Bacterial Dimethylsulfoniopropionate Biosynthesis in the East China Sea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Environmental Parameters
2.3. Enrichments for DMSP-Production
2.4. Bacterial Strain Isolation, Identification and Characterization
2.5. Seawater Incubation Experiments
2.6. DNA and RNA Extraction from Environmental Samples
2.7. Quantitative PCR
2.8. 16S rRNA Gene Amplicon Sequencing and Statistical Analysis
2.9. Metagenomic Sequencing and Analysis
2.10. Clone Library Construction and Sequencing
3. Results
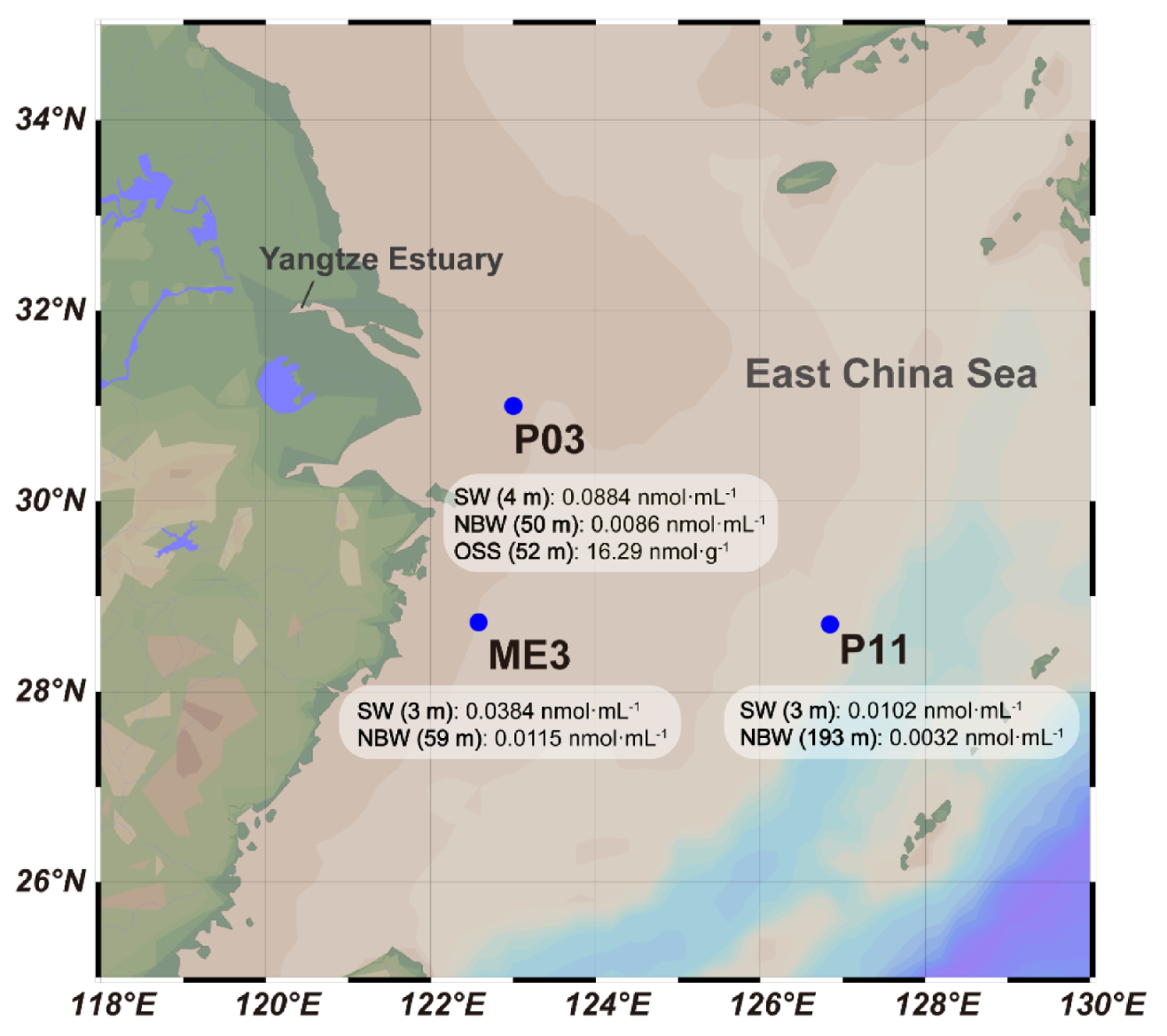
3.1. Sampling of the ECS and Quantification of DMSP
3.2. Enrichment Incubation Experiment for DMSP Producing Bacteria
3.3. qPCR Analysis of DMSP Biosynthesis Genes
3.4. Isolation and Characterization of DMSP-Producing Bacteria from the ECS
3.5. Seawater Incubation Experiment for DMSP Producing Isolates
3.6. Bacterial Communities and DMSP Producers Revealed by 16S rRNA Gene Amplicon Sequencing
3.7. Metagenomic Analysis of DMSP Cycling Genes in ECS Samples
3.8. DsyB and mmtN Diversity Revealed by Clone Library Construction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seymour, J.R.; Simó, R.; Ahmed, T.; Stocker, R. Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science 2010, 329, 342–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curson, A.R.J.; Todd, J.D.; Sullivan, M.J.; Johnston, A.W.B. Catabolism of dimethylsulphoniopropionate: Microorganisms, enzymes and genes. Nat. Rev. Microbiol. 2011, 9, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-H.; Liu, J.; Liu, J.; Yang, G.; Xue, C.X.; Curson, A.R.J.; Todd, J.D. Biogenic production of DMSP and its degradation to DMS—their roles in the global sulfur cycle. Sci. China Life Sci. 2019, 62, 1296–1319. [Google Scholar] [CrossRef] [PubMed]
- Sievert, S.; Kiene, R.; Schulz-Vogt, H. The Sulfur Cycle. Oceanography 2007, 20, 117–123. [Google Scholar] [CrossRef] [Green Version]
- Nevitt, G.A. The neuroecology of dimethyl sulfide: A global-climate regulator turned marine infochemical. Integr. Comp. Biol. 2011, 51, 819–825. [Google Scholar] [CrossRef] [Green Version]
- Stefels, J.; Steinke, M.; Turner, S.; Malin, G.; Belviso, S. Environmental constraints on the production and removal of the climatically active gas dimethylsulphide (DMS) and implications for ecosystem modelling. Biogeochemistry 2007, 245–275. [Google Scholar] [CrossRef] [Green Version]
- Vallina, S.M.; Simo, R. Strong relationship beween DMS and the solar radiation dose over the global surface ocean. Science 2007, 315, 506–508. [Google Scholar] [CrossRef]
- Quinn, P.K.; Bates, T.S. The case against climate regulation via oceanic phytoplankton sulphur emissions. Nature 2011, 480, 51–56. [Google Scholar] [CrossRef]
