Moxifloxacin Activates the SOS Response in Mycobacterium tuberculosis in a Dose- and Time-Dependent Manner
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Cultures
2.2. Isolation of Mutant H37Rv gyrA (D94G)
2.3. RNA Extraction, Reverse Transcription and qPCR
2.4. Statistical Analysis
3. Results
3.1. Characterization of SOS Gene Response to Moxifloxacin
3.2. Mtb Response to Moxifloxacin at Different Growth Phases
3.3. SOS Response in a Moxifloxacin Resistant Strain
3.4. General Transcriptional Response to Moxifloxacin
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. WHO Global Tuberculosis Report; WHO/CDS/TB 201820; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Boshoff, H.I.M.; Reed, M.B.; Barry, C.E.; Mizrahi, V. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell 2003, 113, 183–193. [Google Scholar] [CrossRef]
- Davis, E.O.; Dullaghan, E.M.; Rand, L. Definition of the mycobacterial SOS box and use to identify LexA-regulated genes in Mycobacterium tuberculosis. J. Bacteriol 2002, 184, 3287–3295. [Google Scholar] [CrossRef] [PubMed]
- Dörr, T.; Lewis, K.; Vulić, M. SOS response induces persistence to fluoroquinolones in Escherichia Coli. PLoS Genet. 2009, 5, e1000760. [Google Scholar] [CrossRef] [PubMed]
- Dörr, T.; Vulić, M.; Lewis, K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia Coli. PLoS Biol. 2010, 8, e1000317. [Google Scholar] [CrossRef] [PubMed]
