Identification and Characterisation of pST1023 A Mosaic, Multidrug-Resistant and Mobilisable IncR Plasmid
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria Isolates, Antimicrobial Susceptibility Testing and Mobilisation Experiment
2.2. DNA Sequencing, Assembly and Annotation
2.3. Bioinformatic Analysis
3. Results
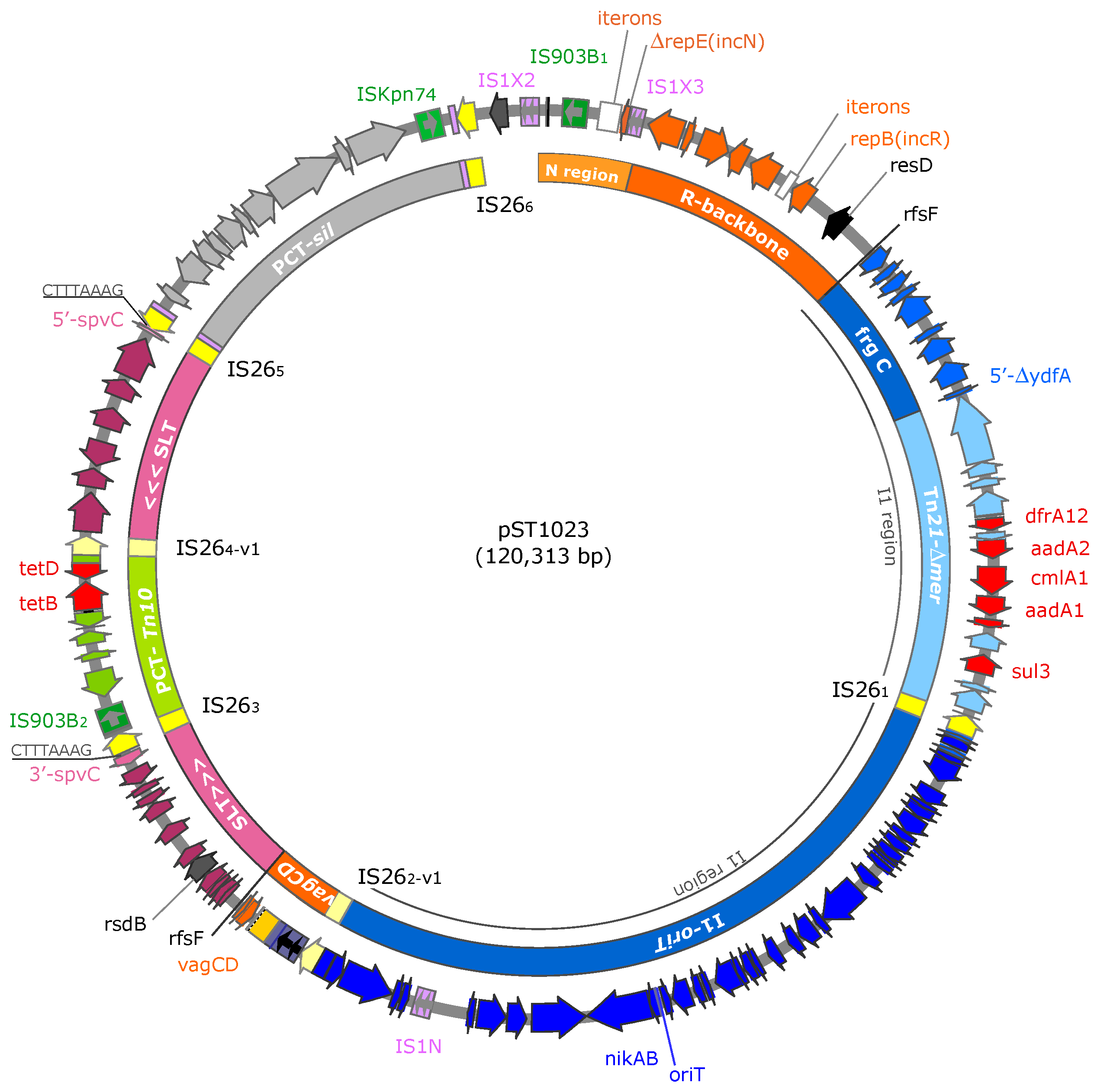
3.1. Genome Sequence of ST1023 and Context of Resistance Genes
3.2. pST1023 Genetic Organisation
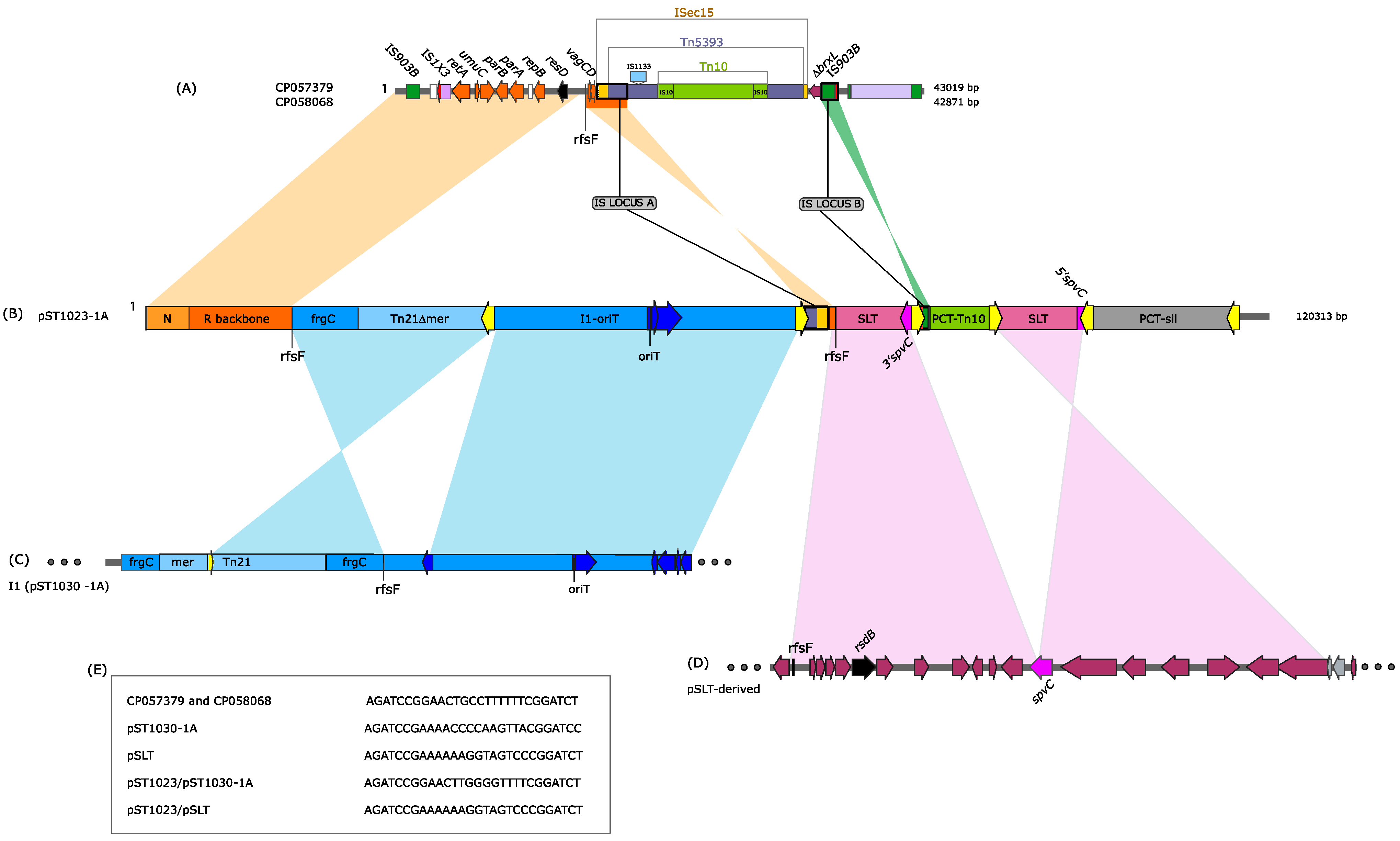
3.3. pST1023 A Mosaic Plasmid
3.4. pST1023 Mobilisation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodriguez-Beltran, J.; DelaFuente, J.; Leon-Sampedro, R.; MacLean, R.C.; San Millan, A. Beyond horizontal gene transfer: The role of plasmids in bacterial evolution. Nat. Rev. Microbiol. 2021, 19, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.I.; Ikuta, K.S.; Sharara, K.S.F.; Swetschinski, L.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; Johnson, S.C.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hulter, N.; Ilhan, J.; Wein, T.; Kadibalban, A.S.; Hammerschmidt, K.; Dagan, T. An evolutionary perspective on plasmid lifestyle modes. Curr. Opin. Microbiol. 2017, 38, 74–80. [Google Scholar] [CrossRef]
- Wein, T.; Dagan, T. Plasmid evolution. Curr. Biol. 2020, 30, R1158–R1163. [Google Scholar] [CrossRef]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threlfall, E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef]
- Garcillan-Barcia, M.P.; Francia, M.V.; de la Cruz, F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 2009, 33, 657–687. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Fernandez, A.; Fortini, D.; Veldman, K.; Mevius, D.; Carattoli, A. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J. Antimicrob. Chemother. 2009, 63, 274–281. [Google Scholar] [CrossRef]
- Jing, Y.; Jiang, X.; Yin, Z.; Hu, L.; Zhang, Y.; Yang, W.; Yang, H.; Gao, B.; Zhao, Y.; Zhou, D.; et al. Genomic diversification of IncR plasmids from China. J. Glob. Antimicrob. Resist. 2019, 19, 358–364. [Google Scholar] [CrossRef]
- Mourao, J.; Machado, J.; Novais, C.; Antunes, P.; Peixe, L. Characterization of the emerging clinically-relevant multidrug-resistant Salmonella enterica serotype 4,[5],12:i:- (monophasic variant of S. Typhimurium) clones. Eur. J. Clin. Microbiol. Infect Dis. 2014, 33, 2249–2257. [Google Scholar] [CrossRef] [PubMed]
