Molecular Characterization of pBOq-IncQ and pBOq-95LK Plasmids of Escherichia coli BOq 01, a New Isolated Strain from Poultry Farming, Involved in Antibiotic Resistance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Antibiotics Susceptibility Test
2.2. Plasmid Analysis
2.3. Plasmid Curing
2.4. Pangenomic and Comparative Plasmid Analysis
3. Results
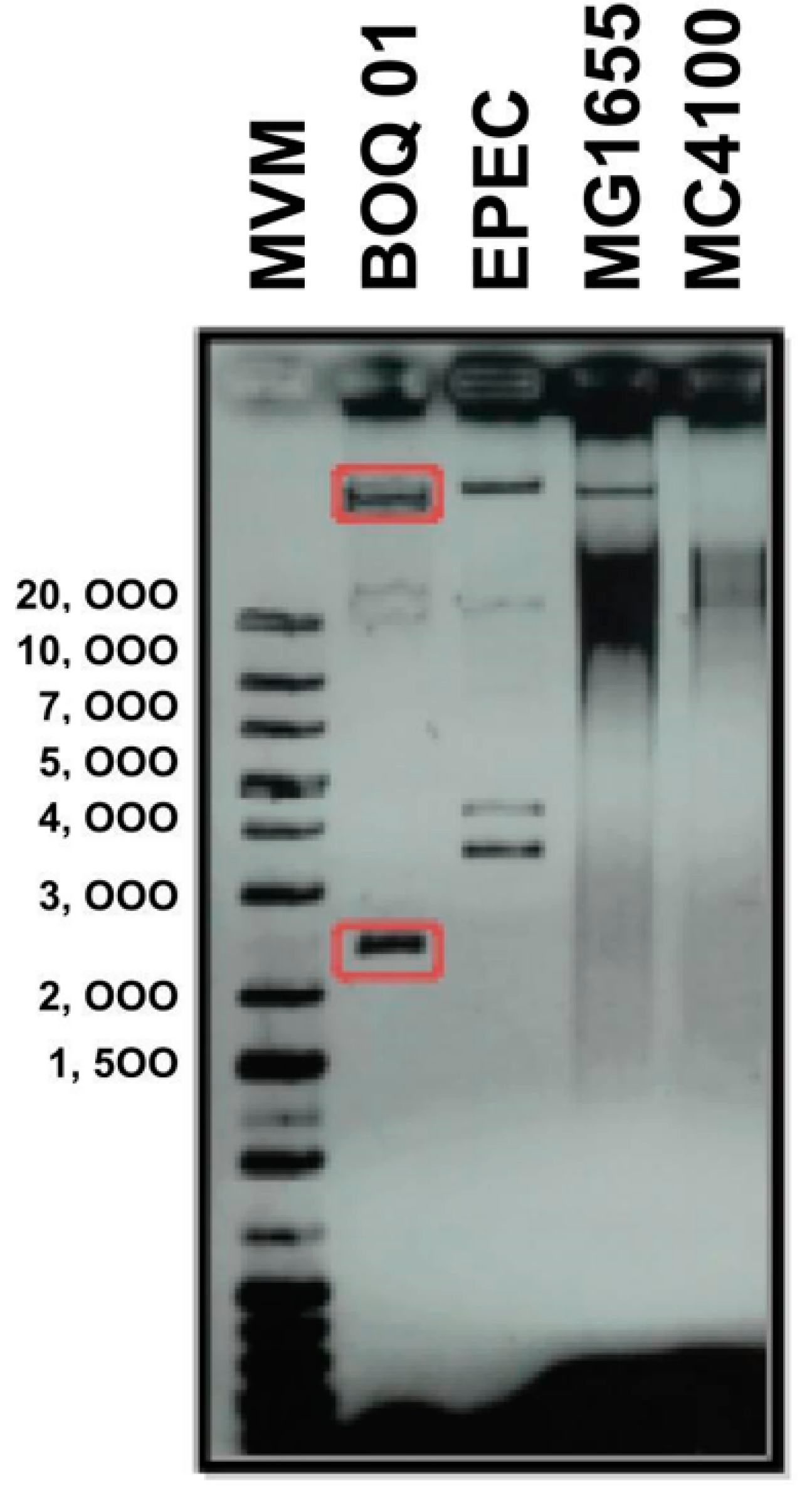
3.1. Plasmid Identification in E. coli Str. BOq 01
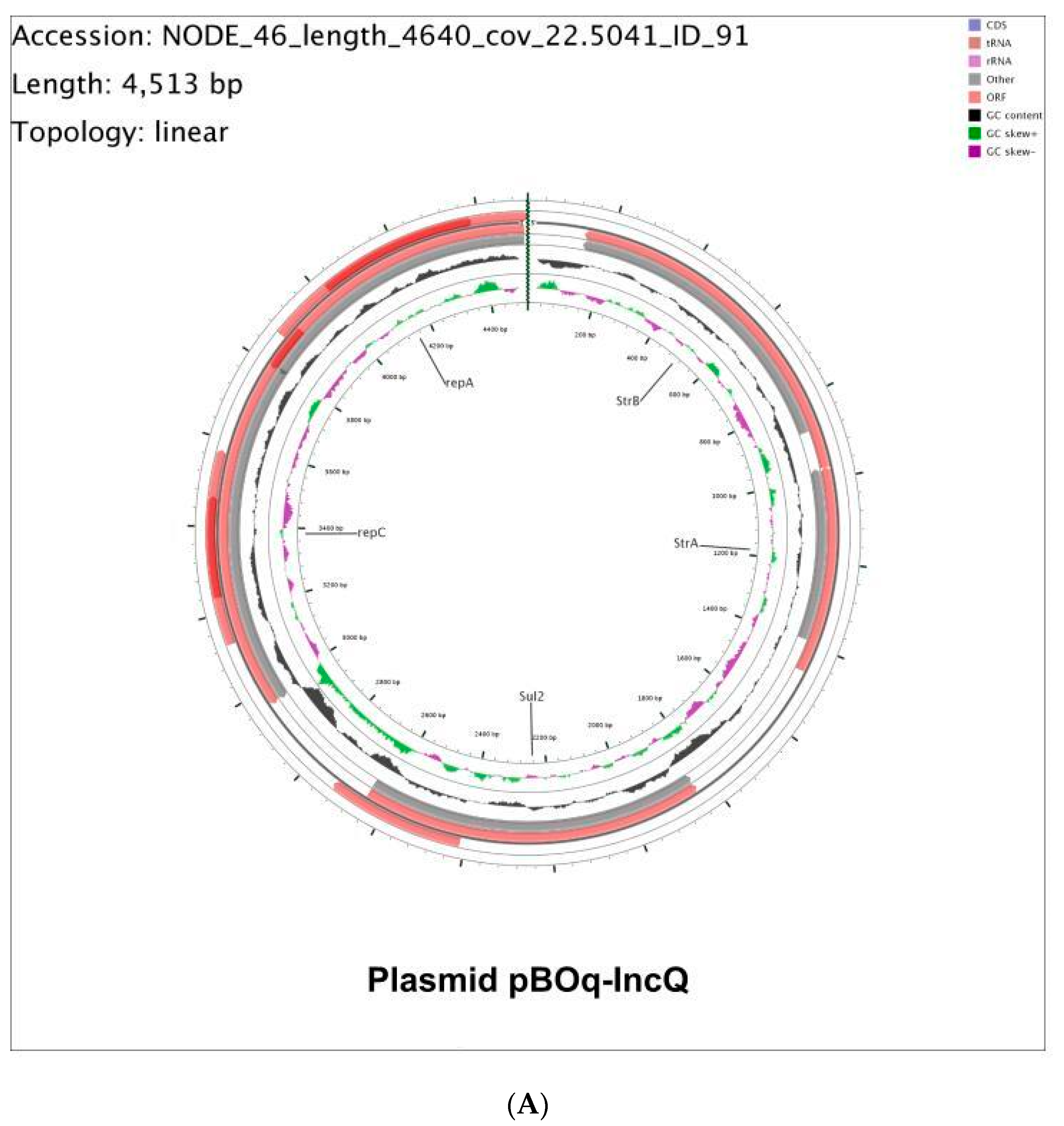
3.2. Molecular Characterization of pBOq-IncQ
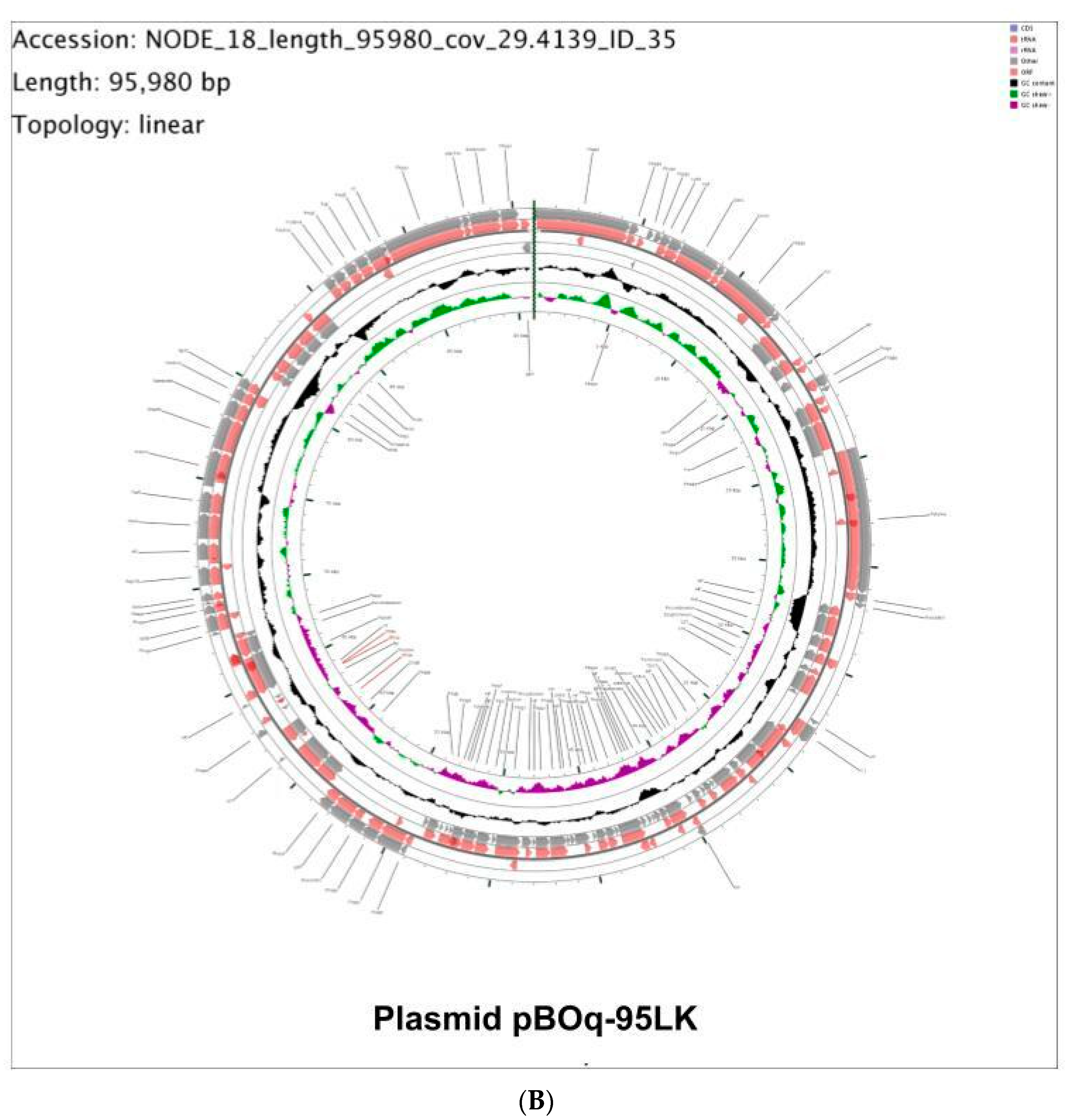
3.3. Molecular Characterization of pBOq-95LK
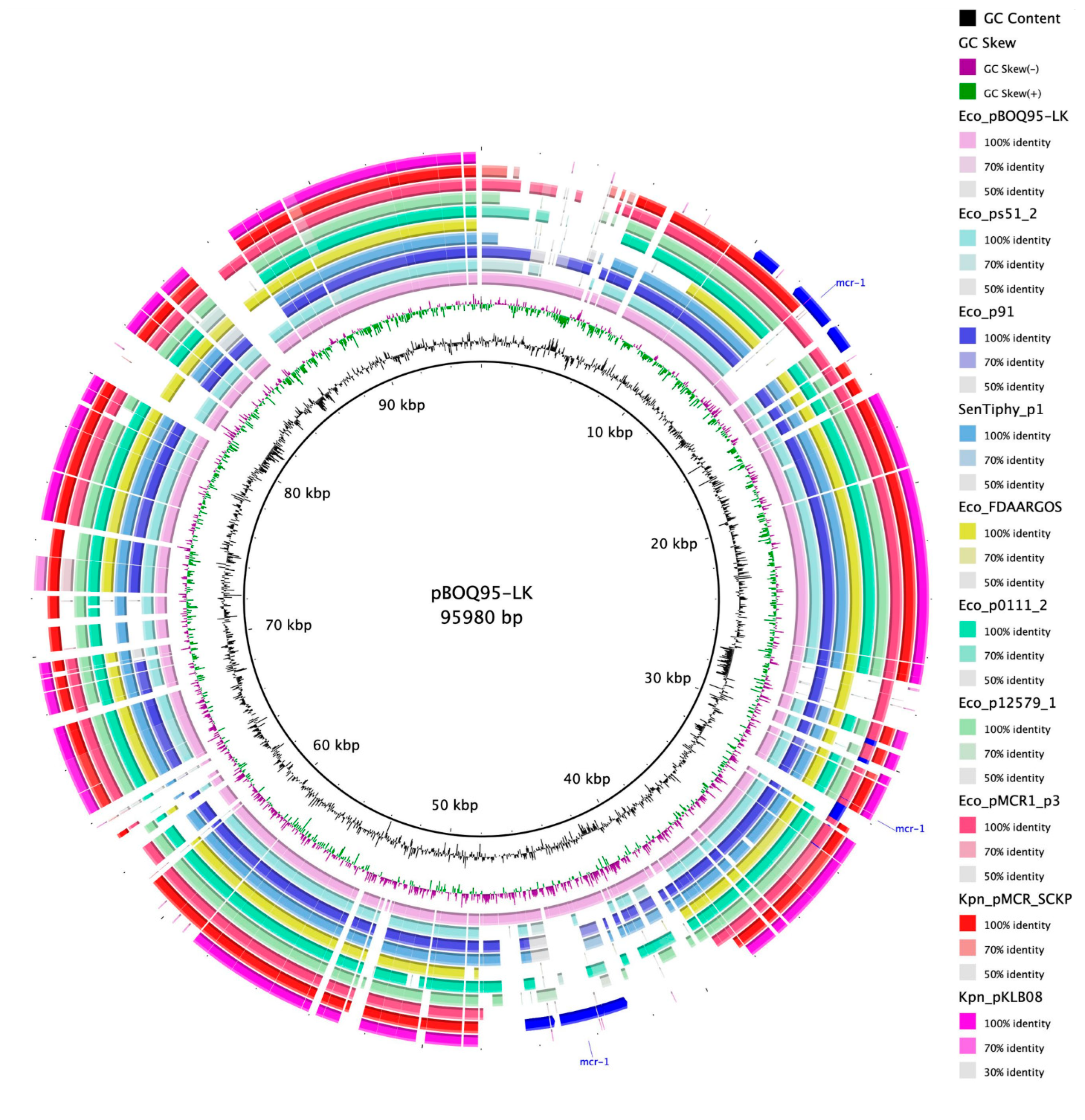
3.4. Pangenomic and Comparative Analysis of Plasmid pBOq-95LK
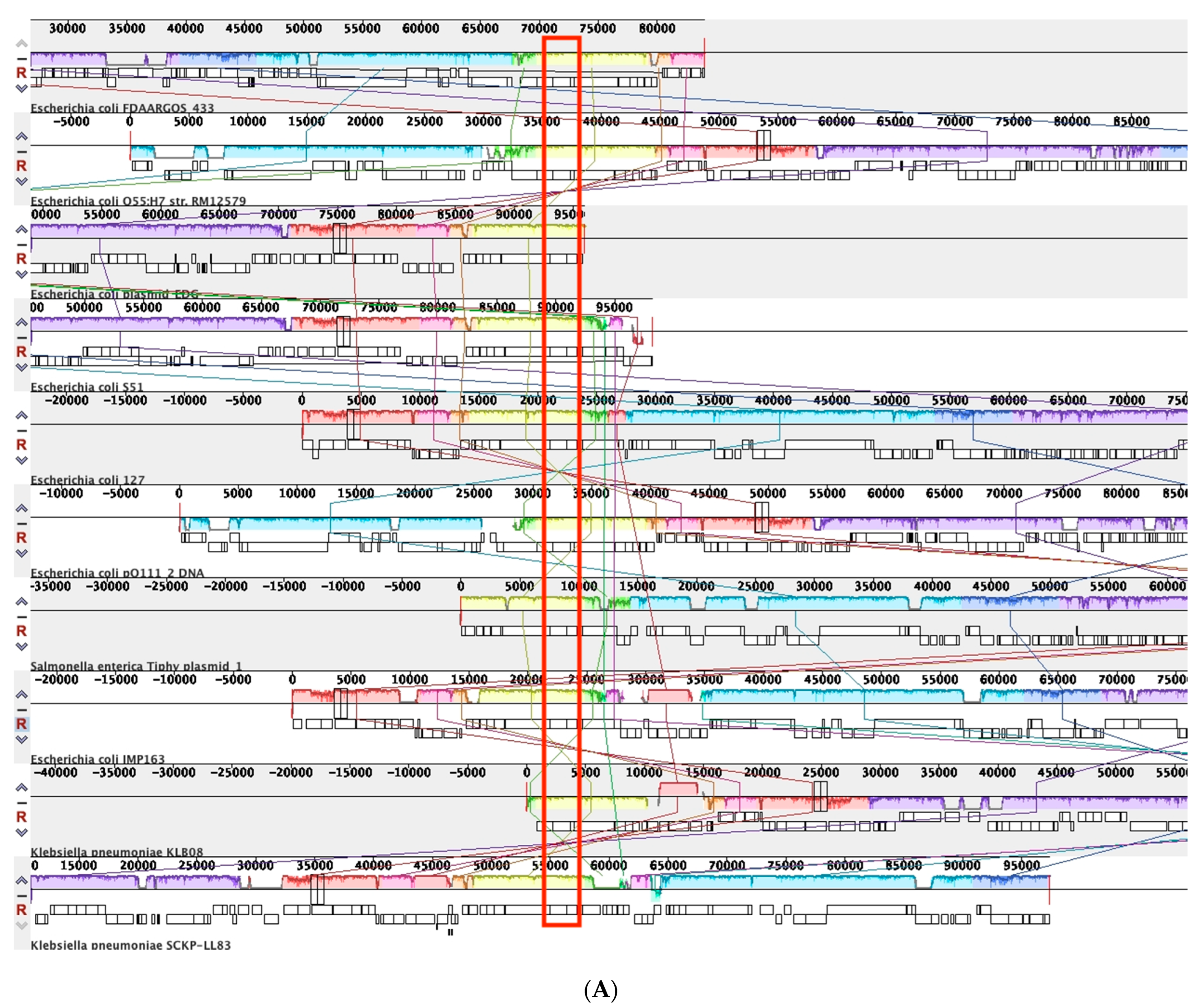
3.4.1. Comparative Analysis of Phage-like Plasmids and pBOq-95LK
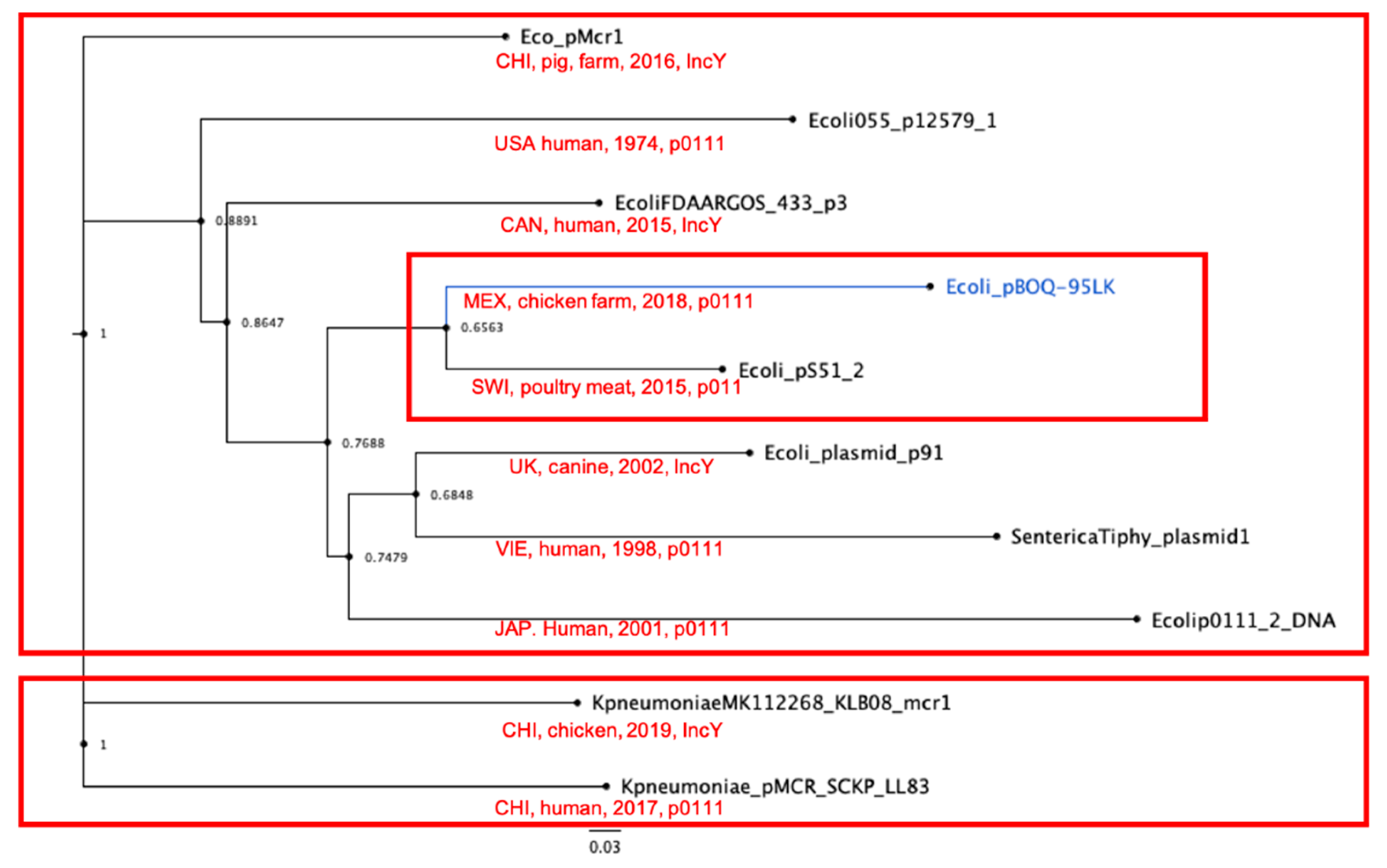
3.4.2. Phylogenetic Analysis of Phage-like Plasmids and pBOq-95LK
3.4.3. Pangenomic Analysis of Phage-like Plasmids and pBOq-95LK