- Gengenbacher, M.; Kaufmann, S.H.E. Mycobacterium tuberculosis: Success through dormancy. Fems. Microbiol. Rev. 2012, 36, 514–532. [Google Scholar] [CrossRef]
- Giannoni, F.; Iona, E.; Sementilli, F.; Brunori, L.; Pardini, M.; Migliori, G.B.; Orefici, G.; Fattorini, L. Evaluation of a New Line Probe Assay for Rapid Identification of gyrA Mutations in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2005, 49, 2928–2933. [Google Scholar] [CrossRef]
- Iacobino, A.; Piccaro, G.; Giannoni, F.; Mustazzolu, A.; Fattorini, L. Mycobacterium tuberculosis is selectively killed by rifampin and rifapentine in hypoxia at neutral pH. Antimicrob. Agents Chemother. 2017, 61, e02296-16. [Google Scholar] [CrossRef]
- Kenn, G.; Christensen, K.S.; Anders, L.-O. Prokaryotic toxin–antitoxin stress response loci. Nat. Rev. Microbiol 2005, 3, 371–382. [Google Scholar]
- Keren, I.; Minami, S.; Rubin, E.; Lewis, K. Characterization and Transcriptome Analysis of Mycobacterium tuberculosis Persisters. Mbio 2011, 2, e00100-11. [Google Scholar] [CrossRef]
- Sala, A.; Bordes, P.; Genevaux, P. Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins 2014, 6, 1002–1020. [Google Scholar] [CrossRef]
- O’Sullivan, D.M.; Hinds, J.; Butcher, P.D.; Gillespie, S.H.; McHugh, T.D. Mycobacterium tuberculosis DNA repair in response to subinhibitory concentrations of ciprofloxacin. J. Antimicrob. Chemother. 2008, 62, 1199–1202. [Google Scholar] [CrossRef] [PubMed]