- Moran, M.A.; Reisch, C.R.; Kiene, R.P.; Whitman, W.B. Genomic insights into bacterial DMSP transformations. Ann. Rev. Mar. Sci. 2012, 4, 523–542. [Google Scholar] [CrossRef]
- Kiene, R.P.; Linn, L.J.; Bruton, J.A. New and important roles for DMSP in marine microbial communities. J. Sea Res. 2000, 43, 209–224. [Google Scholar] [CrossRef]
- Yoch, D.C. Dimethylsulfoniopropionate: Its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl. Environ. Microbiol. 2002, 68, 5804–5815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Alstyne, K.L.; Puglisi, M.P. DMSP in marine macroalgae and macroinvertebrates: Distribution, function, and ecological impacts. Aquat. Sci. 2007, 69, 394–402. [Google Scholar] [CrossRef]
- Raina, J.B.; Tapiolas, D.M.; Forêt, S.; Lutz, A.; Abrego, D.; Ceh, J.; Seneca, F.O.; Clode, P.L.; Bourne, D.G.; Willis, B.L.; et al. DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature 2013, 502, 677–680. [Google Scholar] [CrossRef] [Green Version]
- Curson, A.; Williams, B.; Pinchbeck, B.; Sims, L.; Bermejo Martínez, A.; Rivera, P.; Kumaresan, D.; Mercadé, E.; Spurgin, L.; Carrión, O.; et al. DSYB catalyses the key step of dimethylsulfoniopropionate biosynthesis in many phytoplankton. Nat. Microbiol. 2018, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Gage, D.A.; Rhodes, D.; Nolte, K.D.; Hicks, W.A.; Leustek, T.; Cooper, A.J.L.; Hanson, A.D. A new route for synthesis of dimethylsulphoniopropionate in marine algae. Nature 1997, 387, 891–894. [Google Scholar] [CrossRef]
- Hanson, A.D.; Rivoal, J.; Paquet, L.; Cage, D.A. Biosynthesis of 3-dimethylsulfoniopropionate in Wollastonia biflora (L.). Plant Physiol. 1994, 105, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Curson, A.R.J.; Liu, J.; Bermejo Martínez, A.; Green, R.T.; Chan, Y.; Carrión, O.; Williams, B.T.; Zhang, S.-H.; Yang, G.-P.; Bulman Page, P.C.; et al. Dimethylsulfoniopropionate biosynthesis in marine bacteria and identification of the key gene in this process. Nat. Microbiol. 2017, 2, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Williams, B.T.; Cowles, K.; Bermejo Martínez, A.; Curson, A.R.J.; Zheng, Y.; Liu, J.; Newton-Payne, S.; Hind, A.J.; Li, C.-Y.; Rivera, P.P.L.; et al. Bacteria are important dimethylsulfoniopropionate producers in coastal sediments. Nat. Microbiol. 2019, 4, 1815–1825. [Google Scholar] [CrossRef] [Green Version]
- Kageyama, H.; Tanaka, Y.; Shibata, A.; Waditee-Sirisattha, R.; Takabe, T. Dimethylsulfoniopropionate biosynthesis in a diatom Thalassiosira pseudonana: Identification of a gene encoding MTHB-methyltransferase. Arch. Biochem. Biophys. 2018, 645, 100–106. [Google Scholar] [CrossRef]