- De Vito, D.; Monno, R.; Nuccio, F.; Legretto, M.; Oliva, M.; Coscia, M.F.; Dionisi, A.M.; Calia, C.; Capolongo, C.; Pazzani, C. Diffusion and persistence of multidrug resistant Salmonella Typhimurium strains phage type DT120 in southern Italy. BioMed Res. Int. 2015, 2015, 265042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliva, M.; Monno, R.; D’Addabbo, P.; Pesole, G.; Dionisi, A.M.; Scrascia, M.; Chiara, M.; Horner, D.S.; Manzari, C.; Luzzi, I.; et al. A novel group of IncQ1 plasmids conferring multidrug resistance. Plasmid 2017, 89, 22–26. [Google Scholar] [CrossRef] [PubMed]
- (CLSI), C.a.L.S.I. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 10th ed.; CLSI: Wayne, PA, USA, 2015; Volume 35, p. M07-A10. [Google Scholar]
- Oliva, M.; Calia, C.; Ferrara, M.; D’Addabbo, P.; Scrascia, M.; Mule, G.; Monno, R.; Pazzani, C. Antimicrobial resistance gene shuffling and a three-element mobilisation system in the monophasic Salmonella typhimurium strain ST1030. Plasmid 2020, 111, 102532. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliva, M.; Monno, R.; Addabbo, P.; Pesole, G.; Scrascia, M.; Calia, C.; Dionisi, A.M.; Chiara, M.; Horner, D.S.; Manzari, C.; et al. IS26 mediated antimicrobial resistance gene shuffling from the chromosome to a mosaic conjugative FII plasmid. Plasmid 2018, 100, 22–30. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. A J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome. Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [Green Version]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Moller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siguier, P.; Filee, J.; Chandler, M. Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 2006, 9, 526–531. [Google Scholar] [CrossRef] [Green Version]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Echeita, M.A.; Herrera, S.; Usera, M.A. Atypical, fljB-negative Salmonella enterica subsp. enterica strain of serovar 4,5,12:i:- appears to be a monophasic variant of serovar Typhimurium. J. Clin. Microbiol. 2001, 39, 2981–2983. [Google Scholar] [CrossRef] [Green Version]
- Ingle, D.J.; Ambrose, R.L.; Baines, S.L.; Duchene, S.; Goncalves da Silva, A.; Lee, D.Y.J.; Jones, M.; Valcanis, M.; Taiaroa, G.; Ballard, S.A.; et al. Evolutionary dynamics of multidrug resistant Salmonella enterica serovar 4,[5],12:i:- in Australia. Nat. Commun. 2021, 12, 4786. [Google Scholar] [CrossRef]
