M.; Adnan, M.; Hadi, S. Profiling and Role of Bioactive Molecules from Puntius sophore (Freshwater/Brackish Fish) Skin Mucus with Its Potent Antibacterial, Antiadhesion, and Antibiofilm Activities. Biomolecules 2020, 10, 920. [Google Scholar] [CrossRef] [PubMed]
- Adnan, M.; Patel, M.; Deshpande, S.; Alreshidi, M.; Siddiqui, A.J.; Reddy, M.N.; Emira, N.; De Feo, V. Effect of Adiantum philippense Extract on Biofilm Formation, Adhesion with Its Antibacterial Activities against Foodborne Pathogens, and Characterization of Bioactive Metabolites: An in vitro-in silico Approach. Front. Microbiol. 2020, 11, 823. [Google Scholar] [CrossRef]
- Zhao, X.; Yu, Z.; Ding, T. Quorum-Sensing Regulation of Antimicrobial Resistance in Bacteria. Microorganisms 2020, 8, 425. [Google Scholar] [CrossRef] [Green Version]
- Barzegari, A.; Kheyrolahzadeh, K.; Hosseiniyan Khatibi, S.M.; Sharifi, S.; Memar, M.Y.; Zununi Vahed, S. The Battle of Probiotics and Their Derivatives against Biofilms. Infect. Drug Resist. 2020, 13, 659–672. [Google Scholar] [CrossRef] [Green Version]
- Krzyżek, P. Challenges and Limitations of Anti-quorum Sensing Therapies. Front. Microbiol. 2019, 10, 2473. [Google Scholar] [CrossRef] [Green Version]
- Hawver, L.A.; Jung, S.A.; Ng, W.L. Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol. Rev. 2016, 40, 738–752. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Wu, R.; Zhang, J.; Shang, N.; Li, P. D-Ribose Interferes with Quorum Sensing to Inhibit Biofilm Formation of Lactobacillus paraplantarum L-ZS9. Front. Microbiol. 2017, 8, 1860. [Google Scholar] [CrossRef]
- Jiang, Q.; Chen, J.; Yang, C.; Yin, Y. Quorum Sensing: A Prospective Therapeutic Target for Bacterial Diseases. BioMed Res. Int. 2019, 2019, 2015978. [Google Scholar] [CrossRef] [Green Version]
- Alshammari, E.; Patel, M.; Sachidanandan, M.; Kumar, P.; Adnan, M. Potential evaluation and health fostering intrinsic traits of novel probiotic strain Enterococcus durans F3 isolated from the gut of fresh water fish catla catla. Food Sci. Anim. Resour. 2019, 39, 844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashraf, S.A.; Elkhalifa, A.E.O.; Ahmad, M.F.; Patel, M.; Adnan, M.; Sulieman, A.M.E. Probiotic Fermented Foods and Health Promotion. In African Fermented Food Products-New Trends; Springer: Berlin/Heidelberg, Germany, 2022; pp. 59–88. [Google Scholar]
- Victor, I.U.; Kwiencien, M.; Tripathi, L.; Cobice, D.; McClean, S.; Marchant, R.; Banat, I.M. Quorum sensing as a potential target for increased production of rhamnolipid biosurfactant in Burkholderia thailandensis E264. Appl. Microbiol. Biotechnol. 2019, 103, 6505–6517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polaske, T.J.; Gahan, C.G.; Nyffeler, K.E.; Lynn, D.M.; Blackwell, H.E. Identification of small molecules that strongly inhibit bacterial quorum sensing using a high-throughput lipid vesicle lysis assay. Cell Chem. Biol. 2022, 29, 605–614.e4. [Google Scholar] [CrossRef] [PubMed]
- Mukherji, R.; Prabhune, A. Novel glycolipids synthesized using plant essential oils and their application in quorum sensing inhibition and as antibiofilm agents. Sci. World J. 2014, 2014, 890709. [Google Scholar] [CrossRef]