- Piccaro, G.; Giannoni, F.; Filippini, P.; Mustazzolu, A.; Fattorini, L. Activities of drug combinations against Mycobacterium tuberculosis grown in aerobic and hypoxic acidic conditions. Antimicrob. Agents Chemother. 2013, 57, 1428–1433. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wayne, L.G.; Hayes, L.G. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 1996, 64, 2062–2069. [Google Scholar] [CrossRef] [PubMed]
- Pourahmad Jaktaji, R.; Pasand, S. Overexpression of SOS genes in ciprofloxacin resistant Escherichia coli mutants. Gene 2016, 576, 115–118. [Google Scholar] [CrossRef]
- Zvada, S.P.; Denti, P.; Geldenhuys, H.; Meredith, S.; Van As, D.; Hatherill, M.; Hanekom, W.; Wiesner, L.; Simonsson, U.S.H.; Jindani, A.; et al. Moxifloxacin population pharmacokinetics in patients with pulmonary tuberculosis and the effect of intermittent high-dose rifapentine. Antimicrob. Agents Chemother. 2012, 56, 4471–4473. [Google Scholar] [CrossRef][Green Version]
- Aldred, K.J.; Blower, T.R.; Kerns, R.J.; Berger, J.M.; Osheroff, N. Fluoroquinolone interactions with Mycobacterium tuberculosis gyrase: Enhancing drug activity against wild-type and resistant gyrase. Proc. Natl. Acad. Sci. USA 2016, 113, 839–846. [Google Scholar] [CrossRef]