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Economou, V.; Gousia, P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect. Drug Resist. 2015, 8, 49–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brazas, M.D.; Hancock, R.E.W. Using microarray gene signatures to elucidate mechanisms of antibiotic action and resistance. Drug Discov. Today 2005, 10, 1245–1252. [Google Scholar] [CrossRef]
- Bearson, B.L.; Allen, H.K.; Brunelle, B.W.; Lee, I.S.; Casjens, S.R.; Stanton, T.B. The agricultural antibiotic carbadox induces phage-mediated gene transfer in Salmonella. Front. Microbiol. 2014, 5, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet. Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Gibson, M.K.; Forsberg, K.J.; Dantas, G. Improved Annotation of Antibiotic Resistance Determinants Reveals Microbial Resistomes Cluster by Ecology. ISME J. 2015, 9, 207–216. [Google Scholar] [CrossRef]
- Pitout, J.D. Infections with Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae. Drugs 2010, 70, 313–333. [Google Scholar] [CrossRef]
- Wagner, S.; Lupolova, N.; Gally, D.L.; Argyle, S.A. Convergence of Plasmid Architectures Drives Emergence of Multi-Drug Resistance in a Clonally Diverse Escherichia coli Population from a Veterinary Clinical Care Setting. Vet. Microbiol. 2017, 211, 6–14. [Google Scholar] [CrossRef]
- Croxen, M.A.; Finlay, B.B. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 2010, 8, 26–38. [Google Scholar] [CrossRef]
- Ellis, S.J.; Crossman, L.C.; McGrath, C.J.; Chattaway, M.A.; Hölken, J.M.; Brett, B.; Bundy, L.; Kay, G.L.; Wain, J.; Schüller, S. Identification and characterisation of enteroaggregative Escherichia coli subtypes associated with human disease. Sci. Rep. 2020, 10, 7475. [Google Scholar] [CrossRef]
- Ahmed, S.A.; Awosika, J.; Baldwin, C.; Bishop-Lilly, K.A.; Biswas, B.; Broomall, S.; Chain, P.S.G.; Chertkov, O.; Chokoshvili, O.; Coyne, S.; et al. Genomic Comparison of Escherichia coli O104:H4 Isolates from 2009 and 2011 Reveals Plasmid, and Prophage Heterogeneity, Including Shiga Toxin Encoding Phage Stx2. PLoS ONE 2012, 7, e48228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DuPont, H.L. The Growing Threat of Foodborne Bacterial Enteropathogens of Animal Origin. Clin. Infect. Dis. 2007, 45, 1353–1361. [Google Scholar] [CrossRef] [PubMed]