- Yan, X.; Gu, S.; Cui, X.; Shi, Y.; Wen, S.; Chen, H.; Ge, J. Antimicrobial, anti-adhesive and anti-biofilm potential of biosurfactants isolated from Pediococcus acidilactici and Lactobacillus plantarum against Staphylococcus aureus CMCC26003. Microb. Pathog. 2019, 127, 12–20. [Google Scholar] [CrossRef]
- Roy, A. Review on the biosurfactants: Properties, types and its applications. J. Fundam. Renew. Energy Appl. 2017, 8, 1–14. [Google Scholar] [CrossRef]
- De Giani, A.; Zampolli, J.; Di Gennaro, P. Recent Trends on Biosurfactants with Antimicrobial Activity Produced by Bacteria Associated with Human Health: Different Perspectives on Their Properties, Challenges, and Potential Applications. Front. Microbiol. 2021, 12, 655150. [Google Scholar] [CrossRef]
- Qian, X.; Tian, P.; Zhao, J.; Zhang, H.; Wang, G.; Chen, W. Quorum Sensing of Lactic Acid Bacteria: Progress and Insights. Food Rev. Int. 2022, 1–12. [Google Scholar] [CrossRef]
- Toushik, S.H.; Kim, K.; Ashrafudoulla, M.; Mizan, M.F.R.; Roy, P.K.; Nahar, S.; Kim, Y.; Ha, S.-D. Korean kimchi-derived lactic acid bacteria inhibit foodborne pathogenic biofilm growth on seafood and food processing surface materials. Food Control 2021, 129, 108276. [Google Scholar] [CrossRef]
- Pang, X.; Song, X.; Chen, M.; Tian, S.; Lu, Z.; Sun, J.; Li, X.; Lu, Y.; Yuk, H.G. Combating biofilms of foodborne pathogens with bacteriocins by lactic acid bacteria in the food industry. Compr. Rev. Food Sci. Food Saf. 2022, 21, 1657–1676. [Google Scholar] [CrossRef]
- Patel, M.; Siddiqui, A.J.; Hamadou, W.S.; Surti, M.; Awadelkareem, A.M.; Ashraf, S.A.; Alreshidi, M.; Snoussi, M.; Rizvi, S.M.D.; Bardakci, F. Inhibition of Bacterial Adhesion and Antibiofilm Activities of a Glycolipid Biosurfactant from Lactobacillus rhamnosus with Its Physicochemical and Functional Properties. Antibiotics 2021, 10, 1546. [Google Scholar] [CrossRef] [PubMed]
- Adnan, M.; Siddiqui, A.J.; Hamadou, W.S.; Ashraf, S.A.; Hassan, M.I.; Snoussi, M.; Badraoui, R.; Jamal, A.; Bardakci, F.; Awadelkareem, A.M. Functional and Structural Characterization of Pediococcus pentosaceus-Derived Biosurfactant and Its Biomedical Potential against Bacterial Adhesion, Quorum Sensing, and Biofilm Formation. Antibiotics 2021, 10, 1371. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Husain, F.M.; Ahmad, I.; Al-Thubiani, A.S.; Abulreesh, H.H.; AlHazza, I.M.; Aqil, F. Leaf Extracts of Mangifera indica L. Inhibit Quorum Sensing—Regulated Production of Virulence Factors and Biofilm in Test Bacteria. Front. Microbiol. 2017, 8, 727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blosser, R.S.; Gray, K.M. Extraction of violacein from Chromobacterium violaceum provides a new quantitative bioassay for N-acyl homoserine lactone autoinducers. J. Microbiol. Methods 2000, 40, 47–55. [Google Scholar] [CrossRef]
- Taghadosi, R.; Shakibaie, M.R.; Masoumi, S. Biochemical detection of N-Acyl homoserine lactone from biofilm-forming uropathogenic Escherichia coli isolated from urinary tract infection samples. Rep. Biochem. Mol. Biol. 2015, 3, 56–61. [Google Scholar]
- Essar, D.W.; Eberly, L.; Hadero, A.; Crawford, I.P. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: Interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 1990, 172, 884–900. [Google Scholar] [CrossRef] [Green Version]
