An Overview of the Recent Advances in Antimicrobial Resistance
Abstract
:1. Introduction
- Promoting ASPs to optimize the use of antimicrobial agents;
- Implementing robust IPC strategies;
- Investing in the research and development (R&D) of new antimicrobial agents and alternative therapies;
- Strengthening surveillance and monitoring systems to track the emergence and dissemination of resistant pathogens.
2. Materials and Methods
3. Mechanisms of Antimicrobial Resistance
3.1. Development of Antimicrobial Resistance
3.1.1. Antimicrobial Resistance Mechanisms: Bacteria
3.1.2. Antimicrobial Resistance Mechanisms: Viruses
3.1.3. Antimicrobial Resistance Mechanisms: Fungi
3.1.4. Antimicrobial Resistance Mechanisms: Parasites
3.2. Genetic Mechanisms of Antimicrobial Resistance
3.2.1. Mutations
3.2.2. Horizontal Gene Transfer
3.2.3. Selection Pressure and Its Role in AMR Development
Antimicrobial Use
Suboptimal Dosing
Environmental Factors
3.3. Interplay between Genetic Mechanisms and Selection Pressure
3.4. Clinical and Public Health Implications
4. Epidemiology of Antimicrobial Resistance
4.1. Global Prevalence and Distribution of Antimicrobial Resistance
4.2. Trends in Antimicrobial Resistance over Time and across Different Regions
4.3. Factors Contributing to the Dissemination of Antimicrobial Resistance
4.4. Comprehensive Strategies for Combating Antimicrobial Resistance
5. Impact of Antimicrobial Resistance
5.1. Consequences of Antimicrobial Resistance for Human Health
5.2. Challenges in Treating Resistant Infections
5.3. Economic Impact on Healthcare Systems and Society
5.4. Global Implications and the Need for Coordinated Action
5.5. The Social and Ethical Dimensions of AMR
6. Antimicrobial Stewardship and Infection Prevention Control
6.1. Strategies to Promote Responsible Antimicrobial Use in Healthcare Settings
6.2. Importance of Infection Prevention and Control Strategies
6.3. Global Surveillance Systems for Antimicrobial Resistance
6.4. Novel Approaches to Combat Antimicrobial Resistance
7. The One Health Approach to Antimicrobial Resistance
7.1. Recognition of the Interconnectedness of Human, Animal, and Environmental Health
7.1.1. The Human Health Sector
7.1.2. The Animal Health Sector
7.1.3. The Environmental Health Sector
7.2. Importance of Collaborative Efforts between Healthcare, Veterinary, and Environmental Sectors
7.2.1. Assessing One Health Integration in Antimicrobial Resistance Surveillance Systems
7.2.2. Collaborative Policy Approaches to Combat Antimicrobial Resistance
7.2.3. Joint Research and Innovation
7.2.4. Education and Awareness
7.2.5. Strengthening Infection Prevention and Control
7.2.6. Global Cooperation and Capacity Building
8. Policy and Regulatory Initiatives
8.1. Overview of National and International Policies
8.1.1. International Policies
8.1.2. National Policies
8.2. Regulatory Frameworks to Promote Responsible Antimicrobial Use
8.2.1. Regulatory Frameworks and Antimicrobial Stewardship in Combating Antibiotic Misuse in Human Health
8.2.2. Regulatory Frameworks and Antimicrobial Stewardship in Combating Antibiotic Misuse in Animal Health and Agriculture
8.2.3. Regulatory Frameworks and Antimicrobial Stewardship in Combating Antibiotic Misuse in Environmental Health
8.3. Challenges and Opportunities for Implementation of Policy Interventions
8.3.1. Resource Limitations
8.3.2. Political Will and Regulatory Capacity
8.3.3. Global Coordination and Harmonization
8.3.4. Opportunities for Innovation and Collaboration
9. Future Directions and Challenges
9.1. Emerging Threats in Antimicrobial Resistance
9.1.1. Multidrug-Resistant Pathogens
9.1.2. Novel Resistance Mechanisms
9.2. Research Priorities for New Antimicrobial Agents and Alternative Strategies
9.2.1. Development of Novel Antimicrobials
9.2.2. Alternative Treatment Strategies
9.3. Societal and Ethical Implications of Antimicrobial Resistance Interventions
9.3.1. Access and Equity
9.3.2. Stewardship and Overuse
9.3.3. Environmental Impact
9.3.4. Ethical Dilemmas in Research and Innovation
9.4. Strategic Roadmap and Challenges Ahead
9.4.1. Strengthening Global Surveillance and Data Sharing
9.4.2. Integrating AMR into National and Global Health Agendas
9.4.3. Fostering Collaboration across Sectors
9.4.4. Promoting Public Awareness and Behavioral Change
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Merlino, J. Antimicrobial Resistance a Threat to Public Health. Microbiol. Aust. 2017, 38, 165–167. [Google Scholar] [CrossRef]
- Capozzi, C.; Maurici, M.; Panà, A. Antimicrobial Resistance: It Is a Global Crisis, a Slow Tsunami. Ig. Sanita Pubbl. 2019, 75, 429–450. [Google Scholar] [PubMed]
- Aijaz, M.; Ahmad, M.; Ansari, M.A.; Ahmad, S. Antimicrobial Resistance in a Globalized World: Current Challenges and Future Perspectives. Int. J. Pharm. Drug Des. 2023, 1, 7–22. [Google Scholar]
- Jindal, A.K.; Pandya, K.; Khan, I.D. Antimicrobial Resistance: A Public Health Challenge. Med. J. Armed Forces India 2015, 71, 178–181. [Google Scholar] [CrossRef] [PubMed]
- Larson, E. Community Factors in the Development of Antibiotic Resistance. Annu. Rev. Public Health 2015, 28, 435–447. [Google Scholar] [CrossRef]
- Holmes, A.H.; Moore, L.S.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Piddock, L.J. Understanding the Mechanisms and Drivers of Antimicrobial Resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- Tenover, F.C. Mechanisms of Antimicrobial Resistance in Bacteria. Am. J. Med. 2006, 119, S3–S10. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J. Antibiotic Resistance in Staphylococcus aureus. Current Status and Future Prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef] [PubMed]
- Eliopoulos, G.M.; Cosgrove, S.E.; Carmeli, Y. The Impact of Antimicrobial Resistance on Health and Economic Outcomes. Clin. Infect. Dis. 2003, 36, 1433–1437. [Google Scholar] [CrossRef]
- Cosgrove, S.E. The Relationship between Antimicrobial Resistance and Patient Outcomes: Mortality, Length of Hospital Stay, and Health Care Costs. Clin. Infect. Dis. 2006, 42 (Suppl. S2), S82–S89. [Google Scholar] [CrossRef]
- Maragakis, L.L.; Perencevich, E.N.; Cosgrove, S.E. Clinical and Economic Burden of Antimicrobial Resistance. Expert Rev. Anti-Infect. Ther. 2008, 6, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Marston, H.D.; Dixon, D.M.; Knisely, J.M.; Palmore, T.N.; Fauci, A.S. Antimicrobial Resistance. JAMA 2016, 316, 1193–1204. [Google Scholar] [CrossRef] [PubMed]
- Premanandh, J.; Samara, B.S.; Mazen, A.N. Race against Antimicrobial Resistance Requires Coordinated Action–An Overview. Front. Microbiol. 2016, 6, 1536. [Google Scholar] [CrossRef] [PubMed]
- Stekel, D. First Report of Antimicrobial Resistance Pre-Dates Penicillin. Nature 2018, 562, 192. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, A.J. History of Antibiotics and Evolution of Resistance. Res. J. Pharm. Technol. 2015, 8, 1719–1724. [Google Scholar] [CrossRef]
- Rosenblatt-Farrell, N. The Landscape of Antibiotic Resistance. Environ. Health Perspect. 2009, 117, A244–A250. [Google Scholar] [CrossRef] [PubMed]
- Acar, J.F.; Moulin, G. Antimicrobial Resistance: A Complex Issue. Rev. Sci. Tech. 2012, 31, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, Present, and Future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Vanegas-Múnera, J.M.; Jiménez-Quiceno, J.N. Resistencia Antimicrobiana en el Siglo XXI: ¿Hacia una Era Postantibiótica? Rev. Fac. Nac. Salud Pública 2020, 38, 1–6. [Google Scholar] [CrossRef]
- Medina, E.; Pieper, D.H. Tackling Threats and Future Problems of Multidrug-Resistant Bacteria. In How to Overcome the Antibiotic Crisis; Springer: Singapore, 2016; pp. 3–33. [Google Scholar]
- Donkor, E.S.; Codjoe, F.S. Methicillin-Resistant Staphylococcus aureus and Extended-Spectrum Beta-Lactamase Producing Enterobacteriaceae: A Therapeutic Challenge in the 21st Century. Open Microbiol. J. 2019, 13, 94–100. [Google Scholar] [CrossRef]
- Ynion, G.P.L.; Rosal, C.J.; Ordanel, Z.A.A.; Caipang, C.M. Challenges and Emerging Molecular Approaches in Combating Antimicrobial Resistance. J. Bacteriol. Virol. 2024, 54, 12–39. [Google Scholar] [CrossRef]
- Kumar, S.B.; Arnipalli, S.R.; Ziouzenkova, O. Antibiotics in Food Chain: The Consequences for Antibiotic Resistance. Antibiotics 2020, 9, 688. [Google Scholar] [CrossRef] [PubMed]
- Silbergeld, E.K.; Graham, J.; Price, L.B. Industrial Food Animal Production, Antimicrobial Resistance, and Human Health. Annu. Rev. Public Health 2008, 29, 151–169. [Google Scholar] [CrossRef] [PubMed]
- Jadeja, N.B.; Worrich, A. From Gut to Mud: Dissemination of Antimicrobial Resistance between Animal and Agricultural Niches. Environ. Microbiol. 2022, 28, 3290–3306. [Google Scholar] [CrossRef] [PubMed]
- Khachatourians, G.G. Agricultural Use of Antibiotics and the Evolution and Transfer of Antibiotic-Resistant Bacteria. Can. Med. Assoc. J. 1998, 159, 1129–1136. [Google Scholar]
- Freeland, G.; Atungulu, H.N.G.; Apple, J.; Mukherjee, S. Strategies to Combat Antimicrobial Resistance from Farm to Table. Food Rev. Int. 2023, 39, 27–40. [Google Scholar] [CrossRef]
- Larsson, D.J.; Bengtsson-Palme, A.A.; Brandt, K.K.; de Roda Husman, A.M.; Fagerstedt, P.; Wernersson, A.S. Critical Knowledge Gaps and Research Needs Related to the Environmental Dimensions of Antibiotic Resistance. Environ. Int. 2018, 117, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental Antimicrobial Resistance and Its Drivers: A Potential Threat to Public Health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar]
- Wang, W.; Weng, Y.; Luo, T.; Wang, Q.; Yang, G.; Jin, Y. Antimicrobial and the Resistances in the Environment: Ecological and Health Risks, Influencing Factors, and Mitigation Strategies. Toxics 2023, 11, 185. [Google Scholar] [CrossRef]
- Calvo-Villamañán, A.; Álvarez, S.M.; Carrilero, L. Tackling AMR from a Multidisciplinary Perspective: A Primer from Education and Psychology. Int. Microbiol. 2023, 26, 1–9. [Google Scholar] [CrossRef]
- Hayes, J.F. Fighting Back against Antimicrobial Resistance with Comprehensive Policy and Education: A Narrative Review. Antibiotics 2022, 11, 644. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.J.; Wall, S. The AMR Emergency: Multi-Sector Collaboration and Collective Global Policy Action Is Needed Now. Glob. Health Action 2019, 12 (Suppl. S1), 1855831. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Trejo, A.; Ruiz-Ruiz, J.M.; Gonzalez-Avila, L.U.; Saldaña-Padilla, A.; Hernández-Cortez, C.; Loyola-Cruz, M.A.; Castro-Escarpulli, G. Evasion of Antimicrobial Activity in Acinetobacter baumannii by Target Site Modifications: An Effective Resistance Mechanism. Int. J. Mol. Sci. 2022, 23, 6582. [Google Scholar] [CrossRef] [PubMed]
- Arenz, S.; Wilson, D.N. Bacterial Protein Synthesis as a Target for Antibiotic Inhibition. Cold Spring Harb. Perspect. Med. 2016, 6, a025361. [Google Scholar] [CrossRef]
- Babosan, A.; Fruchard, L.; Carvalho, K.E.A.; Mazel, D.; Baharoglu, Z. Nonessential tRNA and rRNA Modifications Impact the Bacterial Response to Sub-MIC Antibiotic Pressure. Nucleic Acids Res. 2023, 51, 4878–4893. [Google Scholar]
- Sun, J.; Deng, Z.; Yan, A. Bacterial Multidrug Efflux Pumps: Mechanisms, Physiology and Pharmacological Exploitations. Biochem. Biophys. Res. Commun. 2014, 453, 254–267. [Google Scholar] [CrossRef]
- Huang, L.; Wu, C.; Gao, H.; Xu, C.; Dai, M.; Huang, L.; Cheng, G. Bacterial Multidrug Efflux Pumps at the Frontline of Antimicrobial Resistance: An Overview. Antibiotics 2022, 11, 520. [Google Scholar] [CrossRef]
- Zgurskaya, H.I.; Nikaido, H. Multidrug Resistance Mechanisms: Drug Efflux across Two Membranes. Mol. Microbiol. 2000, 37, 219–225. [Google Scholar] [CrossRef]
- Egorov, A.M.; Ulyashova, M.M.; Rubtsova, M.Y. Bacterial Enzymes and Antibiotic Resistance. Acta Naturae 2018, 10, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, R.; Nusrin, K.S. Role of Beta-Lactamases in Antibiotic Resistance: A Review. Int. Res. J. Pharm 2014, 5, 37–41. [Google Scholar] [CrossRef]
- Agarwal, V.; Tiwari, A.; Varadwaj, P. An Extensive Review on β-Lactamase Enzymes and Their Inhibitors. Curr. Med. Chem. 2023, 30, 783–808. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Bradford, P.A. β-Lactams and β-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef] [PubMed]
- Ghai, I.; Ghai, S. Understanding Antibiotic Resistance via Outer Membrane Permeability. Infect. Drug Resist. 2018, 11, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Ghai, I.; Ghai, S. Exploring Bacterial Outer Membrane Barrier to Combat Bad Bugs. Infect. Drug Resist. 2017, 10, 261–273. [Google Scholar] [CrossRef]
- Horne, J.E.; Brockwell, D.J.; Radford, S.E. Role of the Lipid Bilayer in Outer Membrane Protein Folding in Gram-Negative Bacteria. J. Biol. Chem. 2020, 295, 10340–10367. [Google Scholar] [CrossRef]
- Sahoo, J.P.; Mishra, A.P.; Samal, K.C.; Dash, A.K. Insights into the Antibiotic Resistance in Biofilms—A Review. Environ. Conserv. J. 2021, 22, 59–67. [Google Scholar] [CrossRef]
- Bano, S.; Hassan, N.; Rafiq, M.; Hassan, F.; Rehman, M.; Iqbal, N.; Kang, Y.Q. Biofilms as Battlefield Armor for Bacteria against Antibiotics: Challenges and Combating Strategies. Microorganisms 2023, 11, 2595. [Google Scholar] [CrossRef]
- Uruén, C.; Chopo-Escuin, G.; Tommassen, J.; Romero, M.-J.R.C.; Arenas, J. Biofilms as Promoters of Bacterial Antibiotic Resistance and Tolerance. Antibiotics 2020, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Hathroubi, S.; Mekni, M.A.; Domenico, P.; Nguyen, D.; Jacques, M. Biofilms: Microbial Shelters against Antibiotics. Microb. Drug Resist. 2017, 23, 147–156. [Google Scholar] [CrossRef]
- Lou, Z.; Sun, Y.; Rao, Z. Current Progress in Antiviral Strategies. Trends Pharmacol. Sci. 2014, 35, 86–102. [Google Scholar] [CrossRef] [PubMed]
- Irwin, K.K.; Renzette, N.; Kowalik, T.F.; Jensen, J.D. Antiviral Drug Resistance as an Adaptive Process. Virus Evol. 2016, 2, vew014. [Google Scholar] [CrossRef] [PubMed]