- Ronayne, E.A.; Wan, Y.C.S.; Boudreau, B.A.; Landick, R.; Cox, M.M. P1 Ref Endonuclease: A Molecular Mechanism for Phage-Enhanced Antibiotic Lethality. PLoS Genet. 2016, 12, 1–31. [Google Scholar] [CrossRef]
- Sebastian, J.; Swaminath, S.; Nair, R.; Jakkala, K.; Pradhan, A.; Ajitkumar, P. De Novo Emergence of Genetically Resistant Mutants of Mycobacterium tuberculosis from the Persistence Phase Cells Formed against Antituberculosis Drugs In Vitro. Antimicrob. Agents Chemother. 2017, 61, 1–25. [Google Scholar] [CrossRef]
- Dillingham, M.S.; Kowalczykowski, S.C. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 2008, 72, 642–671. [Google Scholar] [CrossRef]
- Brooks, P.C.; Movahedzadeh, F.; Davis, E.O. Identification of some DNA damage-inducible genes of Mycobacterium tuberculosis: Apparent lack of correlation with LexA binding. J. Bacteriol. 2001, 183, 4459–4467. [Google Scholar] [CrossRef]
- Choudhary, E.; Sharma, R.; Kumar, Y.; Agarwal, N. Conditional Silencing by CRISPRi Reveals the Role of DNA Gyrase in Formation of Drug-Tolerant Persister Population in Mycobacterium tuberculosis. Front. Cell Infect. Microbiol. 2019, 9, 70. [Google Scholar] [CrossRef] [PubMed]
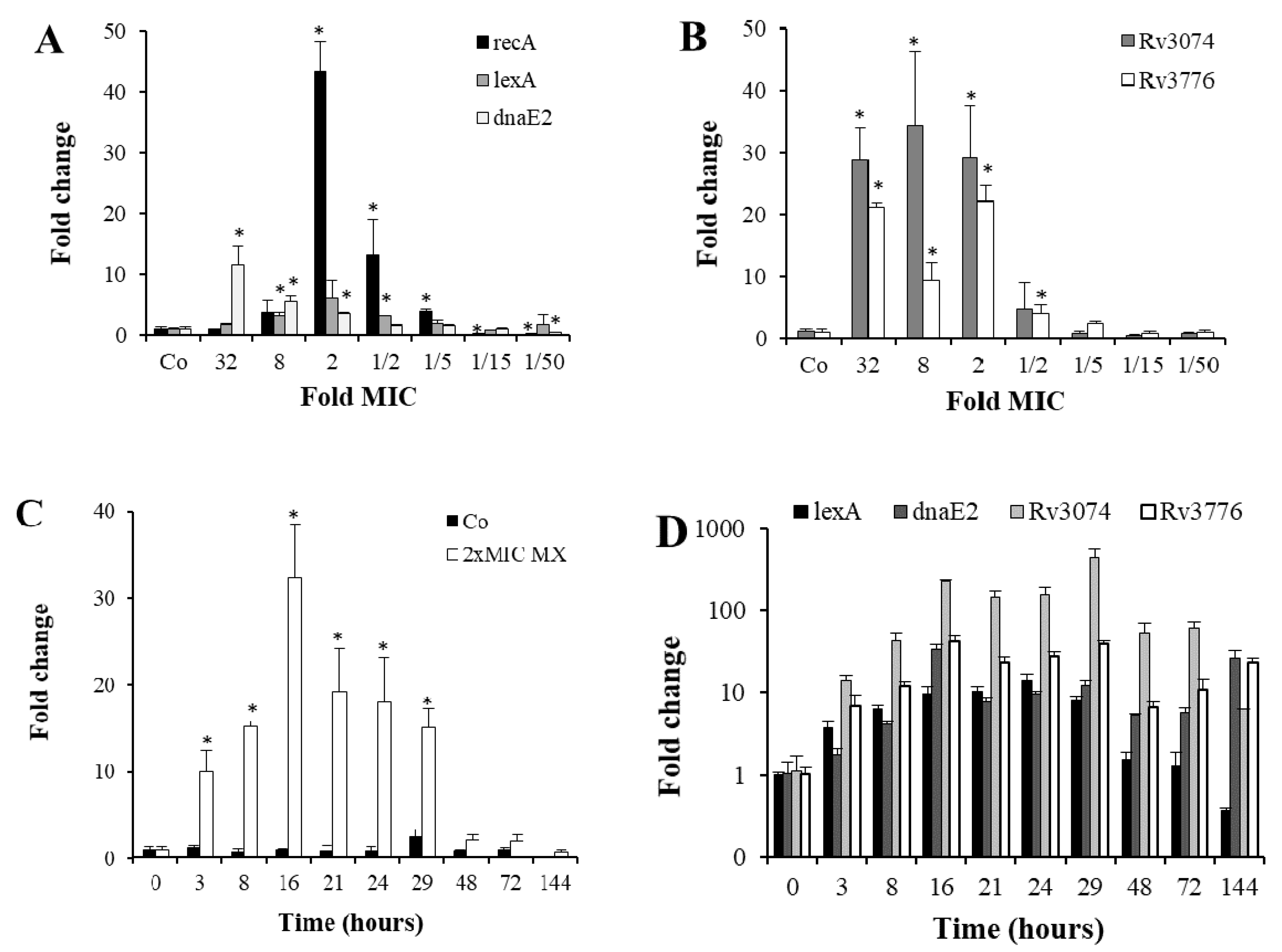
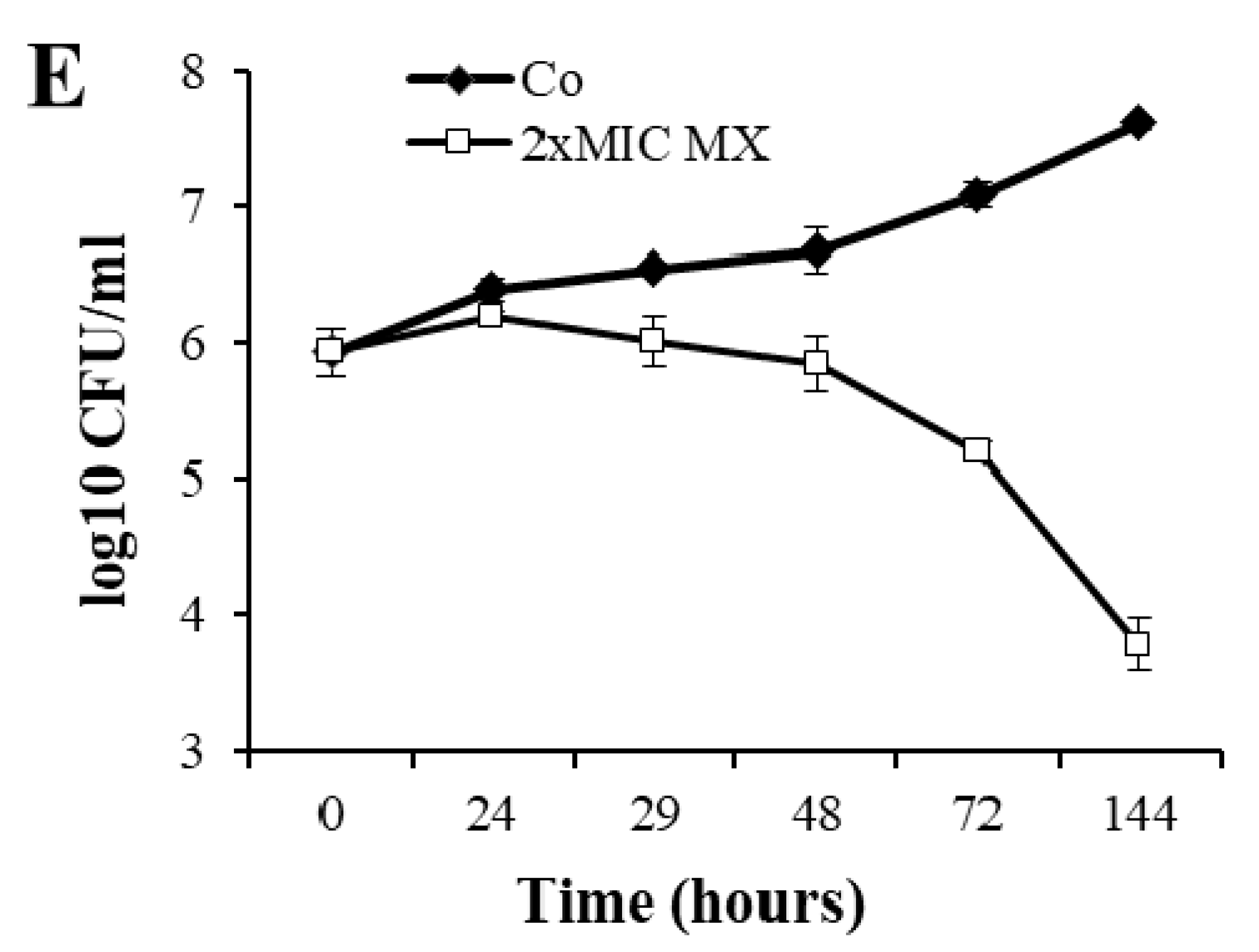
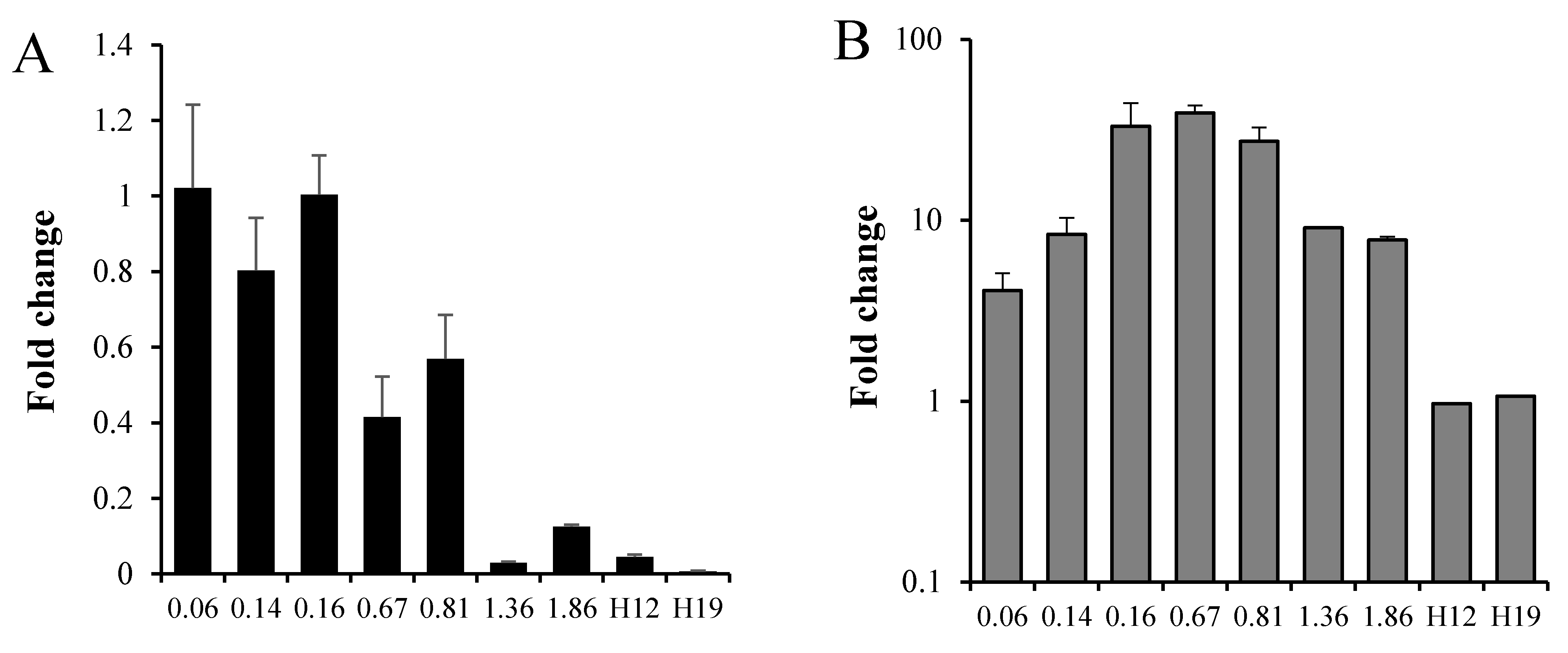
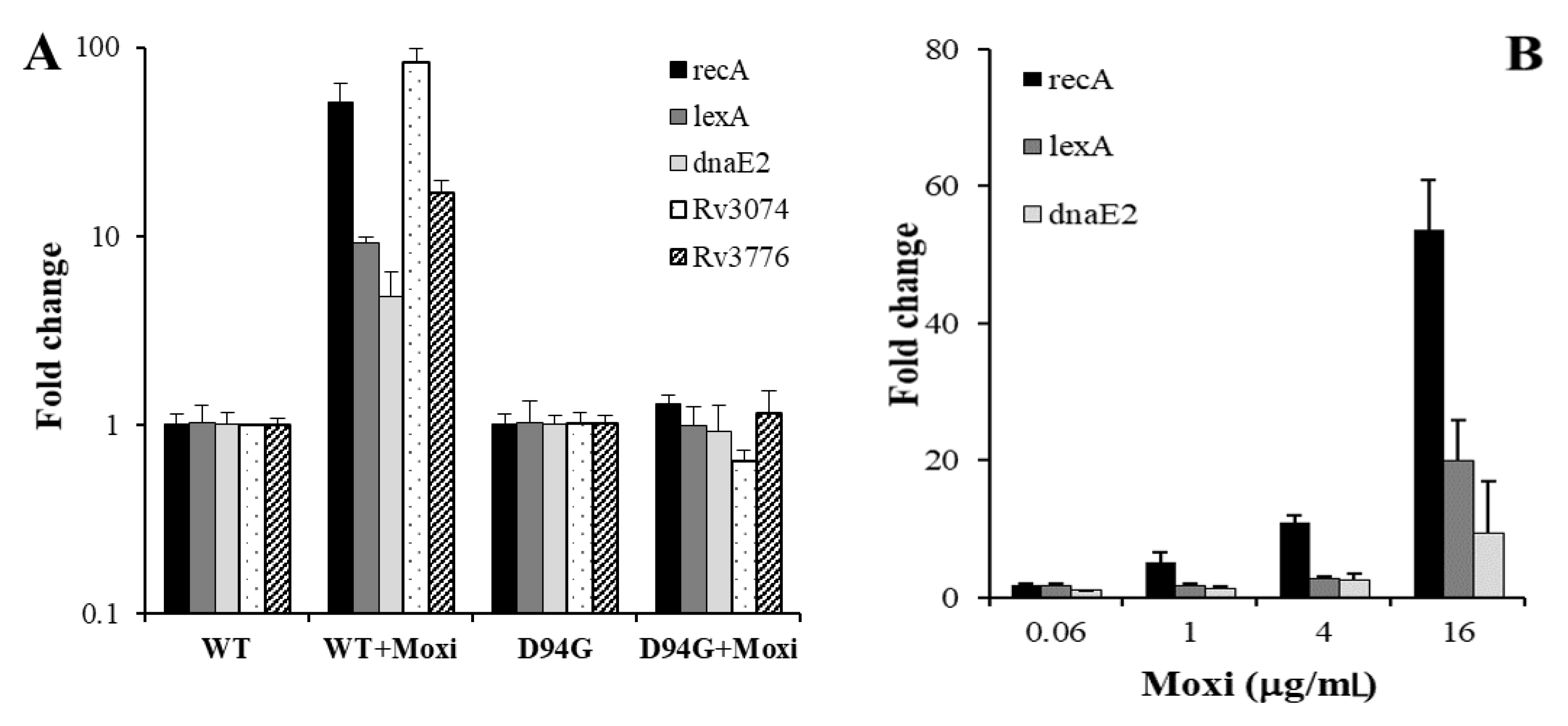
| Category | Rv No. | Gene | FC ± SD | p | F | R |