- Arrieta-Gisasola, A.; Atxaerandio-Landa, A.; Garrido, V.; Grillo, M.J.; Martinez-Ballesteros, I.; Laorden, L.; Garaizar, J.; Bikandi, J. Genotyping Study of Salmonella 4,[5],12:i:- Monophasic Variant of Serovar Typhimurium and Characterization of the Second-Phase Flagellar Deletion by Whole Genome Sequencing. Microorganisms 2020, 8, 2049. [Google Scholar] [CrossRef]
- Harmer, C.J.; Pong, C.H.; Hall, R.M. Structures bounded by directly-oriented members of the IS26 family are pseudo-compound transposons. Plasmid 2020, 111, 102530. [Google Scholar] [CrossRef]
- Guo, Q.; Spychala, C.N.; McElheny, C.L.; Doi, Y. Comparative analysis of an IncR plasmid carrying armA, blaDHA-1 and qnrB4 from Klebsiella pneumoniae ST37 isolates. J. Antimicrob. Chemother. 2016, 71, 882–886. [Google Scholar] [CrossRef] [Green Version]
- Sampei, G.; Furuya, N.; Tachibana, K.; Saitou, Y.; Suzuki, T.; Mizobuchi, K.; Komano, T. Complete genome sequence of the incompatibility group I1 plasmid R64. Plasmid 2010, 64, 92–103. [Google Scholar] [CrossRef]
- Lodwick, D.; Owen, D.; Strike, P. DNA sequence analysis of the imp UV protection and mutation operon of the plasmid TP110: Identification of a third gene. Nucleic Acids Res. 1990, 18, 5045–5050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagdasarian, M.; Bailone, A.; Bagdasarian, M.M.; Manning, P.A.; Lurz, R.; Timmis, K.N.; Devoret, R. An inhibitor of SOS induction, specified by a plasmid locus in Escherichia coli. Proc. Natl. Acad. Sci. USA 1986, 83, 5723–5726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Reuter, M.; Sanghvi, B.; Roberts, G.A.; Cooper, L.P.; Tilling, M.; Blakely, G.W.; Dryden, D.T. ArdA proteins from different mobile genetic elements can bind to the EcoKI Type I DNA methyltransferase of E. coli K12. Biochim. Biophys. Acta 2014, 1844, 505–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuya, N.; Komano, T. Nucleotide sequence and characterization of the trbABC region of the IncI1 Plasmid R64: Existence of the pnd gene for plasmid maintenance within the transfer region. J. Bacteriol. 1996, 178, 1491–1497. [Google Scholar] [CrossRef] [Green Version]