- Kessler, E.; Safrin, M.; Olson, J.C.; Ohman, D.E. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J. Biol. Chem. 1993, 268, 7503–7508. [Google Scholar] [CrossRef]
- Adonizio, A.; Kong, K.F.; Mathee, K. Inhibition of quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa by South Florida plant extracts. Antimicrob. Agents Chemother. 2008, 52, 198–203. [Google Scholar] [CrossRef] [Green Version]
- Kumar, L.; Chhibber, S.; Harjai, K. Zingerone inhibit biofilm formation and improve antibiofilm efficacy of ciprofloxacin against Pseudomonas aeruginosa PAO1. Fitoterapia 2013, 90, 73–78. [Google Scholar] [CrossRef]
- Huston, A.L.; Methe, B.; Deming, J.W. Purification, characterization, and sequencing of an extracellular cold-active aminopeptidase produced by marine psychrophile Colwellia psychrerythraea strain 34H. Appl. Environ. Microbiol. 2004, 70, 3321–3328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkatramanan, M.; Sankar Ganesh, P.; Senthil, R.; Akshay, J.; Veera Ravi, A.; Langeswaran, K.; Vadivelu, J.; Nagarajan, S.; Rajendran, K.; Shankar, E.M. Inhibition of Quorum Sensing and Biofilm Formation in Chromobacterium violaceum by Fruit Extracts of Passiflora edulis. ACS Omega 2020, 5, 25605–25616. [Google Scholar] [CrossRef] [PubMed]
- Musthafa, K.S.; Ravi, A.V.; Annapoorani, A.; Packiavathy, I.S.; Pandian, S.K. Evaluation of anti-quorum-sensing activity of edible plants and fruits through inhibition of the N-acyl-homoserine lactone system in Chromobacterium violaceum and Pseudomonas aeruginosa. Chemotherapy 2010, 56, 333–339. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, D.; Saharan, B. Functional characterization of biomedical potential of biosurfactant produced by Lactobacillus helveticus. Biotechnol. Rep. 2016, 11, 27–35. [Google Scholar] [CrossRef] [Green Version]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. JCMA 2018, 81, 7–11. [Google Scholar] [CrossRef]
- Upadhyay, A.; Upadhyaya, I.; Kollanoor-Johny, A.; Venkitanarayanan, K. Antibiofilm effect of plant derived antimicrobials on Listeria monocytogenes. Food Microbiol. 2013, 36, 79–89. [Google Scholar] [CrossRef]
- Onbas, T.; Osmanagaoglu, O.; Kiran, F. Potential Properties of Lactobacillus plantarum F-10 as a Bio-control Strategy for Wound Infections. Probiotics Antimicrob. Proteins 2019, 11, 1110–1123. [Google Scholar] [CrossRef]
- García-Contreras, R.; Nuñez-López, L.; Jasso-Chávez, R.; Kwan, B.W.; Belmont, J.A.; Rangel-Vega, A.; Maeda, T.; Wood, T.K. Quorum sensing enhancement of the stress response promotes resistance to quorum quenching and prevents social cheating. ISME J. 2015, 9, 115–125. [Google Scholar] [CrossRef]
- Patel, K.; Patel, M. Improving bioremediation process of petroleum wastewater using biosurfactants producing Stenotrophomonas sp. S1VKR-26 and assessment of phytotoxicity. Bioresour. Technol. 2020, 315, 123861. [Google Scholar] [CrossRef]