- Fegan, N.; Gobius, K.S.; Dykes, G.A. Pathogenic Escherichia coli. Encycl. Meat Sci. 2014, 2, 357–361. [Google Scholar] [CrossRef]
- Smalla, K.; Heuer, H.; Götz, A.; Niemeyer, D.; Krögerrecklenfort, E.; Tietze, E. Exogenous isolation of antibiotic resistance plasmids from piggery manure slurries reveals a high prevalence and diversity of IncQ-like plasmids. Appl. Environ. Microbiol. 2000, 66, 4854–4862. [Google Scholar] [CrossRef] [Green Version]
- Rawlings, D.E.; Tietze, E. Comparative biology of IncQ and IncQ-like plasmids. Microbiol. Mol. Biol. Rev. 2001, 65, 481–496. [Google Scholar] [CrossRef] [Green Version]
- Venturini, C.; Zingali, T.; Wyrsch, E.R.; Bowring, B.; Iredell, J.; Partridge, S.R.; Djordjevic, S.P. Diversity of P1 phage-like elements in multidrug resistant Escherichia coli. Sci. Rep. 2019, 9, 18861. [Google Scholar] [CrossRef] [Green Version]
- Octavia, S.; Sara, J.; Lan, R. Characterization of a Large Novel Phage-like Plasmid in Salmonella enterica Serovar Typhimurium. FEMS Microbiol. Lett. 2015, 362, fnv044. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Feng, Y.; Liu, F.; Jiang, H.; Qu, Z.; Lei, M.; Wang, J.; Zhang, B.; Hu, Y.; Ding, J.; et al. A Phage-Like IncY Plasmid Carrying the Mcr-1 Gene in Escherichia coli from a Pig Farm in China. Antimicrob. Agents Chemother. 2017, 61, e02035-16. [Google Scholar] [CrossRef] [Green Version]
- Moura, Q.; Sartori, L.; Silva, K.C.; Cunha, M.P.; Esposito, F.; Lopes, R.; Otutumi, L.K.; Gonçalves, D.D.; Dropa, M. Silent Dissemination of Colistin-Resistant Escherichia coli in South America Could Contribute to the Global Spread of the Mcr-1 Gene. Eurosurveillance 2016, 21, 1–6. [Google Scholar] [CrossRef]
- McGann, P.; Snesrud, E.; Maybank, R.; Corey, B.; Ong, A.C.; Clifford, R.; Hinkle, M.; Whitman, T.; Lesho, E.; Schaecher, K.E. Escherichia coli Harboring Mcr-1 and BlaCTX-M on a Novel IncF Plasmid: First Report of Mcr-1 in the United States. Antimicrob. Agents Chemother. 2016, 60, 4420–4421. [Google Scholar] [CrossRef] [Green Version]