- Sakuma, T.; Tazumi, S.; Furuya, N.; Komano, T. ExcA proteins of IncI1 plasmid R64 and IncIgamma plasmid R621a recognize different segments of their cognate TraY proteins in entry exclusion. Plasmid 2013, 69, 138–145. [Google Scholar] [CrossRef]
- Pong, C.H.; Harmer, C.J.; Ataide, S.F.; Hall, R.M. An IS26 variant with enhanced activity. FEMS Microbiol. Lett. 2019, 366, fnz031. [Google Scholar] [CrossRef]
- Jaffe, A.; Ogura, T.; Hiraga, S. Effects of the ccd function of the F plasmid on bacterial growth. J. Bacteriol. 1985, 163, 841–849. [Google Scholar] [CrossRef] [Green Version]
- Gerdes, K.; Rasmussen, P.B.; Molin, S. Unique type of plasmid maintenance function: Postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 1986, 83, 3116–3120. [Google Scholar] [CrossRef] [Green Version]
- Guiney, D.G.; Fierer, J. The Role of the spv Genes in Salmonella Pathogenesis. Front. Microbiol. 2011, 2, 129. [Google Scholar] [CrossRef] [Green Version]
- Lawley, T.D.; Burland, V.; Taylor, D.E. Analysis of the complete nucleotide sequence of the tetracycline-resistance transposon Tn10. Plasmid 2000, 43, 235–239. [Google Scholar] [CrossRef]
- Gupta, A.; Matsui, K.; Lo, J.F.; Silver, S. Molecular basis for resistance to silver cations in Salmonella. Nat. Med. 1999, 5, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Eger, E.; Heiden, S.E.; Becker, K.; Rau, A.; Geisenhainer, K.; Idelevich, E.A.; Schaufler, K. Hypervirulent Klebsiella pneumoniae Sequence Type 420 with a Chromosomally Inserted Virulence Plasmid. Int. J. Mol. Sci. 2021, 22, 9196. [Google Scholar] [CrossRef] [PubMed]
- Mollet, B.; Clerget, M.; Meyer, J.; Iida, S. Organization of the Tn6-related kanamycin resistance transposon Tn2680 carrying two copies of IS26 and an IS903 variant, IS903. B. J. Bacteriol. 1985, 163, 55–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siguier, P.; Gourbeyre, E.; Varani, A.; Ton-Hoang, B.; Chandler, M. Everyman’s Guide to Bacterial Insertion Sequences. Microbiol. Spectr. 2015, 3, MDNA3-0030-2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahillon, J.; Chandler, M. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998, 62, 725–774. [Google Scholar] [CrossRef] [Green Version]
- Papp, P.P.; Iyer, V.N. Determination of the binding sites of RepA, a replication initiator protein of the basic replicon of the IncN group plasmid pCU1. J. Mol. Biol. 1995, 246, 595–608. [Google Scholar] [CrossRef]