- Amin, S.A.; Hmelo, L.R.; Van Tol, H.M.; Durham, B.P.; Carlson, L.T.; Heal, K.R.; Morales, R.L.; Berthiaume, C.T.; Parker, M.S.; Djunaedi, B.; et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 2015, 522, 98–101. [Google Scholar] [CrossRef]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A.; et al. Structure and function of the global ocean microbiome. Science 2015, 348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiene, R.P.; Linn, L.J. The fate of dissolved dimethylsulfoniopropionate (DMSP) in seawater: Tracer studies using 35S-DMSP. Geochim. Cosmochim. Acta 2000, 17, 2797–2810. [Google Scholar] [CrossRef]
- Cui, Y.; Suzuki, S.; Omori, Y.; Wong, S.K.; Ijichi, M.; Kaneko, R.; Kameyama, S.; Tanimoto, H.; Hamasaki, K. Abundance and distribution of dimethylsulfoniopropionate degradation genes and the corresponding bacterial community structure at dimethyl sulfide hot spots in the tropical and subtropical Pacific Ocean. Appl. Environ. Microbiol. 2015, 81, 4184–4194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowinski, B.; Motard-Côté, J.; Landa, M.; Preston, C.M.; Scholin, C.A.; Birch, J.M.; Kiene, R.P.; Moran, M.A. Microdiversity and temporal dynamics of marine bacterial dimethylsulfoniopropionate genes. Environ. Microbiol. 2019, 21, 1687–1701. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, J.; Zhang, S.-H.; Liang, J.; Lin, H.; Song, D.; Yang, G.-P.; Todd, J.D.; Zhang, X.-H. Novel insights into bacterial dimethylsulfoniopropionate catabolism in the East China Sea. Front. Microbiol. 2018, 9, 3206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.H.; Yang, G.P.; Zhang, H.H.; Yang, J. Spatial variation of biogenic sulfur in the south Yellow Sea and the East China Sea during summer and its contribution to atmospheric sulfate aerosol. Sci. Total Environ. 2014, 488–489, 157–167. [Google Scholar] [CrossRef]
- DeLong, E.F. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 1992, 89, 5685–5689. [Google Scholar] [CrossRef] [Green Version]
- Lane, D.J.; Pace, B.; Olsen, G.J.; Stahl, D.A.; Sogin, M.L.; Pace, N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 1985, 82, 6955–6959. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Carrión, O.; Pratscher, J.; Curson, A.R.J.; Williams, B.T.; Rostant, W.G.; Murrell, J.C.; Todd, J.D. Methanethiol-dependent dimethylsulfide production in soil environments. ISME J. 2017, 11, 2379–2390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Magurran, A.E. Why diversity? In Ecological Diversity and Its Measurement; Springer: Dordrecht, The Netherlands, 1988; pp. 1–5. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Nikolenko, S.I.; Korobeynikov, A.I.; Alekseyev, M.A. BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genom. 2013, 14, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. MetaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef] [Green Version]