|---|---|---|---|---|---|---|
| SOS response | Rv1378c | - | 43.85 ± 8.93 | ** | ACGCCCACCGGGATGTACTA | GAGGGCGACACCGATTCTGG |
| Rv0336 | - | 6.25 ± 0.93 | ** | TGATCACCGCCGAACTGGTG | CCAGCGACACGTCAGATCCC | |
| Rv0515 | - | 6.25 ± 0.93 | ** | TGATCACCGCCGAACTGGTG | CCAGCGACACGTCAGATCCC | |
| Rv1000c | - | 8.09 ± 1.41 | ** | ACATCTACGGCGGCGAACTG | CCGTCGCGGTAGTAGCACAG | |
| Rv2719c | - | 40.58 ± 5.61 | ** | CCGCGGCGATTACTCTCTGG | GGACCGCCACGTCATACAGG | |
| Rv3395c | - | 70.31 ± 4.87 | ** | GGACGGTGGGAGTGCTGTC | ACCGATGCCACCATGCTCAG | |
| Rv0427c | xthA | 4.49 ± 0.18 | ** | ATCGCACTGATGGGCGACTG | AATTGCGCGTCGACAATGGC | |
| Rv3260c | whiB2 | 0.32 ± 0.02 | ** | GGAAGCCACCGACCAATG | CCAGGGCGTACTCCAGAC | |
| DNA repair | Rv0054 | ssb | 15.58 ± 3.70 | * | CGTCAGACCGGCGAATGGAA | TCGATGACGGTGCGCTTCTC |
| Rv1638 | uvrA | 2.55 ± 1.50 | - | CCCGAGATTCTGGCGGTGAC | GCGACCGGATCTCCTTGAGC | |
| Rv2594c | ruvC | 9.90 ± 1.23 | ** | ACGTGGTGTCGACGTGCATT | GACCATCGCGGTGACCTGAG | |
| Rv3585 | radA | 12.18 ± 1.41 | ** | CGGTATCGTTCCCGGTTCGG | CTGACCGGCGGATTCCTCAC | |
| Rv2821c | - | 0.85 ± 0.21 | - | AGCAGGCTGCCGATGATTCC | GAGCTTCGTGTCGCGGAAGA |
| Function | Rv No. | Gene | FC ± SD | p | F | R |
|---|---|---|---|---|---|---|
| SOS Response | Rv2737c | recA | 43.41 ± 5.02 | ** | GCGGTGGAATGAAGCAGGTC | GTTGGCAGCACTTCTCGGATC |