- Weber, I.T. Can We Design Drugs for HIV/AIDS That Are Less Susceptible to Resistance? Future Med. Chem. 2015, 7, 2301–2304. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Sharma, S.; Kumar, R.; Tripathi, B.N.; Barua, S.; Ly, H.; Rouse, B.T. Host-Directed Antiviral Therapy. Clin. Microbiol. Rev. 2020, 33, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Flowers, S.A.; Colón, B.; Whaley, S.G.; Schuler, M.A.; Rogers, P.D. Contribution of Clinically Derived Mutations in ERG11 to Azole Resistance in Candida albicans. Antimicrob. Agents Chemother. 2015, 59, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Engle, K.; Kumar, G. Tackling Multi-Drug Resistant Fungi by Efflux Pump Inhibitors. Biochem. Pharmacol. 2024, 226, 116400. [Google Scholar] [CrossRef]
- Garcia, Í.R.; de Oliveira Garcia, F.A.; Pereira, P.S.; Coutinho, H.D.M.; Siyadatpanah, A.; Norouzi, R.; Rodrigues, F.F.G. Microbial Resistance: The Role of Efflux Pump Superfamilies and Their Respective Substrates. Life Sci. 2022, 295, 120391. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.; Nair, R.; Banerjee, A. Multidrug Transporters of Candida Species in Clinical Azole Resistance. Fungal Genet. Biol. 2019, 132, 103252. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro, R.D.; Portela, F.V.; Pereira, L.M.; de Andrade, A.R.; de Sousa, J.K.; Aguiar, A.L.; Sidrim, J.J. Efflux Pump Inhibition Controls Growth and Enhances Antifungal Susceptibility of Fusarium solani Species Complex. Future Microbiol. 2020, 15, 9–20. [Google Scholar] [CrossRef]
- Aneke, C.I.; Rhimi, W.; Otranto, D.; Cafarchia, C. Synergistic Effects of Efflux Pump Modulators on the Azole Antifungal Susceptibility of Microsporum canis. Mycopathologia 2020, 185, 279–288. [Google Scholar] [CrossRef]
- Cannon, R.D.; Lamping, E.; Holmes, A.R.; Niimi, K.; Baret, P.V.; Keniya, M.V.; Monk, B.C. Efflux-Mediated Antifungal Drug Resistance. Clin. Microbiol. Rev. 2009, 22, 291–321. [Google Scholar] [CrossRef]
- Hori, Y.; Shibuya, K. Role of FKS Gene in the Susceptibility of Pathogenic Fungi to Echinocandins. Med. Mycol. J. 2018, 59, E31–E40. [Google Scholar] [CrossRef] [PubMed]
- Jospe-Kaufman, M.; Ben-Zeev, E.; Mottola, A.; Dukhovny, A.; Berman, J.; Carmeli, S.; Fridman, M. Reshaping Echinocandin Antifungal Drugs to Circumvent Glucan Synthase Point-Mutation-Mediated Resistance. Angew. Chem. Int. Ed. 2024, 63, e202314728. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.D.S.; Prado, A.; Bagon, N.P.; Negri, M.; Svidzinski, T.I.E. Mixed Fungal Biofilms: From Mycobiota to Devices, a New Challenge on Clinical Practice. Microorganisms 2022, 10, 1721. [Google Scholar] [CrossRef] [PubMed]
- de Barros, P.P.; Rossoni, L.; Scorzoni, L.; Fenley, J.D.C.; Junqueira, J.C. Candida Biofilms: An Update on Developmental Mechanisms and Therapeutic Challenges. Mycopathologia 2020, 185, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.F.; Alarcon, E.I. Bacterial Biofilm Formation on Implantable Devices and Approaches to Its Treatment and Prevention. Heliyon 2018, 4, e01067. [Google Scholar] [CrossRef]
- Birnbaum, J.; Scharf, S.; Schmidt, S.; Jonscher, E.; Hoeijmakers, W.A.M.; Flemming, S.; Spielmann, T. A Kelch13-Defined Endocytosis Pathway Mediates Artemisinin Resistance in Malaria Parasites. Science 2020, 367, 51–59. [Google Scholar] [CrossRef]
- Yang, T.; Yeoh, L.M.; Tutor, M.V.; Dixon, M.W.; McMillan, P.J.; Xie, S.C.; Cobbold, S.A. Decreased K13 Abundance Reduces Hemoglobin Catabolism and Proteotoxic Stress, Underpinning Artemisinin Resistance. Cell Rep. 2019, 29, 2917–2928. [Google Scholar] [CrossRef]
- Ward, K.E.; Fidock, D.A.; Bridgford, J.L. Plasmodium falciparum Resistance to Artemisinin-Based Combination Therapies. Curr. Opin. Microbiol. 2022, 69, 102–193. [Google Scholar] [CrossRef]
- Wicht, K.J.; Mok, S.F.D.A. Molecular Mechanisms of Drug Resistance in Plasmodium falciparum Malaria. Annu. Rev. Microbiol. 2020, 74, 431–454. [Google Scholar] [CrossRef]
- Ma, C.I.; Tirtorahardjo, J.A.; Jan, S.; Schweizer, S.S.; Rosario, S.A.; Du, Y.; Andrade, R.M. Auranofin Resistance in Toxoplasma gondii Decreases the Accumulation of Reactive Oxygen Species but Does Not Target Parasite Thioredoxin Reductase. Front. Cell. Infect. Microbiol. 2021, 11, 618994. [Google Scholar] [CrossRef]
- Tekwani, B.L.; Shukla, O.P.; Ghatak, S. Altered Drug Metabolism in Parasitic Diseases. Parasitol. Today 1988, 4, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Wittlin, S.; Mäser, P. From Magic Bullet to Magic Bomb: Reductive Bioactivation of Antiparasitic Agents. ACS Infect. Dis. 2021, 7, 2777–2786. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, S.R.; Taylor, M.C.; Horn, D.; Kelly, J.M.; Cheeseman, I. A Mechanism for Cross-Resistance to Nifurtimox and Benznidazole in Trypanosomes. Proc. Natl. Acad. Sci. USA 2008, 105, 5022–5027. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, C.; Lun, Z.R.; Meshnick, S.R. Unpacking “Artemisinin Resistance”. Trends Pharmacol. Sci. 2017, 38, 506–511. [Google Scholar] [CrossRef]
- Ma, Y.; Frutos-Beltrán, E.; Kang, D.; Pannecouque, C.; De Clercq, E.; Menéndez-Arias, L.; Zhan, P. Medicinal Chemistry Strategies for Discovering Antivirals Effective against Drug-Resistant Viruses. Chem. Soc. Rev. 2021, 45, 4514–4540. [Google Scholar] [CrossRef] [PubMed]
- Nastri, B.M.; Pagliano, P.; Zannella, C.; Folliero, V.; Masullo, A.; Rinaldi, L.; Franci, G. HIV and Drug-Resistant Subtypes. Microorganisms 2023, 11, 221. [Google Scholar] [CrossRef]
- Lefaan, Y.F.M.; Setiadhi, R. Thymidine Kinase as a Causative Factor for Type 1 Herpes Simplex Virus Resistance against Acyclovir. Dentino J. Kedokt. Gigi 2020, 5, 159–164. [Google Scholar] [CrossRef]
- Revie, N.M.; Iyer, K.R.; Robbins, N.; Cowen, L.E. Antifungal Drug Resistance: Evolution, Mechanisms and Impact. Curr. Opin. Microbiol. 2018, 45, 70–76. [Google Scholar] [CrossRef]
- Arastehfar, A.; Lass-Flörl, C.; Garcia-Rubio, R.; Daneshnia, F.; Ilkit, M.; Boekhout, T.; Perlin, D.S. The Quiet and Underappreciated Rise of Drug-Resistant Invasive Fungal Pathogens. J. Fungi 2020, 6, 138. [Google Scholar] [CrossRef]
- Ben-Ami, R.; Kontoyiannis, D.P. Resistance to Antifungal Drugs. Infect. Dis. Clin. N. Am. 2021, 35, 279–311. [Google Scholar] [CrossRef]
- Fissiha, W.; Kinde, M.Z. Anthelmintic Resistance and Its Mechanism: A Review. Heliyon 2021, 7, e08487. [Google Scholar] [CrossRef] [PubMed]
- Alvi, M.A.; Athar Ali, R.M.; Asghar, F.; Saqib, M.; Ashfaq, K.; Rashid, I.; Ahmad, F. Drug Resistance in Parasitic Infections. In Parasitism and Parasitic Control in Animals: Strategies for the Developing World; CABI: Boston, MA, USA, 2023; pp. 111–123. [Google Scholar]
- Heinberg, A.; Kirkman, L. The Molecular Basis of Antifolate Resistance in Plasmodium falciparum: Looking Beyond Point Mutations. Ann. N. Y. Acad. Sci. 2015, 1342, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Reygaert, W.C. An Overview of the Antimicrobial Resistance Mechanisms of Bacteria. AIMS Microbiol. 2018, 4, 482. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Robbins, N.; Cowen, L.E. Molecular Mechanisms Governing Antifungal Drug Resistance. NPJ Antimicrob. Resist. 2023, 1, 5. [Google Scholar] [CrossRef]
- Schroeder, J.W.; Yeesin, P.; Simmons, L.A.; Wang, J.D. Sources of Spontaneous Mutagenesis in Bacteria. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 29–48. [Google Scholar] [CrossRef]
- Fukuda, H.; Hiramatsu, K. Mechanisms of Endogenous Drug Resistance Acquisition by Spontaneous Chromosomal Gene Mutation. Nihon Rinsho 1997, 55, 1185–1190. [Google Scholar] [PubMed]
- Bena, C.E.; Ollion, J.; De Paepe, M.; Ventroux, M.; Robert, L.; Elez, M. Real-Time Monitoring of Replication Errors’ Fate Reveals the Origin and Dynamics of Spontaneous Mutations. Nat. Commun. 2024, 15, 2702. [Google Scholar] [CrossRef]
- Willmott, C.J.; Maxwell, A. A Single Point Mutation in the DNA Gyrase A Protein Greatly Reduces Binding of Fluoroquinolones to the Gyrase-DNA Complex. Antimicrob. Agents Chemother. 1993, 37, 126–127. [Google Scholar] [CrossRef] [PubMed]
- Sarisky, R.T.; Nguyen, T.T.; Duffy, K.E.; Wittrock, R.J.; Leary, J.J. Difference in Incidence of Spontaneous Mutations Between Herpes Simplex Virus Types 1 and 2. Antimicrob. Agents Chemother. 2000, 44, 1524–1529. [Google Scholar] [CrossRef]
- Haver, H.L.; Chua, A.; Ghode, P.; Lakshminarayana, S.B.; Singhal, A.; Mathema, B.; Bifani, P. Mutations in Genes for the F420 Biosynthetic Pathway and a Nitroreductase Enzyme Are the Primary Resistance Determinants in Spontaneous in Vitro-Selected PA-824-Resistant Mutants of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2015, 59, 5316–5323. [Google Scholar] [CrossRef]
- Revitt-Mills, S.A.; Robinson, A. Antibiotic-Induced Mutagenesis: Under the Microscope. Front. Microbiol. 2020, 11, 585175. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, R.S. Antimicrobial-Induced DNA Damage and Genomic Instability in Microbial Pathogens. PLoS Pathog. 2015, 11, e1004678. [Google Scholar] [CrossRef] [PubMed]
- Blázquez, J.; Couce, A.; Rodríguez-Beltrán, J.; Rodríguez-Rojas, A. Antimicrobials as Promoters of Genetic Variation. Curr. Opin. Microbiol. 2012, 15, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Li, X.G.A.Z.; Xie, Z.Y.B.; Li, D.; Chen, J. Sub-Lethal Concentrations of Heavy Metals Induce Antibiotic Resistance via Mutagenesis. J. Hazard. Mater. 2019, 369, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ning, Q.; Deng, Z.; Zhang, M.; You, J. Role of Environmental Stresses in Elevating Resistance Mutations in Bacteria: Phenomena and Mechanisms. Environ. Pollut. 2022, 317, 119603. [Google Scholar] [CrossRef]
- Sevillya, G.; Adato, O.; Snir, S. Detecting Horizontal Gene Transfer: A Probabilistic Approach. BMC Genom. 2020, 21, 106. [Google Scholar] [CrossRef]
- Zhou, H.; Beltrán, J.F.; Brito, I.L. Functions Predict Horizontal Gene Transfer and the Emergence of Antibiotic Resistance. Sci. Adv. 2021, 7, eabj5056. [Google Scholar] [CrossRef]
- Arnold, B.J.; Huang, I.T.; Hanage, W.P. Horizontal Gene Transfer and Adaptive Evolution in Bacteria. Nat. Rev. Microbiol. 2022, 20, 206–218. [Google Scholar] [CrossRef]
- Van Etten, J.; Bhattacharya, D. Horizontal Gene Transfer in Eukaryotes: Not If, but How Much? Trends Genet. 2020, 36, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Fang, G.Y.; Liu, X.Q.; Jiang, Y.J.; Mu, X.J.; Huang, B.W. Horizontal Gene Transfer in Activated Sludge Enhances Microbial Antimicrobial Resistance and Virulence. Sci. Total Environ. 2024, 912, 168908. [Google Scholar] [CrossRef]
- Liu, G.; Thomsen, L.E.; Olsen, J.E. Antimicrobial-Induced Horizontal Transfer of Antimicrobial Resistance Genes in Bacteria: A Mini-Review. J. Antimicrob. Chemother. 2022, 77, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Soler, N.; Forterre, P. Vesiduction: The Fourth Way of HGT. Environ. Microbiol. 2020, 22, 2457–2460. [Google Scholar] [CrossRef] [PubMed]
- Virolle, C.; Goldlust, K.; Djermoun, S.; Bigot, S.; Lesterlin, C. Plasmid Transfer by Conjugation in Gram-Negative Bacteria: From the Cellular to the Community Level. Genes 2020, 11, 1239. [Google Scholar] [CrossRef] [PubMed]
- Waksman, G. From Conjugation to T4S Systems in Gram-Negative Bacteria: A Mechanistic Biology Perspective. EMBO Rep. 2019, 20, e47012. [Google Scholar] [CrossRef] [PubMed]
- Goessweiner-Mohr, N.; Arends, K.; Keller, W.; Grohmann, E. Conjugation in Gram-Positive Bacteria. In Plasmids: Biology and Impact in Biotechnology and Discovery; American Society for Microbiology Press: Washington, DC, USA, 2015; pp. 237–256. [Google Scholar]
- Bañuelos-Vazquez, L.A.; Tejerizo, G.T.; Brom, S. Regulation of Conjugative Transfer of Plasmids and Integrative Conjugative Elements. Plasmid 2017, 91, 82–89. [Google Scholar] [CrossRef]
- Kohler, V.; Keller, W.; Grohmann, E. Regulation of Gram-Positive Conjugation. Front. Microbiol. 2019, 19, 1134. [Google Scholar] [CrossRef]
- Meijer, W.J.; Boer, D.R.; Ares, S.; Alfonso, C.; Rojo, F.; Luque-Ortega, J.R.; Wu, L.J. Multiple Layered Control of the Conjugation Process of the Bacillus subtilis Plasmid pLS20. Front. Mol. Biosci. 2021, 8, 648468. [Google Scholar] [CrossRef]
- Pfeifer, E.; Bonnin, R.A.; Rocha, E.P. Phage-Plasmids Spread Antibiotic Resistance Genes Through Infection and Lysogenic Conversion. mBio 2022, 13, e01851-22. [Google Scholar] [CrossRef]
- Povolo, V.R.; Ackermann, M. Disseminating Antibiotic Resistance During Treatment. Science 2019, 364, 737–738. [Google Scholar] [CrossRef]
- Shen, Z.; Tang, C.M.; Liu, G.Y. Towards a Better Understanding of Antimicrobial Resistance Dissemination: What Can Be Learnt from Studying Model Conjugative Plasmids? Mil. Med. Res. 2022, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Lade, H.; Kim, J.S. Molecular determinants of β-lactam resistance in methicillin-resistant Staphylococcus aureus (MRSA): An updated review. Antibiotics 2023, 12, 1362. [Google Scholar] [CrossRef] [PubMed]
- Manohar, P.; Leptihn, S.; Lopes, B.S.; Nachimuthu, R. Dissemination of carbapenem resistance and plasmids encoding carbapenemases in Gram-negative bacteria isolated in India. JAC-Antimicrob. Resist. 2021, 3, dlab015. [Google Scholar] [CrossRef] [PubMed]
- Bodendoerfer, E.; Marchesi, M.; Imkamp, F.; Courvalin, P.; Böttger, E.C.; Mancini, S. Co-occurrence of aminoglycoside and β-lactam resistance mechanisms in aminoglycoside-non-susceptible Escherichia coli isolated in the Zurich area, Switzerland. Int. J. Antimicrob. Agents 2020, 56, 106019. [Google Scholar] [CrossRef] [PubMed]
- Firmo, E.F.; Beltrão, E.M.B.; da Silva, F.R.F.; Alves, L.C.; Brayner, F.A.; Veras, D.L.; Lopes, A.C.S. Association of blaNDM-1 with blaKPC-2 and aminoglycoside-modifying enzyme genes among Klebsiella pneumoniae, Proteus mirabilis and Serratia marcescens clinical isolates in Brazil. J. Glob. Antimicrob. Resist. 2020, 21, 255–261. [Google Scholar] [CrossRef]
- Chopjitt, P.; Boueroy, P.; Morita, M.; Iida, T.; Akeda, Y.; Hamada, S.; Kerdsin, A. Genetic characterization of multidrug-resistant Escherichia coli harboring colistin-resistant gene isolated from food animals in food supply chain. Front. Cel. Infect. Microbiol. 2024, 14, 1289134. [Google Scholar] [CrossRef] [PubMed]