- Adnan, M.; Alshammari, E.; Ashraf, S.A.; Patel, K.; Lad, K.; Patel, M. Physiological and Molecular Characterization of Biosurfactant Producing Endophytic Fungi Xylaria regalis from the Cones of Thuja plicata as a Potent Plant Growth Promoter with Its Potential Application. BioMed Res. Int. 2018, 2018, 7362148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Singh, S.K.; Kant, C.; Verma, H.; Kumar, D.; Singh, P.P.; Modi, A.; Droby, S.; Kesawat, M.S.; Alavilli, H. Microbial biosurfactant: A new frontier for sustainable agriculture and pharmaceutical industries. Antioxidants 2021, 10, 1472. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Cameotra, S.S. Potential applications of microbial surfactants in biomedical sciences. Trends Biotechnol. 2004, 22, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Singh Saharan, B. Simultaneous production of biosurfactants and bacteriocins by probiotic Lactobacillus casei MRTL3. Int. J. Microbiol. 2014, 2014, 698713. [Google Scholar] [CrossRef] [Green Version]
- Thavasi, R.; Jayalakshmi, S.; Banat, I.M. Effect of biosurfactant and fertilizer on biodegradation of crude oil by marine isolates of Bacillus megaterium, Corynebacterium kutscheri and Pseudomonas aeruginosa. Bioresour. Technol. 2011, 102, 772–778. [Google Scholar] [CrossRef]
- Gudina, E.J.; Teixeira, J.A.; Rodrigues, L.R. Biosurfactant-producing lactobacilli: Screening, production profiles, and effect of medium composition. Appl. Environ. Soil Sci. 2011, 2011, 201254. [Google Scholar] [CrossRef] [Green Version]
- Saravanakumari, P.; Mani, K. Structural characterization of a novel xylolipid biosurfactant from Lactococcus lactis and analysis of antibacterial activity against multi-drug resistant pathogens. Bioresour. Technol. 2010, 101, 8851–8854. [Google Scholar] [CrossRef]
- Falagas, M.E.; Makris, G.C. Probiotic bacteria and biosurfactants for nosocomial infection control: A hypothesis. J. Hosp. Infect. 2009, 71, 301–306. [Google Scholar] [CrossRef]
- Rivera, O.M.P.; Moldes, A.B.; Torrado, A.M.; Domínguez, J.M. Lactic acid and biosurfactants production from hydrolyzed distilled grape marc. Process Biochem. 2007, 42, 1010–1020. [Google Scholar] [CrossRef]
- Rodrigues, L.; Banat, I.M.; Teixeira, J.; Oliveira, R. Biosurfactants: Potential applications in medicine. J. Antimicrob. Chemother. 2006, 57, 609–618. [Google Scholar] [CrossRef]
- Rodrigues, L.; van der Mei, H.C.; Teixeira, J.; Oliveira, R. Influence of biosurfactants from probiotic bacteria on formation of biofilms on voice prostheses. Appl. Environ. Microbiol. 2004, 70, 4408–4410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Servin, A.L. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 2004, 28, 405–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinemann, C.; van Hylckama Vlieg, J.E.; Janssen, D.B.; Busscher, H.J.; van der Mei, H.C.; Reid, G. Purification and characterization of a surface-binding protein from Lactobacillus fermentum RC-14 that inhibits adhesion of Enterococcus faecalis 1131. FEMS Microbiol. Lett. 2000, 190, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Packiavathy, I.A.S.V.; Agilandeswari, P.; Musthafa, K.S.; Pandian, S.K.; Ravi, A.V. Antibiofilm and quorum sensing inhibitory potential of Cuminum cyminum and its secondary metabolite methyl eugenol against gram negative bacterial pathogens. Food Res. Int. 2012, 45, 85–92. [Google Scholar] [CrossRef]