- Zeng, K.; Doi, Y.; Patil, S.; Huang, X.; Tian, G. Emergence of the Plasmid-Mediated Mcr-1 Gene in Colistin-Resistant. Antimicrob. Agents Chemother. 2016, 60, 3862–3863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández-Mendoza, A.; Rivera Mendoza, D.; Moran-Vazquez, A.; Dantán-González, E. Draft Genome Sequences and Complete Sequences of Two Plasmids of Escherichia coli Strain BOq 01, a Multiple-Antibiotic-Resistant Strain Isolated from a Poultry Farm in Mexico. Microbiol. Resour. Announc. 2018, 7, e01104-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, A.W.; Kirby, W.M.M.; Sherris, J.C.; Turck, A.M.; Graevenitz, A. Von 40 Microbiology: A Centenary Perspective 1966 Antibiotic Susceptibility Testing by a Standardized Single Disk Method. Am. J. Clin. Pathol. 1978, 45, 493–496. [Google Scholar] [CrossRef]
- Patel, J.B. Performance Standards for Antimicrobial Susceptibility Testing; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017; ISBN 1562388045. [Google Scholar]
- World Health Organisation. WHO List of Critically Important Antimicrobials for Human Medicine (WHO CIA List); World Health Organization: Geneva, Swizerland, 2019; ISBN 978-92-4-151552-8.