- Ghenea, A.E.; Zlatian, O.M.; Cristea, O.M.; Ungureanu, A.; Mititelu, R.R.; Balasoiu, A.T.; Salan, A.; Iliuta, D.; Popescu, M.; Udriștoiu, A.-L.; et al. TEM, CTX-M, SHV genes in ESBL-producing Escherichia coli and Klebsiella pneumoniae isolated from clinical samples in a county clinical emergency hospital Romania-predominance of CTX-M-15. Antibiotics 2022, 11, 503. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.Y.; Zhou, Z.; Papadopoulos, J.M.; Zuke, J.D.; Falbel, T.G.; Anantharaman, K.; Burton, B.M.; Venturelli, O.S. Efficient plasmid transfer via natural competence in a microbial co-culture. Mol. Syst. Biol. 2023, 19, e11406. [Google Scholar] [CrossRef] [PubMed]
- Winter, M.; Buckling, A.; Harms, K.; Johnsen, P.J.; Vos, M. Antimicrobial Resistance Acquisition via Natural Transformation: Context Is Everything. Curr. Opin. Microbiol. 2021, 64, 133–138. [Google Scholar] [CrossRef]
- Huang, M.; Liu, M.; Huang, L.; Wang, M.; Jia, R.; Zhu, D.; Chen, S.; Zhao, X.; Zhang, S.; Gaio, Q.; et al. The Activation and Limitation of the Bacterial Natural Transformation System: The Function in Genome Evolution and Stability. Microbiol. Res. 2021, 252, 126856. [Google Scholar] [CrossRef]
- Perez, F.; Stiefel, U. The Impact of Natural Transformation on the Acquisition of Antibiotic Resistance Determinants. mBio 2022, 13, e00336-22. [Google Scholar] [CrossRef]
- Carvalho, G.; Fouchet, D.; Danesh, G.; Godeux, A.S.; Laaberki, M.H.; Pontier, D.; Charpentier, X.; Venner, S. Bacterial Transformation Buffers Environmental Fluctuations Through the Reversible Integration of Mobile Genetic Elements. mBio 2020, 11, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.L. Bacteriophage-Mediated Horizontal Gene Transfer: Transduction. In Bacteriophage; Springer: Singapore, 2021; pp. 151–192. [Google Scholar]
- Calero-Cáceres, W.; Ye, M.; Balcázar, J.L. Bacteriophages as Environmental Reservoirs of Antibiotic Resistance. Trends Microbiol. 2019, 27, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Colavecchio, A.; Cadieux, B.; Lo, A.; Goodridge, L.D. Bacteriophages Contribute to the Spread of Antibiotic Resistance Genes Among Foodborne Pathogens of the Enterobacteriaceae Family—A Review. Front. Microbiol. 2017, 8, 1108. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.N.; Penadés, J.R.; Chen, J. Genetic Transduction by Phages and Chromosomal Islands: The New and Noncanonical. PLoS Pathog. 2019, 15, e1007878. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, M.K.; Jagannadham, M.V. Vesicles-Mediated Resistance to Antibiotics in Bacteria. Front. Microbiol. 2015, 6, 758. [Google Scholar] [CrossRef] [PubMed]
- Lehmkuhl, J.; Schneider, J.S.; Werth, K.L.V.; Scherff, N.; Mellmann, A.; Kampmeier, S. Role of Membrane Vesicles in the Transmission of Vancomycin Resistance in Enterococcus faecium. Sci. Rep. 2024, 14, 1895. [Google Scholar] [CrossRef]
- Marrec, L.; Bitbol, A.F. Resist or Perish: Fate of a Microbial Population Subjected to a Periodic Presence of Antimicrobial. PLoS Comput. Biol. 2020, 16, e1007798. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.J. Environmental Factors Influencing the Development and Spread of Antibiotic Resistance. FEMS Microbiol. Rev. 2018, 42, fux053. [Google Scholar] [CrossRef]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Kasimanickam, V.; Kasimanickam, M.; Kasimanickam, R. Antibiotics Use in Food Animal Production: Escalation of Antimicrobial Resistance: Where Are We Now in Combating AMR? Med. Sci. 2021, 9, 14. [Google Scholar] [CrossRef]
- Magouras, I.; Carmo, L.P.; Stärk, K.D.; Schüpbach-Regula, G. Antimicrobial Usage and-Resistance in Livestock: Where Should We Focus? Front. Vet. Sci. 2017, 4, 148. [Google Scholar] [CrossRef] [PubMed]
- Coque, T.M.; Cantón, R.; Pérez-Cobas, A.E.; Fernández-de-Bobadilla, M.D.; Baquero, F. Antimicrobial Resistance in the Global Health Network: Known Unknowns and Challenges for Efficient Responses in the 21st Century. Microorganisms 2023, 11, 105. [Google Scholar] [CrossRef] [PubMed]
- Guidry, C.A.; Davies, S.W.; Metzger, R.; Swenson, B.R.; Sawyer, R.G. Whence Resistance? Surg. Infect. 2015, 16, 716–720. [Google Scholar] [CrossRef] [PubMed]
- Heffernan, A.J.; Lim, S.M.S.; Lipman, J.; Roberts, J.A. A Personalised Approach to Antibiotic Pharmacokinetics and Pharmacodynamics in Critically Ill Patients. Anaesth. Crit. Care Pain Med. 2021, 40, 100970. [Google Scholar] [CrossRef]
- Goneau, L.W.; Delport, J.; Langlois, L.; Poutanen, S.M.; Razvi, H.; Reid, G.; Burton, J.P. Issues Beyond Resistance: Inadequate Antibiotic Therapy and Bacterial Hypervirulence. FEMS Microbes 2020, 1, xtaa004. [Google Scholar] [CrossRef]
- Zhou, S.; Nagel, J.L.K.K.S.; LaPlante, K.L.; Albin, O.R.; Pogue, J.M. Antimicrobial Stewardship and the Infection Control Practitioner: A Natural Alliance. Infect. Dis. Clin. N. Am. 2021, 35, 771–787. [Google Scholar] [CrossRef]
- Blanco, G.; López-Hernández, I.; Morinha, F.; López-Cerero, L. Intensive Farming as a Source of Bacterial Resistance to Antimicrobial Agents in Sedentary and Migratory Vultures: Implications for Local and Transboundary Spread. Sci. Total Environ. 2020, 739, 140356. [Google Scholar] [CrossRef]
- Liguori, K.; Keenum, I.; Davis, B.C.; Calarco, J.; Milligan, E.; Harwood, V.J.; Pruden, A. Antimicrobial Resistance Monitoring of Water Environments: A Framework for Standardized Methods and Quality Control. Environ. Sci. Technol. 2022, 56, 9149–9160. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, C.; Li, F.; Hua, T.; Zhou, Q.; Ho, S.H. Technologies Towards Antibiotic Resistance Genes (ARGs) Removal from Aquatic Environment: A Critical Review. J. Hazard. Mater. 2021, 411, 125148. [Google Scholar] [CrossRef] [PubMed]
- Aljeldah, M.M. Antimicrobial Resistance and Its Spread Is a Global Threat. Antibiotics 2022, 11, 1082. [Google Scholar] [CrossRef] [PubMed]
- Ramamurthy, T.; Ghosh, A.; Chowdhury, G.; Mukhopadhyay, A.K.; Dutta, S.; Miyoshi, S.I. Deciphering the Genetic Network and Programmed Regulation of Antimicrobial Resistance in Bacterial Pathogens. Front. Cell. Infect. Microbiol. 2022, 12, 952491. [Google Scholar] [CrossRef] [PubMed]
- Farquhar, K.S.; Rasouli Koohi, S.; Charlebois, D.A. Does Transcriptional Heterogeneity Facilitate the Development of Genetic Drug Resistance? BioEssays 2021, 43, 2100043. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yu, D.; Chen, L. Antimicrobial Resistance and Mechanisms of Epigenetic Regulation. Front. Cell. Infect. Microbiol. 2023, 13, 1199646. [Google Scholar] [CrossRef] [PubMed]
- Morrison, L.; Zembower, T.R. Antimicrobial Resistance. Gastrointest. Endosc. Clin. N. Am. 2020, 30, 619–635. [Google Scholar] [CrossRef] [PubMed]
- Van Camp, P.J.; Haslam, D.B.; Porollo, A. Bioinformatics Approaches to the Understanding of Molecular Mechanisms in Antimicrobial Resistance. Int. J. Mol. Sci. 2020, 21, 1363. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Singh, A.; Dar, M.A.; Kaur, R.J.; Charan, J.; Iskandar, K.; Haque, M.; Murti, K.; Ravichandiran, V.; Dhingra, S. Menace of Antimicrobial Resistance in LMICs: Current Surveillance Practices and Control Measures to Tackle Hostility. J. Infect. Public Health 2021, 15, 172–181. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. Biomed. Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef]
- Baraka, M.A.; Alboghdadly, A.; Alshawwa, S.; Elnour, A.A.; Alsultan, H.; Alsalman, T.; Alaithan, H.; Islam, A.; El-Fass, K.A.; Mohamed, Y.; et al. Perspectives of Healthcare Professionals Regarding Factors Associated with Antimicrobial Resistance (AMR) and Their Consequences: A Cross Sectional Study in Eastern Province of Saudi Arabia. Antibiotics 2021, 10, 878. [Google Scholar] [CrossRef]
- Bassetti, S.; Tschudin-Sutter, S.; Egli, A.; Osthoff, M. Optimizing Antibiotic Therapies to Reduce the Risk of Bacterial Resistance. Eur. J. Intern. Med. 2022, 99, 7–12. [Google Scholar] [CrossRef]
- Kadri, S.S. Key Takeaways from the US CDC’s 2019 Antibiotic Resistance Threats Report for Frontline Providers. Crit. Care Med. 2020, 48, 939–945. [Google Scholar] [CrossRef]
- Godman, B.; Egwuenu, A.; Haque, M.; Malande, O.O.; Schellack, N.; Kumar, S.; Saleem, Z.; Sneddon, J.; Hoxha, I.; Islam, S.; et al. Strategies to Improve Antimicrobial Utilization with a Special Focus on Developing Countries. Life 2021, 11, 528. [Google Scholar] [CrossRef] [PubMed]
- Pierce, J.; Apisarnthanarak, A.; Schellack, N.; Cornistein, W.; Al Maani, A.; Adnan, S.; Stevens, M.P. Global Antimicrobial Stewardship with a Focus on Low-and Middle-Income Countries: A Position Statement for the International Society for Infectious Diseases. Int. J. Infect. Dis. 2020, 96, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Pokharel, S.; Shrestha, P.; Adhikari, B. Antimicrobial Use in Food Animals and Human Health: Time to Implement ‘One Health’ Approach. Antimicrob. Resist. Infect. Control 2020, 9, 181. [Google Scholar] [CrossRef] [PubMed]
- Micoli, F.; Bagnoli, F.; Rappuoli, R.; Serruto, D. The Role of Vaccines in Combatting Antimicrobial Resistance. Nat. Rev. Microbiol. 2018, 19, 287–302. [Google Scholar] [CrossRef] [PubMed]
- Brazzoli, M.; Piccioli, D.; Marchetti, F. Challenges in Development of Vaccines Directed Toward Antimicrobial Resistant Bacterial Species. Hum. Vaccines Immunother. 2023, 19, 2228669. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.J.; Payne, D.J.; Rappuoli, R.; De Gregorio, E. Technologies to Address Antimicrobial Resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 12887–12895. [Google Scholar] [CrossRef]
- Payne, D.J.; Miller, L.F.; Findlay, D.; Anderson, J.; Marks, L. Time for a Change: Addressing R&D and Commercialization Challenges for Antibacterials. Philos. Trans. R. Soc. B 2015, 370, 20140086. [Google Scholar]
- Watkins, R.R.; Bonomo, R.A. Overview: The Ongoing Threat of Antimicrobial Resistance. Infect. Dis. Clin. N. Am. 2020, 34, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Vercelli, C.; Amadori, M.; Gambino, G.; Re, G. Implications of Veterinary Medicine in the Comprehension and Stewardship of Antimicrobial Resistance Phenomenon. From the Origin Till Nowadays. Vet. Anim. Sci. 2022, 16, 100249. [Google Scholar] [CrossRef]
- Allcock, S.; Gurdasani, Y.E.H.H.M.D.; Dougan, G.; Sandhu, M.S.; Solomon, L.; Török, M.E. Antimicrobial Resistance in Human Populations: Challenges and Opportunities. Glob. Health Epidemiol. Genom. 2017, 2, e4. [Google Scholar] [CrossRef]
- van Leth, F.; Schultsz, C. Unbiased Antimicrobial Resistance Prevalence Estimates Through Population-Based Surveillance. Clin. Microbiol. Infect. 2023, 29, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Iskandar, K.; Molinier, L.; Hallit, S.; Sartelli, M.; Hardcastle, T.C.; Lugova, H.; Dhindra, S.; Sharma, P.; Islam, S.; Mohammed, I.; et al. Surveillance of Antimicrobial Resistance in Low-and Middle-Income Countries: A Scattered Picture. Antimicrob. Resist. Infect. Control 2021, 10, 63. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar, R.; Alam, M. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Evid.-Based Nurs. 2023, 26, ebnurs-2022. [Google Scholar]
- Hendriksen, R.S.; Munk, P.; Njage, P.; Van Bunnik, B.; McNally, L.; Lukjancenko, O.; Röder, T.; Nieuwenhuijse, D.; Pedersen, S.K.; Kjeldgaard, J.; et al. Global Monitoring of Antimicrobial Resistance Based on Metagenomics Analyses of Urban Sewage. Nat. Commun. 2019, 10, 1124. [Google Scholar] [CrossRef] [PubMed]
- Jernigan, J.A.; Hatfield, K.M.; Wolford, H.; Nelson, R.E.; Olubajo, B.; Reddy, S.C.; McCarthy, N.; Prabasaj, P.; McDonald, L.C.; Kallen, A.; et al. Multidrug-Resistant Bacterial Infections in US Hospitalized Patients, 2012–2017. N. Engl. J. Med. 2020, 382, 1309–1319. [Google Scholar] [CrossRef] [PubMed]
- Friedman, N.D.; Carmeli, Y.; Walton, A.L.; Schwaber, M.J. Carbapenem-Resistant Enterobacteriaceae: A Strategic Roadmap for Infection Control. Infect. Control Hosp. Epidemiol. 2017, 38, 580–594. [Google Scholar] [CrossRef]
- Vehreschild, M.J.; Haverkamp, M.; Biehl, L.M.; Lemmen, S.; Fätkenheuer, G. Vancomycin-Resistant Enterococci (VRE): A Reason to Isolate? Infection 2019, 47, 7–11. [Google Scholar] [CrossRef]
- Lee, A.S.; De Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-Resistant Staphylococcus aureus. Nat. Rev. Dis. Primers 2018, 4, 18033. [Google Scholar] [CrossRef] [PubMed]
- Nurjadi, D.; Fleck, R.; Lindner, A.; Schäfer, J.; Gertler, M.; Mueller, A.; Lagler, H.; van Genderen, P.J.J.; Caumes, E.; Boutin, S.; et al. Import of Community-Associated, Methicillin-Resistant Staphylococcus aureus to Europe Through Skin and Soft-Tissue Infection in Intercontinental Travellers, 2011–2016. Clin. Microbiol. Infect. 2019, 25, 739–746. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Cormican, M.; Flamm, R.K.; Mendes, R.E.; Jones, R.N. Temporal and Geographic Variation in Antimicrobial Susceptibility and Resistance Patterns of Enterococci: Results from the SENTRY Antimicrobial Surveillance Program, 1997–2016. Open Forum Infect. Dis. 2019, 6 (Suppl. S1), S54–S62. [Google Scholar] [CrossRef]
- Ansari, W.; Quintana, A.; Mohamed, N.; Patino, N.; Irani, P.; Coyle, K.; Chang, H.; Uyei, J. Impact of CRE Infections on Hospital LOS and Mortality in Asia. Value Health Reg. Issues 2020, 22, S56. [Google Scholar] [CrossRef]
- Sannathimmappa, M.B. Global Escalation in Carbapenem-Resistant Enterobacterales and Carbapenem-Resistant Acinetobacter baumannii Infections: Serious Threat to Human Health from the Pink Corner. Biomed. Biotechnol. Res. J. 2023, 7, 9–16. [Google Scholar] [CrossRef]
- Markovska, R.; Dimitrov, G.; Gergova, R.; Boyanova, L. Clostridioides difficile, a New “Superbug”. Microorganisms 2023, 11, 845. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Monaghan, T.; Yadegar, A.; Louie, T.; Kao, D. Insights into the Evolving Epidemiology of Clostridioides difficile Infection and Treatment: A Global Perspective. Antibiotics 2023, 12, 1141. [Google Scholar] [CrossRef] [PubMed]
- Piret, J.; Boivin, G. Antiviral Resistance in Herpes Simplex Virus and Varicella-Zoster Virus Infections: Diagnosis and Management. Curr. Opin. Infect. Dis. 2016, 29, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Menéndez-Arias, L.; Álvarez, M. Antiretroviral Therapy and Drug Resistance in Human Immunodeficiency Virus Type 2 Infection. Antivir. Res. 2014, 102, 70–86. [Google Scholar] [CrossRef]