- Eikmeyer, F.; Hadiati, A.; Szczepanowski, R.; Wibberg, D.; Schneiker-Bekel, S.; Rogers, L.M.; Brown, C.J.; Top, E.M.; Puhler, A.; Schluter, A. The complete genome sequences of four new IncN plasmids from wastewater treatment plant effluent provide new insights into IncN plasmid diversity and evolution. Plasmid 2012, 68, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Smyshlyaev, G.; Bateman, A.; Barabas, O. Sequence analysis of tyrosine recombinases allows annotation of mobile genetic elements in prokaryotic genomes. Mol. Syst. Biol. 2021, 17, e9880. [Google Scholar] [CrossRef]
- Tavakoli, N.P.; Derbyshire, K.M. Tipping the balance between replicative and simple transposition. Embo J. 2001, 20, 2923–2930. [Google Scholar] [CrossRef] [Green Version]
- Coppo, A.; Colombo, M.; Pazzani, C.; Bruni, R.; Mohamud, K.A.; Omar, K.H.; Mastrandrea, S.; Salvia, A.M.; Rotigliano, G.; Maimone, F. Vibrio cholerae in the horn of Africa: Epidemiology, plasmids, tetracycline resistance gene amplification, and comparison between O1 and non-O1 strains. Am. J. Trop. Med. Hyg. 1995, 53, 351–359. [Google Scholar] [CrossRef]
- Pesesky, M.W.; Tilley, R.; Beck, D.A.C. Mosaic plasmids are abundant and unevenly distributed across prokaryotic taxa. Plasmid 2019, 102, 10–18. [Google Scholar] [CrossRef] [PubMed]
- David, S.; Cohen, V.; Reuter, S.; Sheppard, A.E.; Giani, T.; Parkhill, J.; Rossolini, G.M.; Feil, E.J.; Grundmann, H.; Aanensen, D.M. Integrated chromosomal and plasmid sequence analyses reveal diverse modes of carbapenemase gene spread among Klebsiella pneumoniae. Proc. Natl. Acad. Sci. USA 2020, 117, 25043–25054. [Google Scholar] [CrossRef] [PubMed]
- McMillan, E.A.; Jackson, C.R.; Frye, J.G. Transferable Plasmids of Salmonella enterica Associated With Antibiotic Resistance Genes. Front. Microbiol. 2020, 11, 562181. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Dong, N.; Liu, X.; Yang, C.; Ye, L.; Chan, E.W.; Zhang, R.; Chen, S. Co-conjugation of Virulence Plasmid and KPC Plasmid in a Clinical Klebsiella pneumoniae Strain. Front. Microbiol. 2021, 12, 739461. [Google Scholar] [CrossRef]