- Husain, F.M.; Ahmad, I. Doxycycline interferes with quorum sensing-mediated virulence factors and biofilm formation in gram-negative bacteria. World J. Microbiol. Biotechnol. 2013, 29, 949–957. [Google Scholar] [CrossRef]
- Verstraeten, N.; Braeken, K.; Debkumari, B.; Fauvart, M.; Fransaer, J.; Vermant, J.; Michiels, J. Living on a surface: Swarming and biofilm formation. Trends Microbiol. 2008, 16, 496–506. [Google Scholar] [CrossRef]
- Pesci, E.C.; Pearson, J.P.; Seed, P.C.; Iglewski, B.H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1997, 179, 3127–3132. [Google Scholar] [CrossRef] [Green Version]
- Nithya, C.; Aravindraja, C.; Pandian, S.K. Bacillus pumilus of Palk Bay origin inhibits quorum-sensing-mediated virulence factors in Gram-negative bacteria. Res. Microbiol. 2010, 161, 293–304. [Google Scholar] [CrossRef]
- Winstanley, C.; Fothergill, J.L. The role of quorum sensing in chronic cystic fibrosis Pseudomonas aeruginosa infections. FEMS Microbiol. Lett. 2009, 290, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Vattem, D.A.; Mihalik, K.; Crixell, S.H.; McLean, R.J. Dietary phytochemicals as quorum sensing inhibitors. Fitoterapia 2007, 78, 302–310. [Google Scholar] [CrossRef]
- Kiymaci, M.E.; Altanlar, N.; Gumustas, M.; Ozkan, S.A.; Akin, A. Quorum sensing signals and related virulence inhibition of Pseudomonas aeruginosa by a potential probiotic strain’s organic acid. Microb. Pathog. 2018, 121, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Gómez, N.C.; Ramiro, J.M.; Quecan, B.X.; de Melo Franco, B.D. Use of Potential Probiotic Lactic Acid Bacteria (LAB) Biofilms for the Control of Listeria monocytogenes, Salmonella Typhimurium, and Escherichia coli O157:H7 Biofilms Formation. Front. Microbiol. 2016, 7, 863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Pan, Y.; Ding, L.; Hong, H.; Yan, S.; Wu, B.; Liang, Y. Draft Genome Sequence of Lactobacillus brevis Strain 3M004, a Probiotic with Potential Quorum-Sensing Regulation. Genome Announc. 2017, 5, e00675-17. [Google Scholar] [CrossRef] [Green Version]
- Merghni, A.; Dallel, I.; Noumi, E.; Kadmi, Y.; Hentati, H.; Tobji, S.; Ben Amor, A.; Mastouri, M. Antioxidant and antiproliferative potential of biosurfactants isolated from Lactobacillus casei and their anti-biofilm effect in oral Staphylococcus aureus strains. Microb. Pathog. 2017, 104, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Wasfi, R.; Abd El-Rahman, O.A.; Zafer, M.M.; Ashour, H.M. Probiotic Lactobacillus sp. inhibit growth, biofilm formation and gene expression of caries-inducing Streptococcus mutans. J. Cell. Mol. Med. 2018, 22, 1972–1983. [Google Scholar] [CrossRef] [Green Version]
- Watnick, P.I.; Kolter, R. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 1999, 34, 586–595. [Google Scholar] [CrossRef] [Green Version]
- Pratt, L.A.; Kolter, R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998, 30, 285–293. [Google Scholar] [CrossRef]
- Singh, B.N.; Singh, H.B.; Singh, A.; Singh, B.R.; Mishra, A.; Nautiyal, C.S. Lagerstroemia speciosa fruit extract modulates quorum sensing-controlled virulence factor production and biofilm formation in Pseudomonas aeruginosa. Microbiology 2012, 158, 529–538. [Google Scholar] [CrossRef]