- Rehermann, B.; Bertoletti, A. Immunological Aspects of Antiviral Therapy of Chronic Hepatitis B Virus and Hepatitis C Virus Infections. Hepatology 2015, 61, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Heestermans, T.; Browne, J.L.; Aitken, S.C.; Vervoort, S.C.A.; Klipstein-Grobusch, K. Determinants of Adherence to Antiretroviral Therapy Among HIV-Positive Adults in Sub-Saharan Africa: A Systematic Review. BMJ Glob. Health 2016, 1, e000125. [Google Scholar] [CrossRef] [PubMed]
- Kadia, B.M.; Dimala, C.A.; Fongwen, N.T.; Smith, A.D. Barriers to and Enablers of Uptake of Antiretroviral Therapy in Integrated HIV and Tuberculosis Treatment Programmes in Sub-Saharan Africa: A Systematic Review and Meta-Analysis. AIDS Res. Ther. 2021, 18, 85. [Google Scholar] [CrossRef]
- Bennett, J. Concerning Features of Emerging Fungal Infections. Phys. Assist. Clin. 2023, 8, 433–452. [Google Scholar] [CrossRef]
- Vitiello, A.; Ferrara, F.; Boccellino, M.; Ponzo, A.; Cimmino, C.; Comberiati, E.; Zovi, A.; Clemente, S.; Sabbatucci, M. Antifungal Drug Resistance: An Emergent Health Threat. Biomedicines 2023, 11, 1063. [Google Scholar] [CrossRef] [PubMed]
- Ademe, M.; Girma, F. Candida auris: From Multidrug Resistance to Pan-Resistant Strains. Infect. Drug Resist. 2020, 13, 1287–1294. [Google Scholar] [PubMed]
- Nett, J.E. Candida auris: An Emerging Pathogen “Incognito”? PLoS Pathog. 2019, 15, e1007638. [Google Scholar] [CrossRef] [PubMed]
- Geremia, N.; Brugnaro, P.; Solinas, M.; Scarparo, C.; Panese, S. Candida auris as an Emergent Public Health Problem: A Current Update on European Outbreaks and Cases. Healthcare 2023, 11, 425. [Google Scholar] [CrossRef] [PubMed]
- Thatchanamoorthy, N.; Devi, V.R.; Chandramathi, S.; Tay, S.T. Candida auris: A Mini Review on Epidemiology in Healthcare Facilities in Asia. J. Fungi 2022, 8, 1126. [Google Scholar] [CrossRef]
- Wiederhold, N.P.; Patterson, T.F. Emergence of Azole Resistance in Aspergillus. Semin. Respir. Crit. Care Med. 2015, 36, 673–680. [Google Scholar] [CrossRef]
- Barrett, M.P.; Kyle, D.E.; Sibley, L.D.; Radke, J.B.; Tarleton, R.L. Protozoan Persister-Like Cells and Drug Treatment Failure. Nat. Rev. Microbiol. 2019, 17, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Dhorda, M.; Amaratunga, C.; Dondorp, A.M. Artemisinin and Multidrug-Resistant Plasmodium falciparum—A Threat for Malaria Control and Elimination. Curr. Opin. Infect. Dis. 2021, 34, 432–439. [Google Scholar] [CrossRef]
- Cheema, H.S.; Singh, M.P. Drug Resistance in Plasmodium, Future Malaria Management Strategies and Importance of Medicinal Plants. J. Ayurvedic Herb Med. 2022, 8, 107–112. [Google Scholar] [CrossRef]
- Mita, T.; Venkatesan, M.; Ohashi, J.; Culleton, R.; Takahashi, N.; Tsukahara, T.; Ndounga, M.; Dysoley, L.; Endo, H.; Hombhanje, F.; et al. Limited Geographical Origin and Global Spread of Sulfadoxine-Resistant dhps Alleles in Plasmodium falciparum Populations. J. Infect. Dis. 2011, 204, 1980–1988. [Google Scholar] [CrossRef]
- Anand, S.; Kaur, S.; Khuda, P.; Dudhraj, V.; Rajesh, K.; Bahl, A.; Singh, S.K. A Review of Antimicrobial Surveillance Networks Across the Globe. Epidemiol. Int. 2022, 7, 13–20. [Google Scholar]
- Lee, A.S.; Huttner, B.D.; Catho, G.; Harbarth, S. Methicillin-Resistant Staphylococcus aureus: An Update on Prevention and Control in Acute Care Settings. Infect. Dis. Clin. N. Am. 2021, 35, 931–952. [Google Scholar] [CrossRef] [PubMed]
- Mondal, A.H.; Khare, K.; Saxena, P.; Debnath, P.; Mukhopadhyay, K.; Yadav, D. A Review on Colistin Resistance: An Antibiotic of Last Resort. Microorganisms 2024, 12, 772. [Google Scholar] [CrossRef] [PubMed]
- Allel, K.; Carvajal, L.J.C.; Garcia, P.; Cifuentes, M.; Silva, F.; Munita, J.M.; Undurraga, E.A. Trends and Socioeconomic, Demographic, and Environmental Factors Associated with Antimicrobial Resistance: A Longitudinal Analysis in 39 Hospitals in Chile 2008–2017. Lancet Reg. Health Am. 2023, 21, 100484. [Google Scholar] [CrossRef] [PubMed]
- Sono, T.M.; Yeika, E.; Cook, A.; Kalungia, A.; Opanga, S.A.; Acolatse, J.E.E.; Sefah, I.A.; Jelic, A.G.; Campbell, S.; Lorenzetti, G.; et al. Current Rates of Purchasing of Antibiotics Without a Prescription Across Sub-Saharan Africa; Rationale and Potential Programmes to Reduce Inappropriate Dispensing and Resistance. Expert Rev. Anti Infect. Ther. 2023, 21, 1025–1055. [Google Scholar] [CrossRef] [PubMed]
- Musa, K.; Okoliegbe, I.; Abdalaziz, T.; Aboushady, A.T.; Stelling, J.; Gould, I.M. Laboratory Surveillance, Quality Management, and Its Role in Addressing Antimicrobial Resistance in Africa: A Narrative Review. Antibiotics 2023, 12, 1313. [Google Scholar] [CrossRef] [PubMed]
- Zellweger, R.M.; Carrique-Mas, J.; Limmathurotsakul, D.; Day, N.P.J.; Thwaites, G.E.; Baker, S.; Southeast Asia Antimicrobial Resistance Network. A Current Perspective on Antimicrobial Resistance in Southeast Asia. J. Antimicrob. Chemother. 2017, 72, 2963–2972. [Google Scholar] [CrossRef]
- Nhung, N.T.; Cuong, N.V.; Thwaites, G.; Carrique-Mas, J. Antimicrobial Usage and Antimicrobial Resistance in Animal Production in Southeast Asia: A Review. Antibiotics 2016, 5, 37. [Google Scholar] [CrossRef]
- Malijan, G.M.; Howteerakul, N.; Ali, N.; Siri, S.; Kengganpanich, M.; ODSET Group; Nascimento, R.; Booton, R.D.; Turner, K.M.E.; Cooper, B.S.; et al. A Scoping Review of Antibiotic Use Practices and Drivers of Inappropriate Antibiotic Use in Animal Farms in WHO Southeast Asia Region. One Health 2022, 15, 100412. [Google Scholar] [CrossRef]
- Kakkar, M.; Chatterjee, P.; Chauhan, A.S.; Grace, D.; Lindahl, J.; Beeche, A.; Jing, F.; Chiu, S. Antimicrobial Resistance in South East Asia: Time to Ask the Right Questions. Glob. Health Action 2018, 11, 1483637. [Google Scholar] [CrossRef]
- Tacconelli, E.; Buhl, M.; Humphreys, H.; Malek, V.; Presterl, E.; Rodriguez-Baño, J.; Vos, M.C.; Zingg, W.; Mutters, N.T. Analysis of the Challenges in Implementing Guidelines to Prevent the Spread of Multidrug-Resistant Gram-Negatives in Europe. BMJ Open 2019, 9, e027683. [Google Scholar] [CrossRef] [PubMed]
- Kossow, A.; Stühmer, B.; Schaumburg, F.; Becker, K.; Glatz, B.; Möllers, M.; Kampmeier, S.; Mellmann, A. High Prevalence of MRSA and Multi-Resistant Gram-Negative Bacteria in Refugees Admitted to the Hospital—But No Hint of Transmission. PLoS ONE 2018, 13, e0198103. [Google Scholar] [CrossRef] [PubMed]
- Henderson, A.; Nimmo, G.R. Control of Healthcare- and Community-Associated MRSA: Recent Progress and Persisting Challenges. Br. Med. Bull. 2018, 125, 25–41. [Google Scholar] [CrossRef] [PubMed]
- Machowska, A.; Lundborg, C.S. Drivers of Irrational Use of Antibiotics in Europe. Int. J. Environ. Res. Public Health 2019, 16, 27. [Google Scholar] [CrossRef] [PubMed]
- Wojkowska-Mach, J.; Godman, B.; Glassman, A.; Kurdi, A.; Pilc, A.; Rozanska, A.; Skoczyński, S.; Wałaszek, M.; Bochenek, T. Antibiotic Consumption and Antimicrobial Resistance in Poland; Findings and Implications. Antimicrob. Resist. Infect. Control 2018, 7, 136. [Google Scholar] [CrossRef] [PubMed]
- St Cyr, S.; Kersh, E.; Weinstock, H.; Torrone, E. Trends in Multi-Drug-Resistant Gonorrhea, Gonococcal Isolate Surveillance Project, United States, 1987–2016. Open Forum Infect. Dis. 2018, 5 (Suppl. S1), S352. [Google Scholar] [CrossRef]
- Schlanger, K. Perspectives from North America. Sex. Transm. Infect. 2019, 95 (Suppl. S1), A30. [Google Scholar]
- Marcella, S.; Kanakamedala, H.; Zhou, Y.; Cai, B.; Pogue, J.M. Trends of Carbapenem Resistance in Enterobacterales in the US Between 2015 and 2019. Open Forum Infect. Dis. 2020, 7 (Suppl. S1), S644. [Google Scholar] [CrossRef]
- Livorsi, D.J.; Chorazy, M.L.; Schweizer, M.L.; Balkenende, E.C.; Blevins, A.E.; Nair, R.; Samore, M.H.; Nelson, R.E.; Khander, K.; Perencevich, E.N. A Systematic Review of the Epidemiology of Carbapenem-Resistant Enterobacteriaceae in the United States. Antimicrob. Resist. Infect. Control 2018, 7, 55. [Google Scholar] [CrossRef]
- Labarca, J.A.; Salles, M.J.C.; Seas, C.; Guzmán-Blanco, M. Carbapenem Resistance in Pseudomonas aeruginosa and Acinetobacter baumannii in the Nosocomial Setting in Latin America. Crit. Rev. Microbiol. 2016, 42, 276–292. [Google Scholar]
- Fabre, V.; Cosgrove, S.E.; Lessa, F.C.; Patel, T.S.; Reyes-Morales, G.; Aleman, W.R.; Alvarez, A.A.; Aquiles, B.; Arauz, A.B.; Arguello, F.; et al. Knowledge, Attitudes and Perceptions of Latin American Healthcare Workers Relating to Antibiotic Stewardship and Antibiotic Use: A Cross-Sectional Multi-Country Study. Antimicrob. Resist. Infect. Control 2024, 13, 47. [Google Scholar] [CrossRef] [PubMed]
- Medina-Pizzali, M.L.; Hartinger, S.M.; Salmon-Mulanovich, G.; Larson, A.; Riveros, M.; Mäusezahl, D. Antimicrobial Resistance in Rural Settings in Latin America: A Scoping Review with a One Health Lens. Int. J. Environ. Res. Public Health 2021, 18, 9837. [Google Scholar] [CrossRef] [PubMed]
- Forsythe, S.S.; McGreevey, W.; Whiteside, A.; Shah, M.; Cohen, J.; Hecht, R.; Bollinger, L.A.; Kinghorn, A. Twenty Years of Antiretroviral Therapy for People Living with HIV: Global Costs, Health Achievements, Economic Benefits. Health Aff. 2019, 38, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Apetroaei, M.M.; Velescu, B.Ș.; Nedea, M.I.; Dinu-Pîrvu, C.E.; Drăgănescu, D.; Fâcă, A.I.; Udeanu, D.I.; Arsene, A.L. The Phenomenon of Antiretroviral Drug Resistance in the Context of Human Immunodeficiency Virus Treatment: Dynamic and Ever Evolving Subject Matter. Biomedicines 2024, 12, 915. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Rueda, W.; Rosas-Murrieta, N.H.; Muñoz-Medina, J.E.; González-Bonilla, C.R.; Reyes-Leyva, J.; Santos-López, G. Antiviral Resistance Markers in Influenza Virus Sequences in Mexico, 2000–2017. Infect. Drug Resist. 2018, 11, 1751–1756. [Google Scholar] [CrossRef] [PubMed]
- Hersh, A.L.; Kronman, M.P. Inappropriate Antibiotic Prescribing: Wind at Our Backs or Flapping in the Breeze? Pediatrics 2017, 139, e20170027. [Google Scholar] [CrossRef]
- Weinberg, S.E.; Villedieu, A.; Bagdasarian, N.; Karah, N.; Teare, L.; Elamin, W.F. Control and Management of Multidrug-Resistant Acinetobacter baumannii: A Review of the Evidence and Proposal of Novel Approaches. Infect. Prev. Pract. 2020, 2, 100077. [Google Scholar] [CrossRef] [PubMed]
- Karam, G.; Chastre, J.; Wilcox, M.H.; Vincent, J.L. Antibiotic Strategies in the Era of Multidrug Resistance. Crit. Care 2016, 20, 136. [Google Scholar] [CrossRef]
- Fernando, S.A.; Gray, T.J.; Gottlieb, T. Healthcare-Acquired Infections: Prevention Strategies. Intern. Med. J. 2017, 47, 1341–1351. [Google Scholar] [CrossRef] [PubMed]
- Perez, S.; Innes, G.K.; Walters, M.S.; Mehr, J.; Arias, J.; Greeley, R.; Chew, D. Increase in Hospital-Acquired Carbapenem-Resistant Acinetobacter baumannii Infection and Colonization in an Acute Care Hospital During a Surge in COVID-19 Admissions—New Jersey, February–July 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 1827–1831. [Google Scholar] [CrossRef]
- Slayton, R.B.; Toth, D.; Lee, B.Y.; Tanner, W.; Bartsch, S.M.; Khader, K.; Wonh, K.; Brown, K.; McKinnell, J.A.; Ray, W.; et al. Vital Signs: Estimated Effects of a Coordinated Approach for Action to Reduce Antibiotic-Resistant Infections in Health Care Facilities—United States. Am. J. Transplant. 2015, 15, 3002–3007. [Google Scholar] [CrossRef]
- Zakhour, J.; Haddad, S.F.; Kerbage, A.; Wertheim, H.; Tattevin, P.V.A.; Ünal, S.; Ouedraago, A.S.; Kanj, S.S. Diagnostic Stewardship in Infectious Diseases: A Continuum of Antimicrobial Stewardship in the Fight Against Antimicrobial Resistance. Int. J. Antimicrob. Agents 2023, 62, 106816. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Ning, J.; Ahmed, S.; Huang, J.; Ullah, R.; An, H.H.; Dai, M.; Wang, X.; Yuan, Z. Selection and Dissemination of Antimicrobial Resistance in Agri-Food Production. Antimicrob. Resist. Infect. Control 2019, 8, 158. [Google Scholar] [CrossRef] [PubMed]
- Economou, V.; Gousia, P. Agriculture and Food Animals as a Source of Antimicrobial-Resistant Bacteria. Infect. Drug Resist. 2015, 8, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Steele, J.C.; Meng, X.Z. Usage, Residue, and Human Health Risk of Antibiotics in Chinese Aquaculture: A Review. Environ. Pollut. 2017, 223, 161–169. [Google Scholar] [CrossRef]
- Chen, J.; Sun, R.; Pan, C.; Sun, Y.; Mai, B.; Li, Q.X. Antibiotics and Food Safety in Aquaculture. J. Agric. Food Chem. 2020, 68, 11908–11919. [Google Scholar] [CrossRef]
- Taylor, P.; Reeder, R. Antibiotic Use on Crops in Low and Middle-Income Countries Based on Recommendations Made by Agricultural Advisors. CABI Agric. Biosci. 2020, 1, 1. [Google Scholar] [CrossRef]
- Franklin, A.M.; Aga, D.S.; Cytryn, E.; Durso, L.M.; McLain, J.E.; Pruden, A.; Roberts, M.C.; Rothrock, M.J.; Snow, D.D.; Dungan, R.S. Antibiotics in Agroecosystems: Introduction to the Special Section. J. Environ. Qual. 2016, 45, 377–393. [Google Scholar] [CrossRef]
- Frost, I.; Van Boeckel, T.P.; Pires, J.; Craig, J.; Laxminarayan, R. Global Geographic Trends in Antimicrobial Resistance: The Role of International Travel. J. Travel Med. 2019, 26, taz036. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, S.; Turbett, S.E.; Harris, J.B.; LaRocque, R.C. Antimicrobial-Resistant Bacteria in International Travelers. Curr. Opin. Infect. Dis. 2021, 34, 423–431. [Google Scholar] [CrossRef]
- Thomas, C.M.; Morkeberg, O.H.; Walker, P.F.; Stauffer, W.M. The Cost of Global Connectivity: Faster and More Efficient Spread of Antimicrobial Resistance by International Travelers–A Controversial Commentary. Travel Med. Infect. Dis. 2021, 41, 102045. [Google Scholar] [CrossRef] [PubMed]
- Młynarczyk-Bonikowska, B.; Majewska, A.; Malejczyk, M.; Młynarczyk, G.; Majewski, S. Multiresistant Neisseria gonorrhoeae: A New Threat in Second Decade of the XXI Century. Med. Microbiol. Immunol. 2020, 209, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Pavli, A.; Maltezou, H.C. Infectious Complications Related to Medical Tourism. J. Travel Med. 2021, 28, taaa210. [Google Scholar] [CrossRef] [PubMed]
- Bokhary, H.; Pangesti, K.N.; Rashid, H.; Abd El Ghany, M.; Hill-Cawthorne, G.A. Travel-Related Antimicrobial Resistance: A Systematic Review. Trop. Med. Infect. Dis. 2021, 6, 11. [Google Scholar] [CrossRef]