| Rv2720 | lexA | 6.09 ± 2.94 | ** | GCCGAGGAAGCCGTTGAAG | CGATCACCTTGAGCAGGAACAG | |
| Rv3370c | dnaE2 | 3.59 ± 0.17 | ** | GTTCTACTCGGCGTGGTTCAAG | CAGCGACTGCGGCGAATAG | |
| Rv3074 | - | 29.28 ± 8.38 | ** | CATTAGATGCCGAGTTATGTGG | GCCGAATGGTGACCGTAC | |
| Rv3776 | - | 22.19 ± 2.38 | ** | CTGCGGCTGCGGTAATTC | CGGATAGGCGTGGGTCAG | |
| Transcription | Rv0667 | rpoB | 0.91 ± 0.32 | - | TCGCCGACCTGGATGAGC | CGAAGTGTCGCGCACCTC |
| Rv2703 | sigA | 0.89 ± 0.26 | - | AGTCGGAGGCCCTGCGTCAA | GCCAGCCTCGATCCGCTTGG | |
| Rv2710 | sigB | 0.83 ± 0.22 | - | ATCCGCCAGGCCATCACC | TCCTCATCGGTGGCTTCGC | |
| Rv1221 | sigE | 0.79 ± 0.19 | - | GCCCAACCCCGAGCAGATC | CGTACCGTCCCGAGCTTCAC | |
| Rv3286c | sigF | 0.86 ± 0.12 | - | GTTCCTACCACACCTTGTCCATC | CCCAGGGTGTCTGTGATTGC | |
| Rv0757 | phoP | 3.29 ± 1.00 | - | CGGGATGGACGGCTTTGG | ATCTTGTCCTGTAGCGAGTCAC | |
| Early Genes | Rv3875 | esat-6 | 0.68 ± 0.1 | - | CGCAATCCAGGGAAATGTCACGTC | GTACGCCTCCGAACCGCTACC |
| Rv1886c | fbpB | 0.74 ± 0.08 | * | CTGTAGTCCTTCCGGGCCTGGT | CACCGCTCTGGAACTGAACCTTGA | |
| Dormancy | Rv2031c | acr | 1.47 ± 0.32 | - | CCGAGCGCACCGAGCAGAAG | GCCTTAATGTCGTCCTCGTCAGCA |
| Rv1738c | - | 0.56 ± 0.16 | - | TGCGGCGACCAGTCGGATCA | GCCAGGCCAACACCCACCAATT | |
| Replication | Rv0006 | gyrA | 1.07 ± 0.63 | - | TCGCCCAGGTCATCCAGATT | CAGCAGCAGGTCGTCGC |
| Porins | Rv0431 | - | 0.64 ± 0.37 | - | TTCTCGGCGTCGTCTTCC | TTCTGTGCCTGAGATGTTGTAG |