- Aziz, R.K.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckhardt, T. A Rapid Method for the Identification of Plasmid Desoxyribonucleic Acid in Bacteria. Plasmid 1978, 1, 584–588. [Google Scholar] [CrossRef]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple Alignment of Conserved Genomic Sequence with Rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [Green Version]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple Prokaryote Genome Comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More Models, New Heuristics and High-Performance Computing Europe PMC Funders Group. Nat. Methods 2015, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Posada, D. JModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Oliva, M.; Monno, R.; D’Addabbo, P.; Pesole, G.; Dionisi, A.M.; Scrascia, M.; Chiara, M.; Horner, D.S.; Manzari, C.; Luzzi, I.; et al. A Novel Group of IncQ1 Plasmids Conferring Multidrug Resistance. Plasmid 2017, 89, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Ogura, Y.; Ooka, T.; Asadulghani; Terajima, J.; Nougayrède, J.-P.; Kurokawa, K.; Tashiro, K.; Tobe, T.; Nakayama, K.; Kuhara, S.; et al. Extensive Genomic Diversity and Selective Conservation of Virulence-Determinants in Enterohemorrhagic Escherichia coli Strains of O157 and Non-O157 Serotypes. Genome Biol. 2007, 8, R138. [Google Scholar] [CrossRef] [Green Version]
- Ogura, Y.; Ooka, T.; Iguchi, A.; Toh, H.; Asadulghani, M.; Oshima, K.; Kodama, T.; Abe, H.; Nakayama, K.; Kurokawa, K.; et al. Comparative Genomics Reveal the Mechanism of the Parallel Evolution of O157 and Non-O157 Enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. USA 2009, 106, 17939–17944. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Liu, L.; Feng, Y.; Zong, Z. A P7 Phage-like Plasmid Carrying Mcr-1 in an ST15 Klebsiella pneumoniae Clinical Isolate. Front. Microbiol. 2018, 9, 11. [Google Scholar] [CrossRef]
- Chen, J.; Chen, Y.; He, Q. Action of Bleomycin is Affected by Bleomycin Hydrolase but Not by Caveolin-1. Int. J. Oncol. 2012, 41, 2245–2252. [Google Scholar] [CrossRef] [Green Version]
- Dortet, L.; Girlich, D.; Virlouvet, A.L.; Poirel, L.; Nordmann, P.; Iorga, B.I.; Naas, T. Characterization of BRPMBL, the Bleomycin Resistance Protein Associated with the Carbapenemase NDM. Antimicrob. Agents Chemother. 2017, 61, e02413-16. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Van Dorp, L.; Shaw, L.P.; Bradley, P.; Wang, Q.; Wang, X.; Jin, L.; Zhang, Q.; Liu, Y.; Rieux, A.; et al. The Global Distribution and Spread of the Mobilized Colistin Resistance Gene Mcr-1. Nat. Commun. 2018, 9, 1179. [Google Scholar] [CrossRef] [Green Version]
- Kyle, J.L.; Cummings, C.A.; Parker, C.T.; Quiñones, B.; Vatta, P.; Newton, E.; Huynh, S.; Swimley, M.; Degoricija, L.; Barker, M.; et al. Escherichia coli Serotype O55:H7 Diversity Supports Parallel Acquisition of Bacteriophage at Shiga Toxin Phage Insertion Sites during Evolution of the O157:H7 Lineage. J. Bacteriol. 2012, 194, 1885–1896. [Google Scholar] [CrossRef] [Green Version]