- Martak, D.; Henriot, C.P.; Hocquet, D. Environment, Animals, and Food as Reservoirs of Antibiotic-Resistant Bacteria for Humans: One Health or More? Infect. Dis. Now 2024, 54, 104895. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.R.; Haste, N.M.; Gluckstein, D.P. The Role of Antibiotic Stewardship in Promoting Appropriate Antibiotic Use. Am. J. Lifestyle Med. 2019, 13, 376–383. [Google Scholar] [CrossRef]
- Giamarellou, H.; Galani, L.; Karavasilis, T.; Ioannidis, K.; Karaiskos, I. Antimicrobial Stewardship in the Hospital Setting: A Narrative Review. Antibiotics 2023, 12, 1557. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Burns, K.; Rodríguez Baño, J.; Borg, M.; Daikos, G.; Dumpis, U.; Lucet, J.C.; Moro, M.L.; Tacconelli, E.; Simonse, G.S.; et al. Infection Prevention and Control Measures and Tools for the Prevention of Entry of Carbapenem-Resistant Enterobacteriaceae into Healthcare Settings: Guidance from the European Centre for Disease Prevention and Control. Antimicrob. Resist. Infect. Control 2017, 6, 113. [Google Scholar] [CrossRef]
- Carling, P.C.; Parry, M.F.; Olmstead, R. Environmental Approaches to Controlling Clostridioides difficile Infection in Healthcare Settings. Antimicrob. Resist. Infect. Control 2023, 12, 94. [Google Scholar]
- Pitout, J.D. Transmission Surveillance for Antimicrobial-Resistant Organisms in the Health System. In Microbial Transmission; ASM Press: Washington, DC, USA, 2019; pp. 215–227. [Google Scholar]
- Aarestrup, F.M.; Woolhouse, M.E. Using Sewage for Surveillance of Antimicrobial Resistance. Science 2020, 367, 630–632. [Google Scholar] [CrossRef]
- Pruden, A.; Vikesland, P.J.; Davis, B.C.; de Roda Husman, A.M. Seizing the Moment: Now Is the Time for Integrated Global Surveillance of Antimicrobial Resistance in Wastewater Environments. Curr. Opin. Microbiol. 2021, 64, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, R.; Bose, D.; Gulia, K.; Jaiswal, A. Impact of antimicrobial resistance on sustainable development goals and the integrated strategies for meeting environmental and socio-economic targets. Environ. Prog. Sustain. Energy 2024, 43, e14320. [Google Scholar] [CrossRef]
- Hay, S.I.; Rao, P.C.; Dolecek, C.; Day, N.P.; Stergachis, A.; Lopez, A.D.; Murray, C.J. Measuring and Mapping the Global Burden of Antimicrobial Resistance. BMC Med. 2018, 16, 78. [Google Scholar] [CrossRef]
- Van Hecke, O.; Wang, K.; Lee, J.J.; Roberts, N.W.; Butler, C.C. Implications of Antibiotic Resistance for Patients’ Recovery from Common Infections in the Community: A Systematic Review and Meta-Analysis. Clin. Infect. Dis. 2017, 65, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Huemer, M.; Mairpady Shambat, S.; Brugger, S.D.; Zinkernagel, A.S. Antibiotic Resistance and Persistence—Implications for Human Health and Treatment Perspectives. EMBO Rep. 2020, 21, e51034. [Google Scholar] [CrossRef]
- Colomb-Cotinat, M.; Lacoste, J.; Brun-Buisson, C.; Jarlier, V.; Coignard, B.; Vaux, S. Estimating the Morbidity and Mortality Associated with Infections Due to Multidrug-Resistant Bacteria (MDRB), France, 2012. Antimicrob. Resist. Infect. Control 2016, 5, 56. [Google Scholar] [CrossRef] [PubMed]
- Barrasa-Villar, J.I.; Aibar-Remón, C.; Prieto-Andrés, P.; Mareca-Doñate, R.; Moliner-Lahoz, J. Impact on Morbidity, Mortality, and Length of Stay of Hospital-Acquired Infections by Resistant Microorganisms. Clin. Infect. Dis. 2017, 65, 644–652. [Google Scholar] [CrossRef]
- Founou, R.C.; Founou, L.L.; Essack, S.Y. Clinical and Economic Impact of Antibiotic Resistance in Developing Countries: A Systematic Review and Meta-Analysis. PLoS ONE 2017, 12, e0189621. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, G.S. Antimicrobial Resistance Surveillance in Europe and Beyond. Eurosurveillance 2018, 23, 1800560. [Google Scholar] [CrossRef]
- So, M.; Walti, L. Challenges of Antimicrobial Resistance and Stewardship in Solid Organ Transplant Patients. Curr. Infect. Dis. Rep. 2022, 24, 63–75. [Google Scholar] [CrossRef]
- Menz, B.D.; Charani, E.; Gordon, D.L.L.A.J.; Moonesinghe, S.R.; Phillips, C.J. Surgical Antibiotic Prophylaxis in an Era of Antibiotic Resistance: Common Resistant Bacteria and Wider Considerations for Practice. Infect. Drug Resist. 2021, 10, 5235–5252. [Google Scholar] [CrossRef] [PubMed]
- Cerceo, E.; Deitelzweig, S.B.; Sherman, B.M.; Amin, A.N. Multidrug-Resistant Gram-Negative Bacterial Infections in the Hospital Setting: Overview, Implications for Clinical Practice, and Emerging Treatment Options. Microb. Drug Resist. 2016, 22, 412–431. [Google Scholar] [CrossRef] [PubMed]
- Trotter, A.J.; Aydin, A.; Strinden, M.J.; O’grady, J. Recent and Emerging Technologies for the Rapid Diagnosis of Infection and Antimicrobial Resistance. Curr. Opin. Microbiol. 2019, 51, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Rentschler, S.; Kaiser, L.; Deigner, H.P. Emerging Options for the Diagnosis of Bacterial Infections and the Characterization of Antimicrobial Resistance. Int. J. Mol. Sci. 2021, 22, 456. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, G.; De Gaetano, S.; Midiri, A.; Zummo, S.; Biondo, C. The Challenge of Overcoming Antibiotic Resistance in Carbapenem-Resistant Gram-Negative Bacteria: “Attack on Titan”. Microorganisms 2023, 11, 1912. [Google Scholar] [CrossRef]
- Paterson, D.L. The Challenge of Treating Superbugs. Semin. Respir. Crit. Care Med. 2015, 36, 001–002. [Google Scholar] [CrossRef] [PubMed]
- Rossolini, G.M.; Arena, F.; Pecile, P.; Pollini, S. Update on the Antibiotic Resistance Crisis. Curr. Opin. Pharmacol. 2014, 18, 56–60. [Google Scholar] [CrossRef]
- Morris, S.; Cerceo, E. Trends, Epidemiology, and Management of Multi-Drug Resistant Gram-Negative Bacterial Infections in the Hospitalized Setting. Antibiotics 2020, 9, 196. [Google Scholar] [CrossRef]
- Margolis, E.; Rosch, J.W. Fitness Landscape of the Immune Compromised Favors the Emergence of Antibiotic Resistance. ACS Infect. Dis. 2018, 4, 1275–1277. [Google Scholar] [CrossRef]
- Feuerstadt, P.; Theriault, N.; Tillotson, G. The Burden of CDI in the United States: A Multifactorial Challenge. BMC Infect. Dis. 2023, 23, 132. [Google Scholar] [CrossRef]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [PubMed]
- Broderick, N.; Walsh, K.A.; O’Brien, K.K.; Smith, S.S.; Harrington, P.; O’Neill, M.; Ryan, M.; Fawsitt, C.G. POSC124 Economic Burden of Antimicrobial Resistance: An Analysis of the Additional Bed Day Costs Associated with Treating Resistant Infections in Ireland. Value Health 2022, 25, S111. [Google Scholar] [CrossRef]
- Touat, M.; Opatowski, M.; Brun-Buisson, C.; Cosker, K.; Guillemot, D.; Salomon, J.; Tuppin, P.; de Lagasnerie, G.; Watier, L. A Payer Perspective of the Hospital Inpatient Additional Care Costs of Antimicrobial Resistance in France: A Matched Case–Control Study. Appl. Health Econ. Health Policy 2019, 17, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, K.E.; Joski, P.; Johnston, K.J. Antibiotic-Resistant Infection Treatment Costs Have Doubled Since 2002, Now Exceeding $2 Billion Annually. Health Aff. 2018, 37, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Poudel, A.N.; Zhu, S.; Cooper, N.; Little, P.; Tarrant, C.; Hickman, M.; Yao, G. The Economic Burden of Antibiotic Resistance: A Systematic Review and Meta-Analysis. PLoS ONE 2023, 18, e0285170. [Google Scholar] [CrossRef]
- Ait Ouakrim, D.; Cassini, A.; Cecchini, M.; Plauchoras, D. The Health and Economic Burden of Antimicrobial Resistance. Eur. J. Public Health 2020, 30 (Suppl. S5), ckaa165-1201. [Google Scholar] [CrossRef]
- Shrestha, P.; Cooper, B.S.; Coast, J.; Oppong, R.; Do Thi Thuy, N.; Phodha, T.; Celhay, O.; Guerin, P.J.; Wertheim, H.; Lubell, Y. Enumerating the Economic Cost of Antimicrobial Resistance per Antibiotic Consumed to Inform the Evaluation of Interventions Affecting Their Use. Antimicrob. Resist. Infect. Control 2018, 7, 98. [Google Scholar] [CrossRef]
- Cole, S. Who Will Develop New Antibacterial Agents? Philos. Trans. R. Soc. B 2014, 368, 20130430. [Google Scholar] [CrossRef] [PubMed]
- Tomaras, A.P.; Dunman, P.M. In the Midst of the Antimicrobial Discovery Conundrum: An Overview. Curr. Opin. Microbiol. 2015, 27, 103–107. [Google Scholar] [CrossRef]
- Mattar, C.; Edwards, S.; Baraldi, E.; Hood, J. An Overview of the Global Antimicrobial Resistance Research and Development Hub and the Current Landscape. Curr. Opin. Microbiol. 2020, 57, 56–61. [Google Scholar] [CrossRef]
- Guardabassi, L. Antimicrobial Resistance: A Global Threat with Remarkable Geographical Differences. N. Z. Vet. J. 2017, 65, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Keown, O.P.; Warburton, W.; Davies, S.C.; Darzi, A. Antimicrobial Resistance: Addressing the Global Threat through Greater Awareness and Transformative Action. Health Aff. 2014, 33, 1620–1626. [Google Scholar] [CrossRef] [PubMed]
- Howard, S.J.; Hopwood, S.; Davies, S.C. Antimicrobial Resistance: A Global Challenge. Sci. Transl. Med. 2014, 6, 236ed10. [Google Scholar] [CrossRef]
- Willemsen, A.; Reid, S.; Assefa, Y. A Review of National Action Plans on Antimicrobial Resistance: Strengths and Weaknesses. Antimicrob. Resist. Infect. Control 2022, 11, 90. [Google Scholar] [CrossRef]
- Munkholm, L.; Rubin, O. The Global Governance of Antimicrobial Resistance: A Cross-Country Study of Alignment between the Global Action Plan and National Action Plans. Glob. Health 2020, 16, 109. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.; Schulze, K.; Cassini, A.; Plauchoras, D.; Mossialos, E. Strengthening Implementation of Antimicrobial Resistance National Action Plans. Eur. J. Public Health 2020, 30 (Suppl. S5), ckaa165-1200. [Google Scholar] [CrossRef]
- Littmann, J.; Buyx, A.; Cars, O. Antibiotic Resistance: An Ethical Challenge. Int. J. Antimicrob. Agents 2015, 46, 359–361. [Google Scholar] [CrossRef] [PubMed]
- Saint, V.; Mohsenpour, A.; Mühling, C.; Bozorgmehr, K. Exploring Equity, Gender and Social Determinants Aspects of Antimicrobial Resistance: Scoping Review. Eur. J. Public Health 2020, 30 (Suppl. S5), ckaa166-707. [Google Scholar] [CrossRef]
- Saint, V. Exploring Equity, Social Determinants of Health and Gender Considerations for Antimicrobial Resistance. Eur. J. Public Health 2019, 29 (Suppl. S4), ckz185-799. [Google Scholar] [CrossRef]
- Adebisi, Y.A.; Ogunkola, I.O. The Global Antimicrobial Resistance Response Effort Must Not Exclude Marginalized Populations. Trop. Med. Health 2023, 51, 33. [Google Scholar] [CrossRef]
- Chioro, A.; Coll-Seck, A.M.; Høie, B.; Moeloek, N.; Motsoaledi, A.; Rajatanavin, R.; Touraine, M. Antimicrobial Resistance: A Priority for Global Health Action. Bull. World Health Organ. 2015, 93, 439. [Google Scholar] [CrossRef] [PubMed]
- Pokharel, S.; Adhikari, B.; Johnson, T.; Cheah, P.Y. Interventions to Address Antimicrobial Resistance: An Ethical Analysis of Key Tensions and How They Apply in Low-Income and Middle-Income Countries. BMJ Glob. Health 2024, 9, e012874. [Google Scholar] [CrossRef] [PubMed]
- Adebisi, Y.A. Balancing the Risks and Benefits of Antibiotic Use in a Globalized World: The Ethics of Antimicrobial Resistance. Glob. Health 2023, 19, 27. [Google Scholar] [CrossRef] [PubMed]
- Parsonage, B.; Hagglund, P.K.; Keogh, L.; Wheelhouse, N.; Brown, R.E.; Dancer, S.J. Control of Antimicrobial Resistance Requires an Ethical Approach. Front. Microbiol. 2017, 8, 2124. [Google Scholar] [CrossRef] [PubMed]
- Majumder, M.A.A.; Rahman, S.; Cohall, D.; Bharatha, A.; Singh, K.; Haque, M.; Gittens-St Hilaire, M. Antimicrobial Stewardship: Fighting Antimicrobial Resistance and Protecting Global Public Health. Infect. Drug Resist. 2020, 13, 4713–4738. [Google Scholar] [CrossRef]
- Aryee, A.; Price, N. Antimicrobial Stewardship—Can We Afford to Do without It? Br. J. Clin. Pharmacol. 2015, 79, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Büchler, H.S. Challenges and Success Stories of the Implementation of Infection Control and Antimicrobial Stewardship Strategies. In Proceedings of the 5th Global Ministerial Summit on Patient Safety, Montreux, Switzerland, 17–18 April 2024. [Google Scholar]
- Gitaka, J.; Kamita, M.; Mureithi, D.; Ndegwa, D.; Masika, M.; Omuse, G.; Ngari, M.; Makokha, F.; Mwaura, P.; Mathai, R.; et al. Combating Antibiotic Resistance Using Guidelines and Enhanced Stewardship in Kenya: A Protocol for an Implementation Science Approach. BMJ Open 2020, 10, e030823. [Google Scholar] [CrossRef] [PubMed]
- Hara, G.L. The Role of the ‘A’ Team in the Hospital near You. Int. J. Infect. Dis. 2018, 72, 45. [Google Scholar] [CrossRef]
- Minejima, E.; Wong-Beringer, A. Implementation of Rapid Diagnostics with Antimicrobial Stewardship. Expert Rev. Anti-Infect. Ther. 2016, 14, 1065–1075. [Google Scholar] [CrossRef]
- Banerjee, R.; Patel, R. Molecular Diagnostics for Genotypic Detection of Antibiotic Resistance: Current Landscape and Future Directions. JAC Antimicrob. Resist. 2023, 5, dlad018. [Google Scholar] [CrossRef]
- Lee, C.R.; Lee, J.H.; Kang, L.W.; Jeong, B.C.; Lee, S.H. Educational Effectiveness, Target, and Content for Prudent Antibiotic Use. Biomed. Res. Int. 2015, 2015, 214021. [Google Scholar] [CrossRef] [PubMed]
- McNicholas, M.; Hooper, G. Effects of Patient Education to Reduce Antibiotic Prescribing Rates for Upper Respiratory Infections in Primary Care. Fam. Pract. 2022, 39, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Smiddy, M.P.; Murphy, O.M.; Savage, E.; Fitzgerald, A.P.; O’Sullivan, B.; Murphy, C.; Bernard, M.; Browne, J.P. Efficacy of Observational Hand Hygiene Audit with Targeted Feedback on Doctors’ Hand Hygiene Compliance: A Retrospective Time Series Analysis. J. Infect. Prev. 2019, 20, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Glowicz, J.B.; Landon, E.; Sickbert-Bennett, E.E.; Aiello, A.E.; Dekay, K.; Hoffmann, K.K.; Maragakis, L.; Olmsted, R.N.; Polgreen, P.M.; Trexler, P.A.; et al. SHEA/IDSA/APIC Practice Recommendation: Strategies to Prevent Healthcare-Associated Infections through Hand Hygiene: 2022 Update. Infect. Control Hosp. Epidemiol. 2023, 44, 355–376. [Google Scholar] [CrossRef] [PubMed]
- Donati, D.; Miccoli, G.A.; Cianfrocca, C.; Di Stasio, E.; De Marinis, M.G.; Tartaglini, D. Effectiveness of Implementing Link Nurses and Audits and Feedback to Improve Nurses’ Compliance with Standard Precautions: A Cluster Randomized Controlled Trial. Am. J. Infect. Control 2020, 48, 1204–1210. [Google Scholar] [CrossRef]