- Sarkar, R.; Chaudhary, S.K.; Sharma, A.; Yadav, K.K.; Nema, N.K.; Sekhoacha, M.; Karmakar, S.; Braga, F.C.; Matsabisa, M.G.; Mukherjee, P.K.; et al. Anti-biofilm activity of Marula—A study with the standardized bark extract. J. Ethnopharmacol. 2014, 154, 170–175. [Google Scholar] [CrossRef]
- Kumar, P.; Lee, J.-H.; Beyenal, H.; Lee, J. Fatty acids as antibiofilm and antivirulence agents. Trends Microbiol. 2020, 28, 753–768. [Google Scholar] [CrossRef]
- Davies, D.G.; Marques, C.N.H. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 2009, 191, 1393–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.-G.; Lee, J.-H.; Raorane, C.J.; Oh, S.T.; Park, J.G.; Lee, J. Herring oil and omega fatty acids inhibit Staphylococcus aureus biofilm formation and virulence. Front. Microbiol. 2018, 9, 1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenderska, I.B.; Chong, M.; McNulty, J.; Wright, G.D.; Burrows, L.L. Palmitoyl-DL-carnitine is a multitarget inhibitor of Pseudomonas aeruginosa biofilm development. ChemBioChem 2011, 12, 2759–2766. [Google Scholar] [CrossRef] [PubMed]
- Prasath, K.G.; Sethupathy, S.; Pandian, S.K. Proteomic analysis uncovers the modulation of ergosterol, sphingolipid and oxidative stress pathway by myristic acid impeding biofilm and virulence in Candida albicans. J. Proteom. 2019, 208, 103503. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Song, S.; Yang, C.; Sun, X.; Huang, Y.; Li, K.; Zhao, S.; Zhang, Y.; Deng, Y. Disruption of quorum sensing and virulence in Burkholderia cenocepacia by a structural analogue of the cis-2-dodecenoic acid signal. Appl. Environ. Microbiol. 2019, 85, e00105–e00119. [Google Scholar] [CrossRef] [Green Version]
- Childers, B.M.; Cao, X.; Weber, G.G.; Demeler, B.; Hart, P.J.; Klose, K.E. N-terminal residues of the Vibrio cholerae virulence regulatory protein ToxT involved in dimerization and modulation by fatty acids. J. Biol. Chem. 2011, 286, 28644–28655. [Google Scholar] [CrossRef] [Green Version]
- Lowden, M.J.; Skorupski, K.; Pellegrini, M.; Chiorazzo, M.G.; Taylor, R.K.; Kull, F.J. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc. Natl. Acad. Sci. USA 2010, 107, 2860–2865. [Google Scholar] [CrossRef] [Green Version]
- Plecha, S.C.; Withey, J.H. Mechanism for inhibition of Vibrio cholerae ToxT activity by the unsaturated fatty acid components of bile. J. Bacteriol. 2015, 197, 1716–1725. [Google Scholar] [CrossRef] [Green Version]
- Kabara, J.J.; Swieczkowski, D.M.; Conley, A.J.; Truant, J.P. Fatty acids and derivatives as antimicrobial agents. Antimicrob. Agents Chemother. 1972, 2, 23–28. [Google Scholar] [CrossRef] [Green Version]
- Nicol, M.; Alexandre, S.; Luizet, J.B.; Skogman, M.; Jouenne, T.; Salcedo, S.P.; Dé, E. Unsaturated Fatty Acids Affect Quorum Sensing Communication System and Inhibit Motility and Biofilm Formation of Acinetobacter baumannii. Int. J. Mol. Sci. 2018, 19, 214. [Google Scholar] [CrossRef] [Green Version]









| Strain | Colony Characteristics | Gram’s Reaction | Oil Spreading Test | Drop Collapse Test | BAP Test | %E24 (n-Hexadecane) |
|---|---|---|---|---|---|---|