- Alonso, C.A.; de Toro, M.; de la Cruz, F.; Torres, C. Genomic Insights into Drug Resistance and Virulence Platforms, CRISPR-Cas Systems and Phylogeny of Commensal E. coli from Wildlife. Microorganisms 2021, 9, 999. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Schuyler, J.; Zhang, H.; Shashkina, E.; Du, H.; Fouts, D.E.; Satlin, M.; Kreiswirth, B.N.; Chen, L. Apramycin resistance in epidemic carbapenem-resistant Klebsiella pneumoniae ST258 strains. J. Antimicrob. Chemother. 2021, 76, 2017–2023. [Google Scholar] [CrossRef]
- Sim, W.; Barnard, R.T.; Blaskovich, M.A.T.; Ziora, Z.M. Antimicrobial Silver in Medicinal and Consumer Applications: A Patent Review of the Past Decade (2007(-)2017). Antibiotics 2018, 7, 93. [Google Scholar] [CrossRef] [Green Version]
- Deshmukh, S.P.; Patil, S.M.; Mullani, S.B.; Delekar, S.D. Silver nanoparticles as an effective disinfectant: A review. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 97, 954–965. [Google Scholar] [CrossRef]
- Hosny, A.E.M.; Rasmy, S.A.; Aboul-Magd, D.S.; Kashef, M.T.; El-Bazza, Z.E. The increasing threat of silver-resistance in clinical isolates from wounds and burns. Infect. Drug Resist. 2019, 12, 1985–2001. [Google Scholar] [CrossRef] [Green Version]
- McNeilly, O.; Mann, R.; Hamidian, M.; Gunawan, C. Emerging Concern for Silver Nanoparticle Resistance in Acinetobacter baumannii and Other Bacteria. Front. Microbiol. 2021, 12, 652863. [Google Scholar] [CrossRef]
- Elkrewi, E.; Randall, C.P.; Ooi, N.; Cottell, J.L.; O’Neill, A.J. Cryptic silver resistance is prevalent and readily activated in certain Gram-negative pathogens. J. Antimicrob. Chemother. 2017, 72, 3043–3046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Li, J.; Min, C.; Xia, F.; Tang, M.; Hu, Y.; Zou, M. Characterization of Silver Resistance and Coexistence of sil Operon with Antibiotic Resistance Genes among Gram-Negative Pathogens Isolated from Wound Samples by Using Whole-Genome Sequencing. Infect. Drug Resist. 2022, 15, 1425–1437. [Google Scholar] [CrossRef] [PubMed]
- Varani, A.; He, S.; Siguier, P.; Ross, K.; Chandler, M. The IS6 family, a clinically important group of insertion sequences including IS26. Mob. DNA 2021, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Bielak, E.; Bergenholtz, R.D.; Jorgensen, M.S.; Sorensen, S.J.; Hansen, L.H.; Hasman, H. Investigation of diversity of plasmids carrying the blaTEM-52 gene. J. Antimicrob. Chemother. 2011, 66, 2465–2474. [Google Scholar] [CrossRef] [Green Version]