- Cordeiro, L.; Gnatta, J.R.; Ciofi-Silva, C.L.; Price, A.; de Oliveira, N.A.; Almeida, R.M.; Mainardi, G.M.; Srinivas, S.; Chan, W.; Levin, A.S.S.; et al. Personal Protective Equipment Implementation in Healthcare: A Scoping Review. Am. J. Infect. Control 2022, 50, 898–905. [Google Scholar] [CrossRef]
- Inkster, T.; Walker, J.; Weinbren, M. Water-Free Patient Care, a Narrative Review of the Literature and Discussion of the Pressing Need for a Way Forward. J. Hosp. Infect. 2024, 152, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Low, J.M.; Chan, M.; Low, J.L.; Chua, M.C.W.; Lee, J.H. The Impact of Sink Removal and Other Water-Free Interventions in Intensive Care Units on Water-Borne Healthcare-Associated Infections: A Systematic Review. J. Hosp. Infect. 2024, 150, 61–71. [Google Scholar] [CrossRef]
- Rutala, W.A.; Boyce, J.M.; Weber, D.J. Disinfection, Sterilization and Antisepsis: An Overview. Am. J. Infect. Control 2023, 51, A3–A12. [Google Scholar] [CrossRef]
- Rutala, W.A.; Donskey, C.J.; Weber, D.J. Disinfection and Sterilization: New Technologies. Am. J. Infect. Control 2023, 51, A13–A21. [Google Scholar] [CrossRef]
- Porter, L.; Sultan, O.; Mitchell, B.G.; Jenney, A.; Kiernan, M.; Brewster, D.J.; Russo, P.L. How Long Do Nosocomial Pathogens Persist on Inanimate Surfaces? J. Hosp. Infect. 2024, 147, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Santos-Marques, C.; Ferreira, H.; Pereira, S.G. Infection Prevention and Control Strategies against Carbapenem Resistant Enterobacteriaceae–A Systematic Review. J. Infect. Prev. 2022, 23, 167–185. [Google Scholar] [CrossRef] [PubMed]
- Saliba, R.; Zahar, J.R.; Dabar, G.; Riachy, M.; Karam-Sarkis, D.; Husni, R. Limiting the Spread of Multidrug-Resistant Bacteria in Low-to-Middle-Income Countries: One Size Does Not Fit All. Pathogens 2023, 12, 144. [Google Scholar] [CrossRef] [PubMed]
- Jit, M.; Cooper, B. The Role of Vaccines in Combating Antimicrobial Resistance. Eur. J. Public Health 2020, 30 (Suppl. S5), ckaa165-1204. [Google Scholar] [CrossRef]
- Van Heuvel, L.; Caini, S.; Dückers, M.L.; Paget, J. Assessment of the Inclusion of Vaccination as an Intervention to Reduce Antimicrobial Resistance in AMR National Action Plans: A Global Review. Glob. Health 2022, 18, 85. [Google Scholar] [CrossRef]
- Ba-Nguz, A.; Shah, A.; Bresee, J.S.; Lafond, C.K.E.; Donadel, M.; Seward, J.F. Supporting National Immunization Technical Advisory Groups (NITAGs) in Resource-Constrained Settings: New Strategies and Lessons Learned from the Task Force for Global Health’s Partnership for Influenza Vaccine Introduction. Vaccine 2019, 37, 3446–3653. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, V.; Roviello, G.N. The Potential Role of Vaccines in Preventing Antimicrobial Resistance (AMR): An Update and Future Perspectives. Vaccines 2023, 11, 333. [Google Scholar] [CrossRef]
- Davies, N.G.; Jit, F.S.M.; Atkins, K.E. Modeling the Effect of Vaccination on Selection for Antibiotic Resistance in Streptococcus pneumoniae. Sci. Transl. Med. 2021, 13, eaaz8690. [Google Scholar] [CrossRef] [PubMed]
- Perez, F.; Villegas, M.V. The Role of Surveillance Systems in Confronting the Global Crisis of Antibiotic-Resistant Bacteria. Curr. Opin. Infect. Dis. 2015, 18, 375–383. [Google Scholar] [CrossRef]
- Johnson, A.P. Surveillance of Antibiotic Resistance. Philos. Trans. R. Soc. B 2015, 370, 20140080. [Google Scholar] [CrossRef]
- Cantón, R.; Gottlieb, T.; Coombs, G.W.; Woo, P.C.; Korman, T.M.; Garcia-Castillo, M.; Daley, D.; Bauer, K.A.; Wong, M.; Wolf, D.J.; et al. Antimicrobial Surveillance: A 20-Year History of the SMART Approach to Addressing Global Antimicrobial Resistance into the Future. Int. J. Antimicrob. Agents 2023, 62, 107014. [Google Scholar] [CrossRef] [PubMed]
- Altorf-van der Kuil, W.; Schoffelen, A.F.; de Greeff, S.C.; Thijsen, S.F.; Alblas, H.J.; Notermans, D.W.; Vlek, A.L.M.; van der Sande, M.A.B.; Leenstra, T.; National AMR Surveillance Study Group. National Laboratory-Based Surveillance System for Antimicrobial Resistance: A Successful Tool to Support the Control of Antimicrobial Resistance in the Netherlands. Eurosurveillance 2017, 22, 17–00062. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Yoon, E.J.; Kim, D.; Jeong, S.H.; Shin, J.H.; Shin, J.H.; Kim, Y.A.; Uh, Y.; Park, C.; Lee, K.J. Establishment of the South Korean National Antimicrobial Resistance Surveillance System, Kor-GLASS, in 2016. Eurosurveillance 2018, 23, 1700734. [Google Scholar] [CrossRef]
- Seale, A.C.; Gordon, N.C.I.J.; Peacock, S.J.; Scott, J.A.G. AMR Surveillance in Low and Middle-Income Settings—A Roadmap for Participation in the Global Antimicrobial Surveillance System (GLASS). Wellcome Open Res. 2017, 2, 92. [Google Scholar] [CrossRef] [PubMed]
- Sirijatuphat, R.; Chayangsu, S.; Srisompong, J.; Ruangkriengsin, D.; Thamlikitkul, V.; Tiengrim, S.; Wangchinda, W.; Koomanachai, P.; Rattanaumpawan, P. Feasibility, Challenges, and Benefits of Global Antimicrobial Resistance Surveillance System Implementation: Results from a Multicenter Quasi-Experimental Study. Antibiotics 2022, 11, 348. [Google Scholar] [CrossRef] [PubMed]
- Ajulo, S.; Awosile, B. Global Antimicrobial Resistance and Use Surveillance System (GLASS 2022): Investigating the Relationship between Antimicrobial Resistance and Antimicrobial Consumption Data across the Participating Countries. PLoS ONE 2024, 19, e0297921. [Google Scholar] [CrossRef] [PubMed]
- Sibani, M.; Mazzaferri, F.; Carrara, E.; Pezzani, M.D.; Arieti, F.; Göpel, S.; Paul, M.; Tacconelli, E.; Mutters, N.T.; Vos, A.; et al. White Paper: Bridging the Gap between Surveillance Data and Antimicrobial Stewardship in Long-Term Care Facilities—Practical Guidance from the JPIAMR ARCH and COMBACTE-MAGNET EPI-Net Networks. J. Antimicrob. Chemother. 2020, 75 (Suppl. S2), ii33–ii41. [Google Scholar] [CrossRef]
- Shukla, R.; Lavore, F.; Maity, S.; Derks, M.G.N.; Jones, C.R.; Vermeulen, B.J.A.; Melcrová, A.; Morris, M.A.; Becker, L.M.; Wang, X.; et al. Teixobactin kills bacteria by a two-pronged attack on the cell envelope. Nature 2022, 608, 390–396. [Google Scholar] [CrossRef]
- Domingo-Calap, P.; Delgado-Martínez, J. Bacteriophages: Protagonists of a Post-Antibiotic Era. Antibiotics 2018, 7, 66. [Google Scholar] [CrossRef]
- Garvey, M. Bacteriophages and the One Health Approach to Combat Multidrug Resistance: Is This the Way? Antibiotics 2020, 9, 414. [Google Scholar] [CrossRef]
- Wan, X.; Hendrix, H.; Skurnik, M.; Lavigne, R. Phage-Based Target Discovery and Its Exploitation towards Novel Antibacterial Molecules. Curr. Opin. Biotechnol. 2021, 68, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial Peptides: Application Informed by Evolution. Science 2020, 368, eaau5480. [Google Scholar] [CrossRef] [PubMed]
- Ageitos, J.M.; Sánchez-Pérez, A.; Calo-Mata, P.; Villa, T.G. Antimicrobial Peptides (AMPs): Ancient Compounds That Represent Novel Weapons in the Fight against Bacteria. Biochem. Pharmacol. 2017, 133, 117–138. [Google Scholar] [CrossRef] [PubMed]
- Nuti, R.; Goud, N.S.; Saraswati, A.P.; Alvala, R.; Alvala, M. Antimicrobial Peptides: A Promising Therapeutic Strategy in Tackling Antimicrobial Resistance. Curr. Med. Chem. 2017, 24, 4303–4314. [Google Scholar] [CrossRef] [PubMed]
- Streicher, L.M. Exploring the Future of Infectious Disease Treatment in a Post-Antibiotic Era: A Comparative Review of Alternative Therapeutics. J. Glob. Antimicrob. Resist. 2021, 24, 285–295. [Google Scholar] [CrossRef]
- Ghosh, C.; Sarkar, P.; Issa, R.; Haldar, J. Alternatives to Conventional Antibiotics in the Era of Antimicrobial Resistance. Trends Microbiol. 2019, 27, 323–338. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.; Chan, S.Y.; Song, Q.; Li, P.; Huang, W. The Strategies of Pathogen-Oriented Therapy on Circumventing Antimicrobial Resistance. Research 2020, 2020, 2016201. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, B.A.; Bhat, B.A.; Mir, M.A. Antimicrobial resistance: New insights and therapeutic implications. App. Microbiol. Biotechnol. 2022, 106, 6427–6440. [Google Scholar] [CrossRef]
- Wang, C.H.; Hsieh, Y.H.; Powers, Z.M.; Kao, C.Y. Defeating Antibiotic-Resistant Bacteria: Exploring Alternative Therapies for a Post-Antibiotic Era. Int. J. Mol. Sci. 2020, 21, 1061. [Google Scholar] [CrossRef]
- Weldick, P.J.; Wang, A.; Halbus, A.F.; Paunov, V.N. Emerging Nanotechnologies for Targeting Antimicrobial Resistance. Nanoscale 2022, 14, 4018–4041. [Google Scholar] [CrossRef]
- Kumar, M.; Curtis, A.; Hoskins, C. Application of Nanoparticle Technologies in the Combat against Antimicrobial Resistance. Pharmaceutics 2018, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Deng, S.; Zhang, L. A Review of Artificial Intelligence Applications for Antimicrobial Resistance. Biosaf. Health 2021, 3, 22–31. [Google Scholar] [CrossRef]
- Melo, M.C.; Maasch, J.R.; de la Fuente-Nunez, C. Accelerating Antibiotic Discovery through Artificial Intelligence. Commun. Biol. 2021, 4, 1050. [Google Scholar] [CrossRef]
- Cella, E.; Giovanetti, M.; Benedetti, F.; Scarpa, F.; Johnston, C.; Borsetti, A.; Azarian, T.; Zella, D.; Ciccozzi, M. Joining Forces against Antibiotic Resistance: The One Health Solution. Pathogens 2023, 12, 1074. [Google Scholar] [CrossRef] [PubMed]
- Nardulli, P.; Ballini, A.; Zamparella, M.; De Vito, D. The Role of Stakeholders’ Understandings in Emerging Antimicrobial Resistance: A One Health Approach. Microorganisms 2023, 11, 2797. [Google Scholar] [CrossRef]
- Kamenshchikova, A.; Wolffs, P.F.; Hoebe, C.J.; Horstman, K. Anthropocentric Framings of One Health: An Analysis of International Antimicrobial Resistance Policy Documents. Crit. Public Health 2021, 31, 306–315. [Google Scholar] [CrossRef]
- Velazquez-Meza, M.E.; Galarde-López, M.; Carrillo-Quiróz, B.; Alpuche-Aranda, C.M. Antimicrobial Resistance: One Health Approach. Vet. World 2022, 15, 743. [Google Scholar] [CrossRef]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell. Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef]
- Van Duin, D.; Paterson, D.L. Multidrug-Resistant Bacteria in the Community: Trends and Lessons Learned. Infect. Dis. Clin. N. Am. 2016, 30, 377–390. [Google Scholar] [CrossRef]
- Davis, M.E.; Liu, T.L.; Taylor, Y.J.; Davidson, L.; Schmid, M.; Yates, T.; Scotton, J.; Spencer, M.D. Exploring Patient Awareness and Perceptions of the Appropriate Use of Antibiotics: A Mixed-Methods Study. Antibiotics 2017, 6, 23. [Google Scholar] [CrossRef]
- Kohut, M.R.; Keller, S.C.; Linder, J.A.; Tamma, P.D.; Cosgrove, S.E.; Speck, K.; Ahn, R.; Dullabh, P.; Miller, M.A.; Szymczak, J.E. The Inconvincible Patient: How Clinicians Perceive Demand for Antibiotics in the Outpatient Setting. Fam. Pract. 2019, 37, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Magin, P.; Davey, A.R.; Davis, J. Evidence-Based Strategies for Better Antibiotic Prescribing. Aust. J. Gen. Pract. 2022, 51, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Lekshmi, M.; Ammini, P.; Kumar, S.; Varela, M.F. The Food Production Environment and the Development of Antimicrobial Resistance in Human Pathogens of Animal Origin. Microorganisms 2017, 5, 11. [Google Scholar] [CrossRef]
- Kim, J.; Ahn, J. Emergence and Spread of Antibiotic-Resistant Foodborne Pathogens from Farm to Table. Food Sci. Biotechnol. 2022, 31, 1481–1499. [Google Scholar] [CrossRef] [PubMed]
- Vora, N.M.; Hannah, L.; Walzer, C.; Vale, M.M.; Lieberman, S.; Emerson, A.; Jennings, J.; Bonds, M.H.; Evans, J.; Chilukuri, B.; et al. Interventions to Reduce Risk for Pathogen Spillover and Early Disease Spread to Prevent Outbreaks, Epidemics, and Pandemics. Emerg. Infect. Dis. 2023, 3, e221079. [Google Scholar] [CrossRef]
- Iwu, C.D.; Korsten, L.; Okoh, A.I. The Incidence of Antibiotic Resistance within and Beyond the Agricultural Ecosystem: A Concern for Public Health. Microbiol. Open 2020, 9, e1035. [Google Scholar] [CrossRef] [PubMed]
- Collineau, L.; Bourély, C.; Rousset, L.; Berger-Carbonne, A.; Ploy, M.C.; Pulcini, C.; Colomb-Cotinat, M. Towards One Health Surveillance of Antibiotic Resistance: Characterisation and Mapping of Existing Programmes in Humans, Animals, Food and the Environment in France, 2021. Eurosurveillance 2023, 28, 2200804. [Google Scholar] [CrossRef]
- Oberin, M.; Badger, S.; Faverjon, C.; Cameron, A.; Bannister-Tyrrell, M. Electronic Information Systems for One Health Surveillance of Antimicrobial Resistance: A Systematic Scoping Review. BMJ Glob. Health 2022, 7, e007388. [Google Scholar] [CrossRef] [PubMed]
- Aenishaenslin, C.; Häsler, B.; Ravel, A.; Parmley, E.J.; Mediouni, S.; Bennani, H.; Stärk, K.D.C.; Buckeridge, D.L. Evaluating the Integration of One Health in Surveillance Systems for Antimicrobial Use and Resistance: A Conceptual Framework. Front. Vet. Sci. 2021, 8, 611931. [Google Scholar] [CrossRef]
- Joshi, M.P.; Hafner, T.; Twesigye, G.; Ndiaye, A.; Kiggundu, R.; Mekonnen, N.; Kusu, N.; Berthé, S.; Lusaya, E.P.; Acho, A. Strengthening Multisectoral Coordination on Antimicrobial Resistance: A Landscape Analysis of Efforts in 11 Countries. J. Pharm. Policy Pract. 2021, 21, 27. [Google Scholar] [CrossRef]
- Falkenberg, T. Applying a One Health Approach to Inter- and Trans-Disciplinary Research on Antimicrobial Resistance. Eur. J. Public Health 2019, 29 (Suppl. S4), ckz185-798. [Google Scholar] [CrossRef]
- Rhouma, M.; Archambault, M.; Butaye, P. Antimicrobial Use and Resistance in Animals from a One Health Perspective. Vet. Sci. 2023, 10, 319. [Google Scholar] [CrossRef] [PubMed]
- Marvasi, M.; Casillas, L.; Vassallo, A.; Purchase, D. Educational Activities for Students and Citizens Supporting the One Health Approach on Antimicrobial Resistance. Antibiotics 2021, 10, 1519. [Google Scholar] [CrossRef] [PubMed]
- Pearson, M.; Chandler, C. Knowing Antimicrobial Resistance in Practice: A Multi-Country Qualitative Study with Human and Animal Healthcare Professionals. Glob. Health Action 2019, 12 (Suppl. S1), 1599560. [Google Scholar] [CrossRef] [PubMed]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. In Antimicrobial Resistance in Bacteria from Livestock and Companion Animals; ASM Press: Washington, DC, USA, 2018; pp. 521–547. [Google Scholar]