| Rv1352 | - | 1.1 ± 0.13 | - | CGATCACGCTCGCACCTG | GGACACCCTGCCGAACAC | |
| Rv1698 | - | 0.35 ± 0.18 | - | CGGCAAGTCGGTGGTCATC | CTCGGCGGAGTTGGCTTC | |
| Pumps | Rv1217c | - | 0.52 ± 0.16 | - | GGTTGTTATCGGCGGTGAC | CCAGGTTGAGCAGCATCTG |
| Rv1410c | - | 0.64 ± 0.11 | - | GTTCATCATCGGCTCGGTAGTG | CCAGCGTGATCGGCAATAGC | |
| Rv2846c | efpA | 1.15 ± 0.31 | - | CTTGACCACGCCTACACCTAC | GATCGCTTCCTTGACCTCCTG | |
| Rv3065 | mmr | 1.78 ± 0.18 | - | AAGCACGGAAGGGTTCACTC | TACGGCGACCAGCACAATG | |
| Toxins | Rv2583c | relA | 0.45 ± 0.18 | * | GCAGCAGTTCGTGGTGTCG | CTCGCGGGCCATCGCATC |
| Rv1246c | relE | 0.71 ± 0.49 | - | GAGCGACGACCATCCCTACC | CGGCTTGCCCAACCTATGC | |
| Rv2866 | relG | 0.66 ± 0.17 | - | CCGTGCGGTTCACCACAAC | TACAGCAGGCGGTACGTTCC | |
| Rv3358 | relK | 5.86 ± 1.10 | ** | AGGTGAGTTGTCGGGATACTGG | GCCTTCAGCATCGTGACTTCG | |
| Rv1102c | mazF3 | 0.68 ± 0.07 | - | GAATCAACCGTCAGTCGTCAGC | GCGAGCAGGTAGCCGATTTG | |
| Rv1495 | mazF4 | 0.49 ± 0.02 | - | GTTGCGTGGTCAGGTCTATCG | GGTTGCGGGCGTTGTTGG | |
| Rv1991c | mazF6 | 1.13 ± 0.20 | - | GGTGCTCGTAATCCAGTCAGATC | GTCGGTGAGGTCAGTCTTGTTG | |
| Rv2801c | mazF9 | 0.66 ± 0.32 | - | CGAGGTAGCGAAGCGAACAAC | CGGATAGACCTTGGCGATGTTG | |
| Persistence | Rv0251c | acr2 | 1 ± 0.55 | - | CGACGAGCACACGCAAGACG | CGGAACGAGCGGCGGAATG |
| Rv2517c | - | 2.64 ± 0.18 | ** | GCTACACCCGCTTCTACAACC | CCGCCGTTCCTTCTTGCTC | |