- Hyatt, D.; Chen, G.L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef] [Green Version]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Börnigen, D.; Morgan, X.C.; Huttenhower, C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 2013, 4, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Andreae, M.O.; Andreae, T.W.; Meyerdierks, D.; Thiel, C. Marine sulfur cycling and the atmospheric aerosol over the springtime North Atlantic. Chemosphere 2003, 52, 1321–1343. [Google Scholar] [CrossRef]
- Pinhassi, J.; Simó, R.; González, J.M.; Vila, M.; Alonso-Sáez, L.; Kiene, R.P.; Moran, M.A.; Pedrós-Alió, C. Dimethylsulfoniopropionate turnover is linked to the composition and dynamics of the bacterioplankton assemblage during a microcosm phytoplankton bloom. Appl. Environ. Microbiol. 2005, 71, 7650–7660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, G.P.; Jing, W.W.; Kang, Z.Q.; Zhang, H.H.; Song, G.S. Spatial variations of dimethylsulfide and dimethylsulfoniopropionate in the surface microlayer and in the subsurface waters of the South China Sea during springtime. Mar. Environ. Res. 2008, 65, 85–97. [Google Scholar] [CrossRef] [Green Version]
- Varaljay, V.A.; Gifford, S.M.; Wilson, S.T.; Sharma, S.; Karl, D.M.; Moran, M.A. Bacterial dimethylsulfoniopropionate degradation genes in the oligotrophic North Pacific subtropical gyre. Appl. Environ. Microbiol. 2012, 78, 2775–2782. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Kang, Y.C.; Fuhrman, J.A. Imperfect retention of natural bacterioplankton cells by glass fiber filters. Mar. Ecol. Prog. Ser. 1995, 119, 285–290. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Yang, G.P.; Kieber, D.J.; Motard-Coˆté, J.; Kiene, R.P. Assessment of DMSP turnover reveals a non-bioavailable pool of dissolved DMSP in coastal waters of the Gulf of Mexico. Environ. Chem. 2016, 13, 266–279. [Google Scholar] [CrossRef]
- Gonzalez, J.M.; Moran, M.A. Numerical dominance of a group of marine bacteria in the alpha-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 1997, 63, 4237–4242. [Google Scholar] [CrossRef] [Green Version]
- Biers, E.J.; Sun, S.; Howard, E.C. Prokaryotic genomes and diversity in surface ocean waters: Interrogating the global ocean sampling metagenome. Appl. Environ. Microbiol. 2009, 75, 2221–2229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, S.M.; Malin, G.; Liss, P.S.; Harbour, D.S.; Holligan, P.M. The seasonal variation of dimethyl sulfide and dimethylsulfoniopropionate concentrations in nearshore waters. Limnol. Oceanogr. 1988, 33, 364–375. [Google Scholar] [CrossRef]