| L. plantarum-MBP001 | Creamy, small, circular, slightly raised, entire, smooth | Gram-positive, rod shaped | Positive | Positive | Positive | 64.38 ± 1.47 |
| No. | RT | % Area | Compound Name | Class | Structure |
|---|---|---|---|---|---|
| 1 | 5.784 | 0.30 | Octane, 2,4,6-trimethyl- | Fatty acyl |  |
| 2 | 6.686 | 0.32 | Undecane | Fatty acyl |  |
| 3 | 7.500 | 0.90 | Dodecane | Fatty acyl |  |
| 4 | 8.936 | 0.27 | Hexadecane | Fatty acyl |  |
| 5 | 12.146 | 7.29 | n-Hexadecanoic acid | Fatty acid |  |
| 6 | 13.015 | 6.88 | Oleic Acid | Fatty acid |  |
| 7 | 13.149 | 3.30 | Ethyl Oleate | Fatty acid |  |
| 8 | 13.256 | 0.40 | Octadecanoic acid, ethyl ester | Fatty acid |  |
| 9 | 13.350 | 0.30 | 3,7,11,15-Tetramethylhexadec-2-en-1-yl acetate | Prenol lipid |  |
| 10 | 14.256 | 1.55 | 1H-indene, 1-hexadecyl-2,3-dihydro- | Fatty acyl |  |
| 11 | 14.848 | 2.23 | Hexadecanoic acid, 2-hydroxy-1-(hydroxymethyl) ethyl ester | Glycerolipid |  |
| 12 | 16.297 | 0.74 | Octadecanoic acid, 2,3-dihydroxypropyl ester | Glycerolipid |  |
| Compound Name | 3QP5 | 1RO5 | 3IT7 |
|---|---|---|---|
| Chlorolactone (standard QS inhibitor) | −8.1 | −7.6 | −7.8 |
| Octane, 2,4,6-trimethyl- | −6.1 | −6.3 | −5.6 |
| Undecane | −5.5 | −5.6 | −5.0 |
| Dodecane | −5.6 | −5.7 | −4.8 |
| Hexadecane | −6.0 | −5.2 | −5.0 |
| n-Hexadecanoic acid | −7.9 | −5.1 | −4.6 |
| Oleic Acid | −6.6 | −7.3 | −5.1 |
| 3,7,11,15-Tetramethylhexadec-2-en-1-yl acetate | −6.7 | −5.9 | −5.9 |
| 1H-indene, 1-hexadecyl-2,3-dihydro- | −7.6 | −6.4 | −7.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patel, M.; Siddiqui, A.J.; Ashraf, S.A.; Surti, M.; Awadelkareem, A.M.; Snoussi, M.; Hamadou, W.S.; Bardakci, F.; Jamal, A.; Jahan, S.; et al. Lactiplantibacillus plantarum-Derived Biosurfactant Attenuates Quorum Sensing-Mediated Virulence and Biofilm Formation in Pseudomonas aeruginosa and Chromobacterium violaceum. Microorganisms 2022, 10, 1026. https://doi.org/10.3390/microorganisms10051026
Patel M, Siddiqui AJ, Ashraf SA, Surti M, Awadelkareem AM, Snoussi M, Hamadou WS, Bardakci F, Jamal A, Jahan S, et al. Lactiplantibacillus plantarum-Derived Biosurfactant Attenuates Quorum Sensing-Mediated Virulence and Biofilm Formation in Pseudomonas aeruginosa and Chromobacterium violaceum. Microorganisms. 2022; 10(5):1026. https://doi.org/10.3390/microorganisms10051026
Chicago/Turabian StylePatel, Mitesh, Arif Jamal Siddiqui, Syed Amir Ashraf, Malvi Surti, Amir Mahgoub Awadelkareem, Mejdi Snoussi, Walid Sabri Hamadou, Fevzi Bardakci, Arshad Jamal, Sadaf Jahan, and et al. 2022. "Lactiplantibacillus plantarum-Derived Biosurfactant Attenuates Quorum Sensing-Mediated Virulence and Biofilm Formation in Pseudomonas aeruginosa and Chromobacterium violaceum" Microorganisms 10, no. 5: 1026. https://doi.org/10.3390/microorganisms10051026
APA StylePatel, M., Siddiqui, A. J., Ashraf, S. A., Surti, M., Awadelkareem, A. M., Snoussi, M., Hamadou, W. S., Bardakci, F., Jamal, A., Jahan, S., Sachidanandan, M., & Adnan, M. (2022). Lactiplantibacillus plantarum-Derived Biosurfactant Attenuates Quorum Sensing-Mediated Virulence and Biofilm Formation in Pseudomonas aeruginosa and Chromobacterium violaceum. Microorganisms, 10(5), 1026. https://doi.org/10.3390/microorganisms10051026