| Rv1152 | - | 0.37 ± 0.06 | * | GGACGCTTCGGCACTTTC | CGCATCGCATCGGACTTC | |
| Rv2497c | pdhA | 0.69 ± 0.45 | - | ACGCCTGGACGAGGACTC | ACGCACGGTGTGGTGAAC | |
| Rv3290c | lat | 3.83 ± 0.41 | ** | CGTGAAGTCCGTCGCTCTTG | CCGAGGAGGCAACGAATGTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iacobino, A.; Piccaro, G.; Pardini, M.; Fattorini, L.; Giannoni, F. Moxifloxacin Activates the SOS Response in Mycobacterium tuberculosis in a Dose- and Time-Dependent Manner. Microorganisms 2021, 9, 255. https://doi.org/10.3390/microorganisms9020255
Iacobino A, Piccaro G, Pardini M, Fattorini L, Giannoni F. Moxifloxacin Activates the SOS Response in Mycobacterium tuberculosis in a Dose- and Time-Dependent Manner. Microorganisms. 2021; 9(2):255. https://doi.org/10.3390/microorganisms9020255
Chicago/Turabian StyleIacobino, Angelo, Giovanni Piccaro, Manuela Pardini, Lanfranco Fattorini, and Federico Giannoni. 2021. "Moxifloxacin Activates the SOS Response in Mycobacterium tuberculosis in a Dose- and Time-Dependent Manner" Microorganisms 9, no. 2: 255. https://doi.org/10.3390/microorganisms9020255
APA StyleIacobino, A., Piccaro, G., Pardini, M., Fattorini, L., & Giannoni, F. (2021). Moxifloxacin Activates the SOS Response in Mycobacterium tuberculosis in a Dose- and Time-Dependent Manner. Microorganisms, 9(2), 255. https://doi.org/10.3390/microorganisms9020255