| Strain | Resistance(s) a | Resistance Genes | Plasmid | Transconjugant (Plasmid) | Resistance Genes Transferred by Conjugation | Frequency of Conjugation (SD) d |
|---|---|---|---|---|---|---|
| ST1023 | CmSmSuTcTp | dfrA12-aadA2-cmlA1-aadA1-sul3-tetB-tetC | pST1023 | ND | none | none |
| BA3A | CmSmSuTcTp | dfrA12-aadA2-cmlA1-aadA1-sul3-tetB-tetC | pST1023 | BA3C | ||
| ApSmSu | blaTEM-strAB-sul2 | pST1007-1D (b) | (pST1007-1D) | blaTEM-strAB-sul2 | 3.0 (±3.9) × 10−1 | |
| BA3D | ||||||
| (pST1007-1D) | blaTEM-strAB-sul2; tetB | 8.0 (±0.2) × 10−5 | ||||
| (pST1023) | dfrA12-aadA2-cmlA1-aadA1-sul3-tetB-tetC | |||||
| BA3B | CmSmSuTcTp | dfrA12-aadA2-cmlA1-aadA1-sul3-tetB-tetC | pST1023 | BA3E | blaTEM-aphaI-strAB-sul2-tetD | 1.6 (±0.0) × 10−1 |
| ApKnSmSuTc | blaTEM-aphaI-strAB-sul2-tetD | pVC1035 (c) | (pVC1035) | |||
| BA3F | ||||||
| (pVC1035) | blaTEM-aphaI-strAB-sul2-tetD | 1.6 (±0.0) × 10−4 | ||||
| (pST1023) | dfrA12-aadA2-cmlA1-aadA1-sul3-tetB-tetC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calia, C.; Oliva, M.; Ferrara, M.; Minervini, C.F.; Scrascia, M.; Monno, R.; Mulè, G.; Cumbo, C.; Marzella, A.; Pazzani, C. Identification and Characterisation of pST1023 A Mosaic, Multidrug-Resistant and Mobilisable IncR Plasmid. Microorganisms 2022, 10, 1592. https://doi.org/10.3390/microorganisms10081592
Calia C, Oliva M, Ferrara M, Minervini CF, Scrascia M, Monno R, Mulè G, Cumbo C, Marzella A, Pazzani C. Identification and Characterisation of pST1023 A Mosaic, Multidrug-Resistant and Mobilisable IncR Plasmid. Microorganisms. 2022; 10(8):1592. https://doi.org/10.3390/microorganisms10081592
Chicago/Turabian StyleCalia, Carla, Marta Oliva, Massimo Ferrara, Crescenzio Francesco Minervini, Maria Scrascia, Rosa Monno, Giuseppina Mulè, Cosimo Cumbo, Angelo Marzella, and Carlo Pazzani. 2022. "Identification and Characterisation of pST1023 A Mosaic, Multidrug-Resistant and Mobilisable IncR Plasmid" Microorganisms 10, no. 8: 1592. https://doi.org/10.3390/microorganisms10081592
APA StyleCalia, C., Oliva, M., Ferrara, M., Minervini, C. F., Scrascia, M., Monno, R., Mulè, G., Cumbo, C., Marzella, A., & Pazzani, C. (2022). Identification and Characterisation of pST1023 A Mosaic, Multidrug-Resistant and Mobilisable IncR Plasmid. Microorganisms, 10(8), 1592. https://doi.org/10.3390/microorganisms10081592