- Matamoros, S.; Van Hattem, J.M.; Arcilla, M.S.; Willemse, N.; Melles, D.C.; Penders, J.; Vinh, T.N.; Thi Hoa, N.; Bootsma, M.C.J.; Van Genderen, P.J.; et al. Global Phylogenetic Analysis of Escherichia coli and Plasmids Carrying the Mcr-1 Gene Indicates Bacterial Diversity but Plasmid Restriction. Sci. Rep. 2017, 7, 15364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snesrud, E.; He, S.; Chandler, M.; Dekker, J.P.; Hickman, A.B.; McGann, P.; Dyda, F. A Model for Transposition of the Colistin Resistance Gene Mcr-1 by ISApl1. Antimicrob. Agents Chemother. 2016, 60, 6973–6976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfeifer, E.; Moura De Sousa, J.A.; Touchon, M.; Rocha, E.P.C. Bacteria Have Numerous Distinctive Groups of Phage-Plasmids with Conserved Phage and Variable Plasmid Gene Repertoires. Nucleic Acids Res. 2021, 49, 2655–2673. [Google Scholar] [CrossRef]
- Joshua-Tor, L.; Xu, H.E.; Johnston, S.A.; Rees, D.C. Crystal Structure of a Conserved Protease That Binds DNA: The Bleomycin Hydrolase, Gal6. Science 1995, 269, 945–950. [Google Scholar] [CrossRef]
- Dolejska, M.; Papagiannitsis, C.C. Plasmid-Mediated Resistance is Going Wild. Plasmid 2018, 99, 99–111. [Google Scholar] [CrossRef] [PubMed]


| Family | Antibiotic | Disk Content (µg/Disk) | Inhibition Zone Diameter (mm) | |||
|---|---|---|---|---|---|---|
| R | I | S | Results E. coli Strain BOq 01 | |||
| Penicillins | AMP | 10 | ≤13 | 14–16 | ≥17 | 6 |
| CB | 100 | - | - | - | 6 | |
| Phenicols | CHL | 30 | ≤12 | 13–17 | ≥18 | 8 |
| Aminoglycosides | GEN | 10 | ≤12 | 13–14 | ≥15 | 14 |
| KAN | 30 | ≤13 | 14–17 | ≥18 | 13 | |
| STR | 10 | ≤15 | 12–14 | ≥11 | 6 | |
| HYG | 50 | - | - | - | 9 | |
| Tetracyclines | TET | 30 | ≤11 | 12–14 | ≥15 | 10 |
| Fosfomicyns | FOS | 200 | ≤12 | 13–15 | ≥16 | 19 |
| Quinolones and fluoroquinolones | NAL | 30 | ≤13 | 14–18 | ≥19 | 16 |
| ENR | 5 | ≤15 | 16–20 | ≥21 | 17 | |
| Cephalosporin | CLX | 30 | ≤19 | 20–22 | ≥23 | 18 |
| Diaminopyrimidine | TMP | 5 | ≤10 | 11–15 | ≥16 | 25 |
| Sulfonamides | SDX | 250 | ≤12 | 13–16 | ≥17 | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Mendoza, A.; Salgado-Morales, R.; Morán-Vázquez, A.; López-Torres, D.; García-Gómez, B.I.; Dantán-González, E. Molecular Characterization of pBOq-IncQ and pBOq-95LK Plasmids of Escherichia coli BOq 01, a New Isolated Strain from Poultry Farming, Involved in Antibiotic Resistance. Microorganisms 2022, 10, 1509. https://doi.org/10.3390/microorganisms10081509
Hernández-Mendoza A, Salgado-Morales R, Morán-Vázquez A, López-Torres D, García-Gómez BI, Dantán-González E. Molecular Characterization of pBOq-IncQ and pBOq-95LK Plasmids of Escherichia coli BOq 01, a New Isolated Strain from Poultry Farming, Involved in Antibiotic Resistance. Microorganisms. 2022; 10(8):1509. https://doi.org/10.3390/microorganisms10081509
Chicago/Turabian StyleHernández-Mendoza, Armando, Rosalba Salgado-Morales, Abimael Morán-Vázquez, David López-Torres, Blanca Inés García-Gómez, and Edgar Dantán-González. 2022. "Molecular Characterization of pBOq-IncQ and pBOq-95LK Plasmids of Escherichia coli BOq 01, a New Isolated Strain from Poultry Farming, Involved in Antibiotic Resistance" Microorganisms 10, no. 8: 1509. https://doi.org/10.3390/microorganisms10081509
APA StyleHernández-Mendoza, A., Salgado-Morales, R., Morán-Vázquez, A., López-Torres, D., García-Gómez, B. I., & Dantán-González, E. (2022). Molecular Characterization of pBOq-IncQ and pBOq-95LK Plasmids of Escherichia coli BOq 01, a New Isolated Strain from Poultry Farming, Involved in Antibiotic Resistance. Microorganisms, 10(8), 1509. https://doi.org/10.3390/microorganisms10081509