- Pieri, A.; Aschbacher, R.; Fasani, G.; Mariella, J.; Brusetti, L.; Pagani, E.; Sartelli, M.; Pagani, L. Country Income Is Only One of the Tiles: The Global Journey of Antimicrobial Resistance among Humans, Animals, and Environment. Antibiotics 2020, 9, 473. [Google Scholar] [CrossRef] [PubMed]
- Gandra, S.; Alvarez-Uria, G.; Turner, P.; Joshi, J.; Limmathurotsakul, D.; van Doorn, H.R. Antimicrobial Resistance Surveillance in Low- and Middle-Income Countries: Progress and Challenges in Eight South Asian and Southeast Asian Countries. Clin. Microbiol. Rev. 2020, 33, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Nadimpalli, M.; Delarocque-Astagneau, E.; Love, D.C.; Price, L.B.; Huynh, B.T.; Collard, J.M.; Lay, K.S.; Borand, L.; Ndir, A.; Walsh, T.R.; et al. Combating Global Antibiotic Resistance: Emerging One Health Concerns in Lower- and Middle-Income Countries. Clin. Infect. Dis. 2018, 66, 963–969. [Google Scholar] [CrossRef]
- Wernli, D.; Harbarth, S.; Levrat, N.; Pittet, D. A ‘Whole of United Nations Approach’ to Tackle Antimicrobial Resistance? A Mapping of the Mandate and Activities of International Organisations. BMJ Glob. Health 2022, 7, e008181. [Google Scholar] [CrossRef]
- Pinto Ferreira, J.; Gochez, D.; Jeannin, M.; Magongo, M.W.; Loi, C.; Bucher, K.; Moulin, G.; Erlacher-Vindel, E. From OIE Standards to Responsible and Prudent Use of Antimicrobials: Supporting Stewardship for the Use of Antimicrobial Agents in Animals. JAC-Antimicrob. Resist. 2022, 4, dlac017. [Google Scholar] [CrossRef]
- Pinto Ferreira, J.; Battaglia, D.; Dorado García, A.; Tempelman, K.; Bullon, C.; Motriuc, N.; Caudell, M.; Cahill, S.; Song, J.; LeJeune, J. Achieving Antimicrobial Stewardship on the Global Scale: Challenges and Opportunities. Microorganisms 2022, 10, 1599. [Google Scholar] [CrossRef]
- Skandalis, N.; Maeusli, M.; Papafotis, D.; Miller, S.; Lee, B.; Theologidis, I.; Luna, B. Environmental Spread of Antibiotic Resistance. Antibiotics 2021, 10, 640. [Google Scholar] [CrossRef] [PubMed]
- Orubu, E.S.F.; Sutradhar, I.; Zaman, M.H.; Wirtz, V.J. Benchmarking National Action Plans on Antimicrobial Resistance in Eight Selected LMICs: Focus on the Veterinary Sector Strategies. J. Glob. Health 2020, 10, 020414. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.P.; Ashiru-Oredope, D.; Beech, E. Antibiotic Stewardship Initiatives as Part of the UK 5-Year Antimicrobial Resistance Strategy. Antibiotics 2015, 4, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Manning, M.L.; Pfeiffer, J.; Larson, E.L. Combating Antibiotic Resistance: The Role of Nursing in Antibiotic Stewardship. Am. J. Infect. Control 2016, 44, 1454–1457. [Google Scholar] [CrossRef]
- Wang, S.; Hu, Y.J.; Little, P.; Wang, Y.; Chang, Q.; Zhou, X.; Moore, M.; Harwell, J.I. The Impact of the National Action Plan on the Epidemiology of Antibiotic Resistance among 352,238 Isolates in a Teaching Hospital in China from 2015 to 2018. Antimicrob. Resist. Infect. Control 2019, 8, 22. [Google Scholar] [CrossRef]
- Sangeda, R.Z.; Kibona, J.; Munishi, C.; Arabi, F.; Manyanga, V.P.; Mwambete, K.D.; Horumpende, P.G. Assessment of Implementation of Antimicrobial Resistance Surveillance and Antimicrobial Stewardship Programs in Tanzanian Health Facilities a Year after Launch of the National Action Plan. Front. Public Health 2020, 8, 454. [Google Scholar] [CrossRef]
- Kuupiel, D.; Bawontuo, V.; Mashamba-Thompson, T.P. Improving the Accessibility and Efficiency of Point-of-Care Diagnostics Services in Low- and Middle-Income Countries: Lean and Agile Supply Chain Management. Diagnostics 2017, 7, 58. [Google Scholar] [CrossRef]
- Roth, L.; Bempong, D.; Babigumira, J.B.; Banoo, S.; Cooke, E.; Jeffreys, D.; Kasonde, L.H.G.M.; Lin, J.C.W.; Lumpkin, M. Expanding Global Access to Essential Medicines: Investment Priorities for Sustainably Strengthening Medical Product Regulatory Systems. Glob. Health 2018, 14, 102. [Google Scholar] [CrossRef] [PubMed]
- Porter, G.; Kotwani, A.; Bhullar, L.; Joshi, J. Over-the-Counter Sales of Antibiotics for Human Use in India: The Challenges and Opportunities for Regulation. Med. Law Int. 2012, 21, 147–173. [Google Scholar] [CrossRef]
- Adhikari, B.; Pokharel, S.; Raut, S.; Adhikari, J.; Thapa, S.; Paudel, K.; Narayan, G.C.; Neupane, S.; Neupane, R.; Yadav, R. Why Do People Purchase Antibiotics Over-the-Counter? A Qualitative Study with Patients, Clinicians and Dispensers in Central, Eastern and Western Nepal. BMJ Glob. Health 2021, 6, e005829. [Google Scholar] [CrossRef]
- Sommanustweechai, A.; Chanvatik, S.; Sermsinsiri, V.; Sivilaikul, S.; Patcharanarumol, W.; Yeung, S.; Tangcharoensathien, V. Antibiotic Distribution Channels in Thailand: Results of Key-Informant Interviews, Reviews of Drug Regulations and Database Searches. Bull. World Health Organ. 2018, 96, 101. [Google Scholar] [CrossRef] [PubMed]
- Yevutsey, S.K.; Buabeng, K.O.; Aikins, M.; Anto, B.P.; Biritwum, R.B.; Frimodt-Møller, N.; Gyansa-Lutterodt, M. Situational Analysis of Antibiotic Use and Resistance in Ghana: Policy and Regulation. BMC Public Health 2017, 17, 896. [Google Scholar] [CrossRef]
- Kalungia, A.C.; Burger, J.; Godman, B.; Costa, J.D.O.; Simuwelu, C. Non-Prescription Sale and Dispensing of Antibiotics in Community Pharmacies in Zambia. Expert Rev. Anti-Infect. Ther. 2016, 14, 1215–1223. [Google Scholar] [CrossRef] [PubMed]
- Batista, A.D.; Rodrigues, D.; Figueiras, A.; Zapata-Cachafeiro, M.; Roque, F.; Herdeiro, M.T. Antibiotic Dispensation Without a Prescription Worldwide: A Systematic Review. Antibiotics 2020, 9, 786. [Google Scholar] [CrossRef] [PubMed]
- Simões, A.S.; Alves, D.A.; Gregório, J.; Couto, I.; Dias, S.; Póvoa, P.; Viveiros, M.; Gonçalves, L.; Lapão, L.V. Fighting Antibiotic Resistance in Portuguese Hospitals: Understanding Antibiotic Prescription Behaviours to Better Design Antibiotic Stewardship Programmes. J. Glob. Antimicrob. Resist. 2018, 13, 226–230. [Google Scholar] [CrossRef]
- Bauer, K.A.; Kullar, R.; Gilchrist, M.; File, T.M., Jr. Antibiotics and Adverse Events: The Role of Antimicrobial Stewardship Programs in ‘Doing No Harm’. Curr. Opin. Infect. Dis. 2019, 32, 553–558. [Google Scholar] [CrossRef]
- Wagenaar, J.A.; Scott, H.M. Reducing Antimicrobial Usage in Agriculture and Aquaculture: Beyond Regulatory Policy. Zoonoses Public Health 2015, 62, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Low, C.X.; Tan, L.T.H.; Ab Mutalib, N.S.; Pusparajah, P.; Goh, B.H.; Chan, K.G.; Letchumanan, V.; Lee, L.H. Unveiling the Impact of Antibiotics and Alternative Methods for Animal Husbandry: A Review. Antibiotics 2021, 10, 578. [Google Scholar] [CrossRef]
- Dewulf, J.; Sternberg-Lewerin, S.; Ryan, M. Tackling Antimicrobial Resistance in the Food and Livestock Sector. In Challenges to Tackling Antimicrobial Resistance: Economic and Policy Responses; Anderson, M., Cecchini, M., Mossialos, E., Eds.; Cambridge University Press: Cambridge, UK, 2020; pp. 99–124, (European Observatory on Health Systems and Policies). [Google Scholar]
- Kuehn, B.M. FDA Moves to Curb Antibiotic Use in Livestock. JAMA 2014, 311, 347–348. [Google Scholar] [CrossRef]
- Patel, S.J.; Wellington, M.; Shah, R.M.; Ferreira, M.J. Antibiotic Stewardship in Food-Producing Animals: Challenges, Progress, and Opportunities. Clin. Ther. 2020, 42, 1649–1658. [Google Scholar] [CrossRef]
- Miller, R.A.; Salmon, P.; Sharkey, M. Approaches to Developing Judicious Uses of Veterinary Antibacterial Drugs. J. Vet. Pharmacol. Ther. 2021, 44, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Singer, A.C.; Shaw, H.; Rhodes, V.; Hart, A. Review of Antimicrobial Resistance in the Environment and Its Relevance to Environmental Regulators. Front. Microbiol. 2016, 17, 1728. [Google Scholar] [CrossRef] [PubMed]
- Rayan, R.A. Pharmaceutical Effluent Evokes Superbugs in the Environment: A Call to Action. Biosaf. Health 2023, 50, 363–371. [Google Scholar] [CrossRef]
- Ågerstrand, M.; Josefsson, H.; Wernersson, A.S.; Larsson, D.J. Opportunities to Tackle Antibiotic Resistance Development in the Aquatic Environment through the Water Framework Directive. Ambio 2023, 52, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Okoye, C.O.; Nyaruaba, R.; Ita, R.E.; Okon, S.U.; Addey, C.I.; Ebido, C.C.; Opabunmi, A.O.; Okeke, E.S.; Chukwudozie, K.I. Antibiotic Resistance in the Aquatic Environment: Analytical Techniques and Interactive Impact of Emerging Contaminants. Environ. Toxicol. Pharmacol. 2022, 96, 103995. [Google Scholar] [CrossRef] [PubMed]
- Hijazi, K.; Joshi, C.; Gould, I.M. Challenges and Opportunities for Antimicrobial Stewardship in Resource-Rich and Resource-Limited Countries. Expert Rev. Anti-Infect. Ther. 2019, 17, 621–634. [Google Scholar] [CrossRef]
- Khan, M.S.; Durrance-Bagale, A.; Mateus, A.; Sultana, Z.; Hasan, R.; Hanefeld, J. What Are the Barriers to Implementing National Antimicrobial Resistance Action Plans? A Novel Mixed-Methods Policy Analysis in Pakistan. Health Policy Plan. 2020, 35, 973–982. [Google Scholar] [CrossRef]
- Frumence, G.; Mboera, L.E.; Katale, B.Z.; Sindato, C.; Kimera, S.; Durrance-Bagale, A.; Mshana, S.E.; Clark, T.G.; Rweyemamu, M.M.; Legido-Quigley, H.; et al. Policy Actors and Human and Animal Health Practitioners’ Perceptions of Antimicrobial Use and Resistance in Tanzania: A Qualitative Study. J. Glob. Antimicrob. Resist. 2021, 25, 40–47. [Google Scholar] [CrossRef]
- Tang, K.; Millar, B.; Moore, J. Antimicrobial Resistance (AMR). Br. J. Biomed. Sci. 2023, 80, 11387. [Google Scholar] [CrossRef]
- Umair, M.; Mohsin, M.; Sönksen, U.W.; Walsh, T.R.; Kreienbrock, L.; Laxminarayan, R. Measuring Antimicrobial Use Needs Global Harmonization. Glob. Chall. 2021, 5, 2100017. [Google Scholar] [CrossRef]
- Alm, R.A.; Gallant, K. Innovation in Antimicrobial Resistance: The CARB-X Perspective. ACS Infect. Dis. 2020, 6, 1317–1322. [Google Scholar] [CrossRef] [PubMed]
- Altarac, D.; Gutch, M.; Mueller, J.; Ronsheim, M.; Tommasi, R.; Perros, M. Challenges and Opportunities in the Discovery, Development, and Commercialization of Pathogen-Targeted Antibiotics. Drug Discov. Today 2021, 26, 2084–2089. [Google Scholar] [CrossRef] [PubMed]
- Barrass, L.; Roberts, A.; Arisa, O.; Robertson, E.; Oliver, A.; Sharma, O.; Kinnersley, J.; Craig, D.; Oyewole, A. OP57: The Identification of Technological Innovations to Address the Challenge of Antimicrobial Resistance Using Horizon Scanning Approaches. Int. J. Technol. Assess. Health Care 2022, 38 (Suppl. S1), S22. [Google Scholar] [CrossRef]
- Sharma, S.; Chauhan, A.; Ranjan, A.; Mathkor, D.M.H.S.; Ramniwas, S.; Tuli, H.S.; Jindal, T.; Yadav, V. Emerging Challenges in Antimicrobial Resistance: Implications for Pathogenic Microorganisms, Novel Antibiotics, and Their Impact on Sustainability. Front. Microbiol. 2024, 15, 1403168. [Google Scholar] [CrossRef]
- Feng, Y. Transferability of MCR-1/2 Polymyxin Resistance: Complex Dissemination and Genetic Mechanism. ACS Infect. Dis. 2018, 4, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Talebi, G.; Hashemi, A. Metallo-β-Lactamase, Extended Spectrum β-Lactamase, and mcr-1 Gene as Major Therapeutic Challenges. Rev. Res. Med. Microbiol. 2022, 33, e40–e47. [Google Scholar] [CrossRef]
- Son, S.J.; Huang, R.; Squire, C.J.; Leung, I.K. MCR-1: A Promising Target for Structure-Based Design of Inhibitors to Tackle Polymyxin Resistance. Drug Discov. Today 2019, 24, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Murugaiyan, J.; Kumar, P.A.; Rao, G.S.; Iskandar, K.; Hawser, S.H.J.P.; Mohsen, Y.; Awual, W.A.; Alvarez, R.; Nanono, S. Progress in Alternative Strategies to Combat Antimicrobial Resistance: Focus on Antibiotics. Antibiotics 2022, 11, 200. [Google Scholar] [CrossRef]
- Karas, J.A.; Chen, F.; Schneider-Futschik, E.K.; Kang, Z.; Hussein, M.S.J.; Hoyer, D.; Giltrap, A.; Payne, R.J.; Li, J.; Velkov, T. Synthesis and Structure−Activity Relationships of Teixobactin. Ann. N. Y. Acad. Sci. 2020, 1459, 86–105. [Google Scholar] [CrossRef] [PubMed]
- Bhavnani, S.M.; Krause, K.M.; Ambrose, P.G. A Broken Antibiotic Market: Review of Strategies to Incentivize Drug Development. Open Forum Infect. Dis. 2020, 7, ofaa083. [Google Scholar] [CrossRef]
- Beyer, P.; Paulin, S. Priority Pathogens and the Antibiotic Pipeline: An Update. Bull. World Health Organ. 2020, 98, 151. [Google Scholar] [CrossRef] [PubMed]
- Breijyeh, Z.; Karaman, R. Design and Synthesis of Novel Antimicrobial Agents. Antibiotics 2023, 12, 628. [Google Scholar] [CrossRef] [PubMed]
- Hitchcock, N.M.; Gomes Nunes, D.D.; Shiach, J.; Saraiva Hodel, K.V.; Barbosa, J.D.V.; Rodrigues, L.A.P.; Colerm, S.; Soares, M.B.P.; Badaró, R. Current Clinical Landscape and Global Potential of Bacteriophage Therapy. Microorganisms 2023, 15, 1020. [Google Scholar] [CrossRef] [PubMed]
- Sieiro, C.; Areal-Hermida, L.; Pichardo-Gallardo, Á.; Almuiña-González, R.; De Miguel, T.; Sánchez, S.; Sánchez-Pérez, A.; Villa, T.G. A Hundred Years of Bacteriophages: Can Phages Replace Antibiotics in Agriculture and Aquaculture? Antibiotics 2020, 9, 493. [Google Scholar] [CrossRef]
- Alomari, M.M.M.; Dec, M.; Urban-Chmiel, R. Bacteriophages as an Alternative Method for Control of Zoonotic and Foodborne Pathogens. Viruses 2021, 13, 2348. [Google Scholar] [CrossRef] [PubMed]
- Adesanya, O.; Oduselu, T.; Akin-Ajani, O.; Adewumi, O.M.; Ademowo, O.G. An Exegesis of Bacteriophage Therapy: An Emerging Player in the Fight against Anti-Microbial Resistance. AIMS Microbiol. 2020, 6, 204. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582770. [Google Scholar] [CrossRef]
- Matthyssen, T.; Li, W.; Holden, J.A.; Lenzo, J.C.; Hadjigol, S.; O’Brien-Simpson, N.M. The Potential of Modified and Multimeric Antimicrobial Peptide Materials as Superbug Killers. Front. Chem. 2022, 9, 795433. [Google Scholar] [CrossRef]
- Bellotti, D.; Remelli, M. Lights and Shadows on the Therapeutic Use of Antimicrobial Peptides. Molecules 2022, 27, 4584. [Google Scholar] [CrossRef]
- Teixeira, M.C.; Carbone, C.; Sousa, M.C.; Espina, M.; Garcia, M.L.; Sanchez-Lopez, E.; Souto, E.B. Nanomedicines for the Delivery of Antimicrobial Peptides (AMPs). Nanomaterials 2020, 10, 560. [Google Scholar] [CrossRef]