- Vila-Costa, M.; Rinta-Kanto, J.M.; Sun, S.; Sharma, S.; Poretsky, R.; Moran, M.A. Transcriptomic analysis of a marine bacterial community enriched with dimethylsulfoniopropionate. ISME J. 2010, 4, 1410–1420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, D.; Zhang, Y.; Liu, J.; Zhong, H.; Zheng, Y.; Zhou, S.; Yu, M.; Todd, J.D.; Zhang, X.H. Metagenomic insights into the cycling of dimethylsulfoniopropionate and related molecules in the Eastern China Marginal Seas. Front. Microbiol. 2020, 11, 157. [Google Scholar] [CrossRef] [Green Version]
- Curson, A.R.J.; Sullivan, M.J.; Todd, J.D.; Johnston, A.W.B. DddY, a periplasmic dimethylsulfoniopropionate lyase found in taxonomically diverse species of Proteobacteria. ISME J. 2011, 5, 1191–1200. [Google Scholar] [CrossRef]
- Howard, E.C.; Sun, S.; Biers, E.J.; Moran, M.A. Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ. Microbiol. 2008, 10, 2397–2410. [Google Scholar] [CrossRef]
- Uchida, A.; Ooguri, T.; Ishida, T.; Kitaguchi, H.; Ishida, Y. Biosynthesis of dimethylsulfoniopropionate in Crypthecodinium cohnii (Dinophyceae). In Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds; Springer: Boston, MA, USA, 1996; pp. 97–107. [Google Scholar]
- Sun, H.; Zhang, Y.; Tan, S.; Zheng, Y.; Zhou, S.; Ma, Q.Y.; Yang, G.P.; Todd, J.D.; Zhang, X.H. DMSP-producing bacteria are more abundant in the surface microlayer than subsurface seawater of the East China Sea. Microb. Ecol. 2020, 80, 350–365. [Google Scholar] [CrossRef]
- Kiene, R.P. Turnover of dissolved DMSP in estuarine and shelf waters of the Northern Gulf of Mexico. In Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds; Springer: Boston, MA, USA, 1996; pp. 337–349. [Google Scholar]
- Zubkov, M.V.; Fuchs, B.M.; Archer, S.D.; Kiene, R.P.; Amann, R.; Burkill, P.H. Rapid turnover of dissolved DMS and DMSP by defined bacterioplankton communities in the stratified euphotic zone of the North Sea. Deep. Res. Part. II Top. Stud. Oceanogr. 2002, 49, 3017–3038. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Zhang, Y.; Liu, J.; Zhong, H.; Williams, B.T.; Zheng, Y.; Curson, A.R.J.; Sun, C.; Sun, H.; Song, D.; et al. Bacterial Dimethylsulfoniopropionate Biosynthesis in the East China Sea. Microorganisms 2021, 9, 657. https://doi.org/10.3390/microorganisms9030657
Liu J, Zhang Y, Liu J, Zhong H, Williams BT, Zheng Y, Curson ARJ, Sun C, Sun H, Song D, et al. Bacterial Dimethylsulfoniopropionate Biosynthesis in the East China Sea. Microorganisms. 2021; 9(3):657. https://doi.org/10.3390/microorganisms9030657
Chicago/Turabian StyleLiu, Ji, Yunhui Zhang, Jingli Liu, Haohui Zhong, Beth T. Williams, Yanfen Zheng, Andrew R. J. Curson, Chuang Sun, Hao Sun, Delei Song, and et al. 2021. "Bacterial Dimethylsulfoniopropionate Biosynthesis in the East China Sea" Microorganisms 9, no. 3: 657. https://doi.org/10.3390/microorganisms9030657