- Bergman, P.; Raqib, R.; Rekha, R.S.; Agerberth, B.; Gudmundsson, G.H. Host Directed Therapy against Infection by Boosting Innate Immunity. Front. Immunol. 2020, 11, 1209. [Google Scholar] [CrossRef] [PubMed]
- Owen, A.M.; Fults, J.B.; Patil, N.K.; Hernandez, A.; Bohannon, J.K. TLR Agonists as Mediators of Trained Immunity: Mechanistic Insight and Immunotherapeutic Potential to Combat Infection. Front. Immunol. 2021, 11, 622614. [Google Scholar] [CrossRef] [PubMed]
- Latreille, E.; Lee, W.L. Modulation of the Host Response as a Therapeutic Strategy in Severe Lung Infections. Viruses 2023, 15, 1462. [Google Scholar] [CrossRef] [PubMed]
- Kelly, A.M.; McLoughlin, R.M. Target the Host, Kill the Bug; Targeting Host Respiratory Immunosuppressive Responses as a Novel Strategy to Improve Bacterial Clearance during Lung Infection. Front. Immunol. 2020, 11, 767. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Jin, W.; Liu, S.J.; Jiao, Z.; Li, X. Probiotics, Prebiotics, and Postbiotics in Health and Disease. MedComm 2023, 4, e420. [Google Scholar] [CrossRef]
- Quigley, E.M.; Gajula, P. Recent Advances in Modulating the Microbiome. F1000Research 2020, 9, F1000 Faculty Rev-46. [Google Scholar] [CrossRef]
- Schwabkey, Z.I.; Jenq, R.R. Microbiome Anomalies in Allogeneic Hematopoietic Cell Transplantation. Annu. Rev. Med. 2020, 71, 137–148. [Google Scholar] [CrossRef]
- Wuethrich, I.; Pelzer, B.W.; Khodamoradi, Y.; Vehreschild, M.J. The Role of the Human Gut Microbiota in Colonization and Infection with Multidrug-Resistant Bacteria. Gut Microbes 2021, 13, 1911279. [Google Scholar] [CrossRef] [PubMed]
- Broom, A.; Kenny, K.; Prainsack, B.; Broom, J. Antimicrobial Resistance as a Problem of Values? Views from Three Continents. Crit. Public Health 2021, 31, 451–463. [Google Scholar] [CrossRef]
- Sariola, S.; Butcher, A. In Critique of Anthropocentrism: A More-than-Human Ethical Framework for Antimicrobial Resistance. Med. Humanit. 2022, 48, e16. [Google Scholar]
- Tarrant, C.; Colman, A.M.; Jenkins, D.R.; Chattoe-Brown, E.; Perera, N.; Mehtar, S.; Nakkawita, W.M.I.D.; Bolscher, M.; Krockow, E. Drivers of Broad-Spectrum Antibiotic Overuse across Diverse Hospital Contexts—A Qualitative Study of Prescribers in the UK, Sri Lanka, and South Africa. Antibiotics 2021, 19, 94. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J.; Melby, M.K.; Lock, M.; Nichter, M. Accounting for Variation in and Overuse of Antibiotics among Humans. BioEssays 2021, 43, 2000163. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, R.A.; Taing, L.; Bhatia, H. Antimicrobial Resistance and Environmental Health: A Water Stewardship Framework for Global and National Action. Antibiotics 2022, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Anyaegbunam, N.J.; Anekpo, C.C.; Anyaegbunam, Z.K.G.; Doowuese, Y.; Chinaka, C.B.; Odo, O.J.; Sharndama, H.C.; Mba, I.E. The Resurgence of Phage-Based Therapy in the Era of Increasing Antibiotic Resistance: From Research Progress to Challenges and Prospects. Microbiol. Res. 2022, 264, 127155. [Google Scholar] [CrossRef] [PubMed]
- Luong, T.; Salabarria, A.C.; Roach, D.R. Phage Therapy in the Resistance Era: Where Do We Stand and Where Are We Going? Clin. Ther. 2020, 42, 1659–1680. [Google Scholar] [CrossRef]
- Dunachie, S.J.; Day, N.P.; Dolecek, C. The Challenges of Estimating the Human Global Burden of Disease of Antimicrobial Resistant Bacteria. Curr. Opin. Microbiol. 2020, 57, 95–101. [Google Scholar] [CrossRef]
- Gajdács, M.; Urbán, E.; Stájer, A.; Baráth, Z. Antimicrobial Resistance in the Context of the Sustainable Development Goals: A Brief Review. Eur. J. Investig. Health Psychol. Educ. 2021, 11, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Thornber, K.; Kirchhelle, C. Hardwiring Antimicrobial Resistance Mitigation into Global Policy. JAC-Antimicrob. Resist. 2022, 4, dlac083. [Google Scholar] [CrossRef]
- Asaaga, F.A.; Young, J.C.; Oommen, M.A.; Chandarana, R.; August, J.; Joshi, J.; Chanda, M.M.; Vanak, A.T.; Srinivas, P.N.; Hoti, S.L.; et al. Operationalising the “One Health” Approach in India: Facilitators of and Barriers to Effective Cross-Sector Convergence for Zoonoses Prevention and Control. BMC Public Health 2021, 21, 1517. [Google Scholar] [CrossRef]
- Achi, C.R.; Ayobami, O.; Mark, G.; Egwuenu, A.; Ogbolu, D.; Kabir, J. Operationalising One Health in Nigeria: Reflections from a High-Level Expert Panel Discussion Commemorating the 2020 World Antibiotics Awareness Week. Front. Public Health 2021, 9, 673504. [Google Scholar] [CrossRef]
- Mariappan, V.; Vellasamy, K.M.; Mohamad, N.A.; Subramaniam, S.; Vadivelu, J. OneHealth Approaches Contribute Towards Antimicrobial Resistance: Malaysian Perspective. Front. Microbiol. 2021, 12, 718774. [Google Scholar] [CrossRef] [PubMed]
- Parveen, S.; Garzon-Orjuela, N.; Amin, D.; McHugh, P.; Vellinga, A. Public Health Interventions to Improve Antimicrobial Resistance Awareness and Behavioural Change Associated with Antimicrobial Use: A Systematic Review Exploring the Use of Social Media. Antibiotics 2022, 11, 669. [Google Scholar] [CrossRef] [PubMed]
- Fletcher-Miles, H.; Gammon, J.; Williams, S.; Hunt, J. A Scoping Review to Assess the Impact of Public Education Campaigns to Affect Behavior Change Pertaining to Antimicrobial Resistance. Am. J. Infect. Control 2020, 48, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, C.; Batura, N.; Wulandari, L.P.L.; Khan, M.; Wiseman, V. Improving Antibiotic Use through Behaviour Change: A Systematic Review of Interventions Evaluated in Low-and Middle-Income Countries. Health Policy Plan. 2021, 36, 754–773. [Google Scholar] [CrossRef] [PubMed]
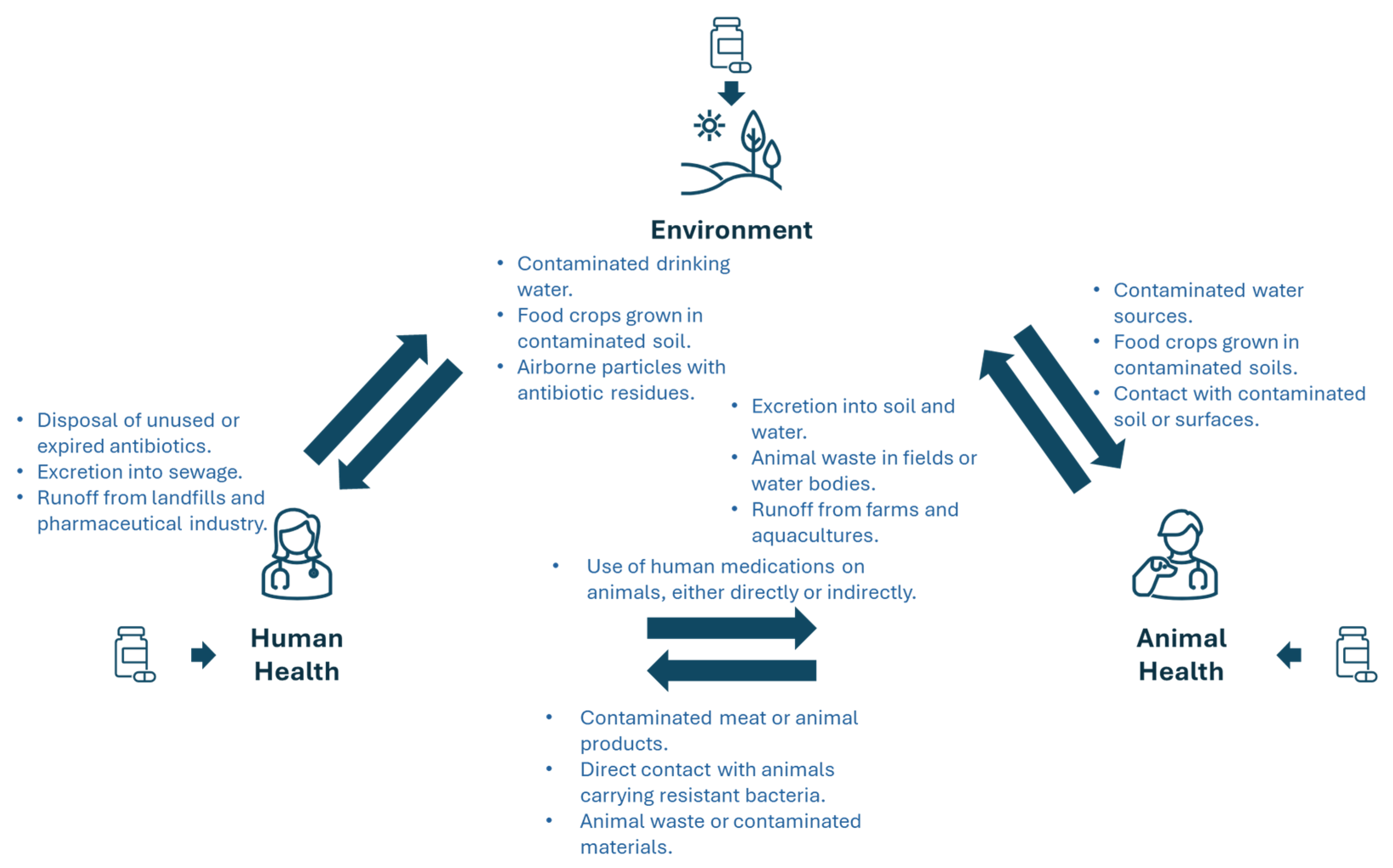


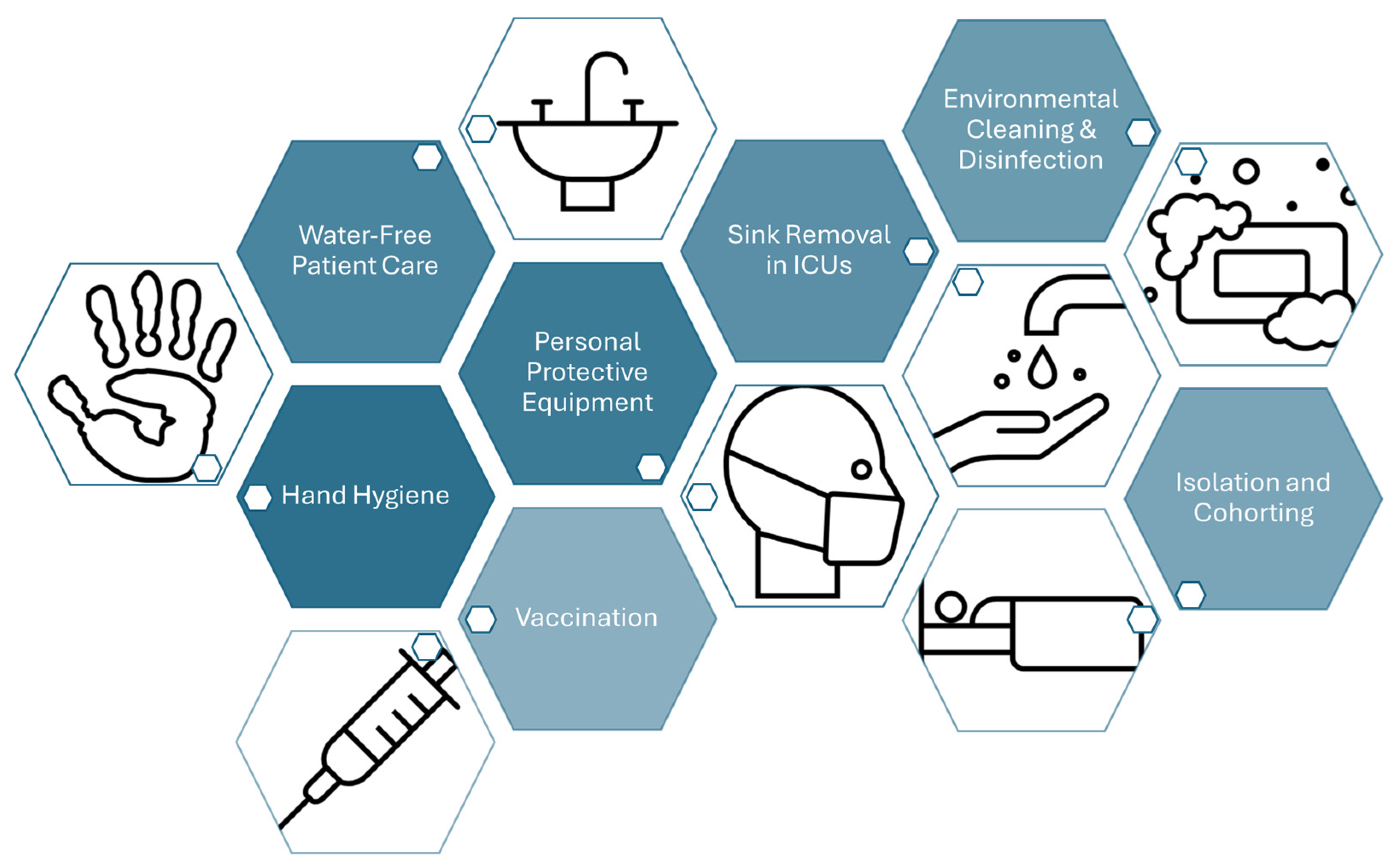
| Organism | Mechanism | Details | Example | Ref. |
|---|---|---|---|---|
| Bacteria | Target Modification | Antibiotic’s molecular target alteration inhibits drug binding | rRNA mutations hinder aminoglycoside binding | [34,35,36] |
| Efflux Pumps | Membrane proteins expel antibiotics and reduce their intracellular concentrations | Efflux of tetracyclines, fluoroquinolones, and macrolides by specific bacterial pumps | [37,38,39] | |
| Enzymatic Degradation | Enzymes chemically modify or degrade antibiotics and neutralize their effects | β-lactamases hydrolyze the β-lactam ring (penicillin, cephalosporin) | [40,41,42,43] | |
| Membrane Permeability | Outer membrane permeability reduction limits antibiotic entry | Gram-negative bacteria alter outer membrane proteins to resist drug influx | [44,45,46] | |
| Biofilm Formation | Biofilms limit antibiotic penetration and enhance resistance | Biofilm-associated resistance in P. aeruginosa infections | [47,48,49,50] | |
| Viruses | Mutation | Mutations in viral genes encoding enzymes reduce the binding affinity of antiviral drugs | HIV resistance due to mutations in reverse transcriptase and protease | [51,52] |
| Changes in Viral Entry Proteins | Surface protein mutations prevent antiviral drugs from achieving host cell entry | Resistance to entry inhibitors in HIV treatment | [53] | |
| Altered Drug Activation | Mutations impair the activation of antiviral drugs within the virus or host cells | Acyclovir resistance in herpes simplex virus due to TK gene mutations | [54] | |
| Fungi | Target Modification | Antifungal drugs’ molecular target alteration reduces drug efficacy | Azole resistance in Candida spp. due to ERG11 gene mutations | [55] |
| Efflux Pumps | Membrane proteins expel antifungal drugs and reduce their intracellular concentrations | Azole resistance in C. albicans due to ABC transporter overexpression | [56,57,58,59,60,61] | |
| Cell Wall Alteration | Cell wall composition modifications decrease drug binding or penetration | Echinocandin resistance in Candida spp. due to FKS gene mutations | [62,63] | |
| Biofilm Formation | Biofilms protect cells from antifungal drugs and difficult infection treatment | Biofilm-related resistance in Candida infections associated with medical devices | [64,65,66] | |
| Parasites | Target Modification | Parasites acquire mutations in genes encoding drug targets, leading to reduced drug efficacy | Antifolate resistance in P. falciparum due to DHFR gene mutations | [67,68,69,70,71] |
| Efflux pumps | Membrane proteins actively remove drugs from cells and reduce their intracellular concentrations | Chloroquine resistance in P. falciparum due to transporter protein expression | [70] | |
| Metabolic Pathway Alteration | Metabolic pathway alterations to bypass drug effects | Glycolytic pathway alterations confer resistance to drugs in trypanosomes | [72] | |
| Reduced Drug Activation | Mutations reduce the activation of drugs within parasites, leading to resistance | Metronidazole resistance in G. lamblia due to NTR mutations | [73,74,75] |
| Approach | Description | Advantages | Challenges | Examples | Ref. |
|---|---|---|---|---|---|
| New Antibiotics with Novel Mechanisms | Development of antibiotics targeting non-traditional bacterial processes. | Target novel aspects of bacterial physiology, useful for resistant bacteria. | Difficulty in the discovery and development of novel classes. | Teixobactin—targets bacterial cell wall synthesis in a new way, with no observed resistance yet. | [324] |
| Bacteriophage Therapy | Utilizes viruses that specifically infect and kill bacteria. | High specificity, ecological safety, and potential beyond clinical applications. | Large-scale production, stability, and bacterial resistance to phages. | Phage therapy for P. aeruginosa infections in cystic fibrosis patients. | [325,326] |
| Phage-Based Small Molecule Development | Exploits phage–bacteria interactions to identify new antibacterial targets and inhibitory proteins. | Novel targets for drug development, potentially more specific and effective treatments. | Development and scaling challenges. | Phage-derived enzymes (endolysins) are used to treat S. aureus infections. | [327] |
| Antimicrobial Peptides (AMPs) | Naturally occurring peptides with broad-spectrum antibacterial, antiviral, and antifungal properties. | Broad-spectrum activity, lower risk of resistance development, several FDA-approved drugs. | Requires more research on mechanisms of action and clinical optimization. | Daptomycin is used to treat infections by resistant Gram-positive bacteria (e.g., MRSA). | [328,329,330] |
| Immunotherapies | Monoclonal antibodies target and neutralize bacterial toxins and virulence factors. | High specificity, lower risk of resistance development. | High production costs and the need for targeted pathogen identification. | Bezlotoxumab used to neutralize C. difficile toxins. | [331,332] |
| Probiotics and Fecal Transplant Therapy | Enhances human microbiota to prevent infections. | Restores healthy microbiomes, potentially reducing antibiotic use. | Efficacy varies and needs more standardized research. | FMT used to treat recurrent C. difficile infections. | [333] |
| Pathogen-Oriented Therapy (POT) | Targeted approach using antibiotic conjugates, nanotechnologies, and CRISPR-Cas systems. | Highly specific to pathogens, avoids non-targeted antimicrobial use. | Complex development, regulatory challenges, and scaling issues. | CRISPR-Cas antimicrobials targeting resistant bacteria, antibiotic–antibody conjugates. | [334,335] |
| Combination Therapies | Use of multiple drugs to prevent or overcome resistance. | Enhances efficacy and prevents resistance development. | Risk of drug interactions and complex treatment regimens. | Ceftazidime-avibactam is used to treat multidrug-resistant Gram-negative bacteria. | [336] |
| Repurposing Existing Drugs | Using known drugs for new antibacterial purposes. | Cost-effective, faster clinical adoption due to existing safety profiles. | Limited applicability, potential to develop resistance to repurposed drugs. | Colistin is used to treat MDR Acinetobacter and Pseudomonas species. | [336] |
| Nanotechnology | Use of nanoparticles to enhance antibiotic efficacy or act as antimicrobial agents. | Enhances antibiotic activity and overcomes biofilm-related resistance. | Safety, bioavailability, and large-scale production. | Silver nanoparticles used in combination with antibiotics to enhance their efficacy. | [337,338] |
| Artificial Intelligence | AI-driven approaches for drug discovery, resistance prediction, and treatment optimization. | Accelerates drug discovery, predicts resistance, optimizes antimicrobial regimens. | Data quality, model accuracy, and integration into clinical workflows. | IBM’s AI-driven drug discovery used to identify new antibiotics like halicin. | [339,340] |
| One Health Approach | Multisectoral approach linking human, animal, and environmental health to combat AMR. | Promotes cross-sector collaboration and addresses complex AMR drivers globally. | Requires significant international cooperation and systemic change across multiple industries. | WHO Global Action Plan on AMR, One Health AMR Surveillance Programs (WHO, FAO, OIE). | [341,342,343,344] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, M.; Antunes, W.; Mota, S.; Madureira-Carvalho, Á.; Dinis-Oliveira, R.J.; Dias da Silva, D. An Overview of the Recent Advances in Antimicrobial Resistance. Microorganisms 2024, 12, 1920. https://doi.org/10.3390/microorganisms12091920
Oliveira M, Antunes W, Mota S, Madureira-Carvalho Á, Dinis-Oliveira RJ, Dias da Silva D. An Overview of the Recent Advances in Antimicrobial Resistance. Microorganisms. 2024; 12(9):1920. https://doi.org/10.3390/microorganisms12091920
Chicago/Turabian StyleOliveira, Manuela, Wilson Antunes, Salete Mota, Áurea Madureira-Carvalho, Ricardo Jorge Dinis-Oliveira, and Diana Dias da Silva. 2024. "An Overview of the Recent Advances in Antimicrobial Resistance" Microorganisms 12, no. 9: 1920. https://doi.org/10.3390/microorganisms12091920
APA StyleOliveira, M., Antunes, W., Mota, S., Madureira-Carvalho, Á., Dinis-Oliveira, R. J., & Dias da Silva, D. (2024). An Overview of the Recent Advances in Antimicrobial Resistance. Microorganisms, 12(9), 1920. https://doi.org/10.3390/microorganisms12091920










