Differential Labeling of Chemically Modified Peptides and Lipids among Cyanobacteria Planktothrix and Microcystis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Organisms
2.2. Growth Conditions
2.3. Cell Fixation and Labeling
2.4. Peptide Extraction
2.5. HPLC-MS Analysis
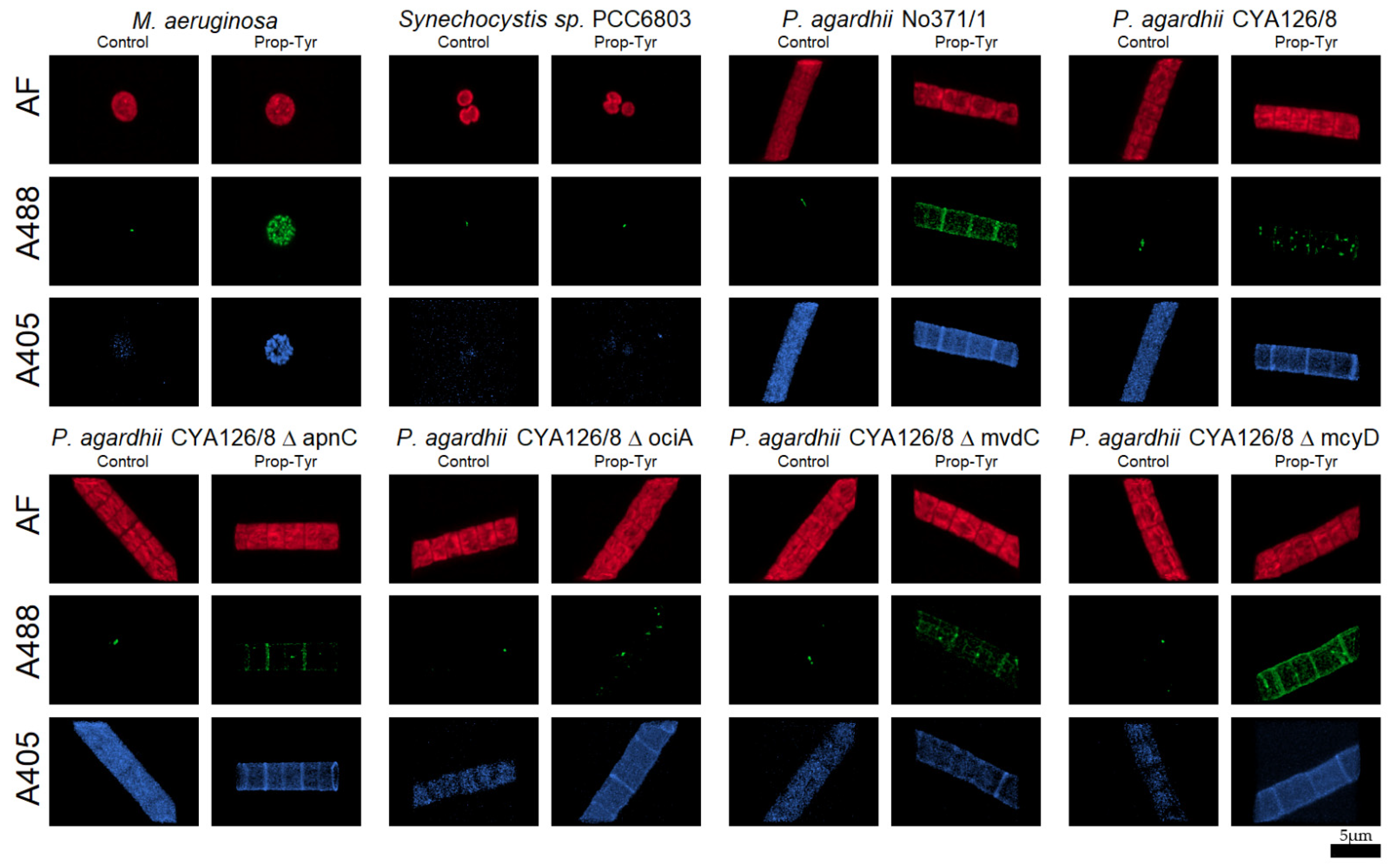
2.6. Microscopic Analysis
2.7. Colocalization Coefficient
3. Results
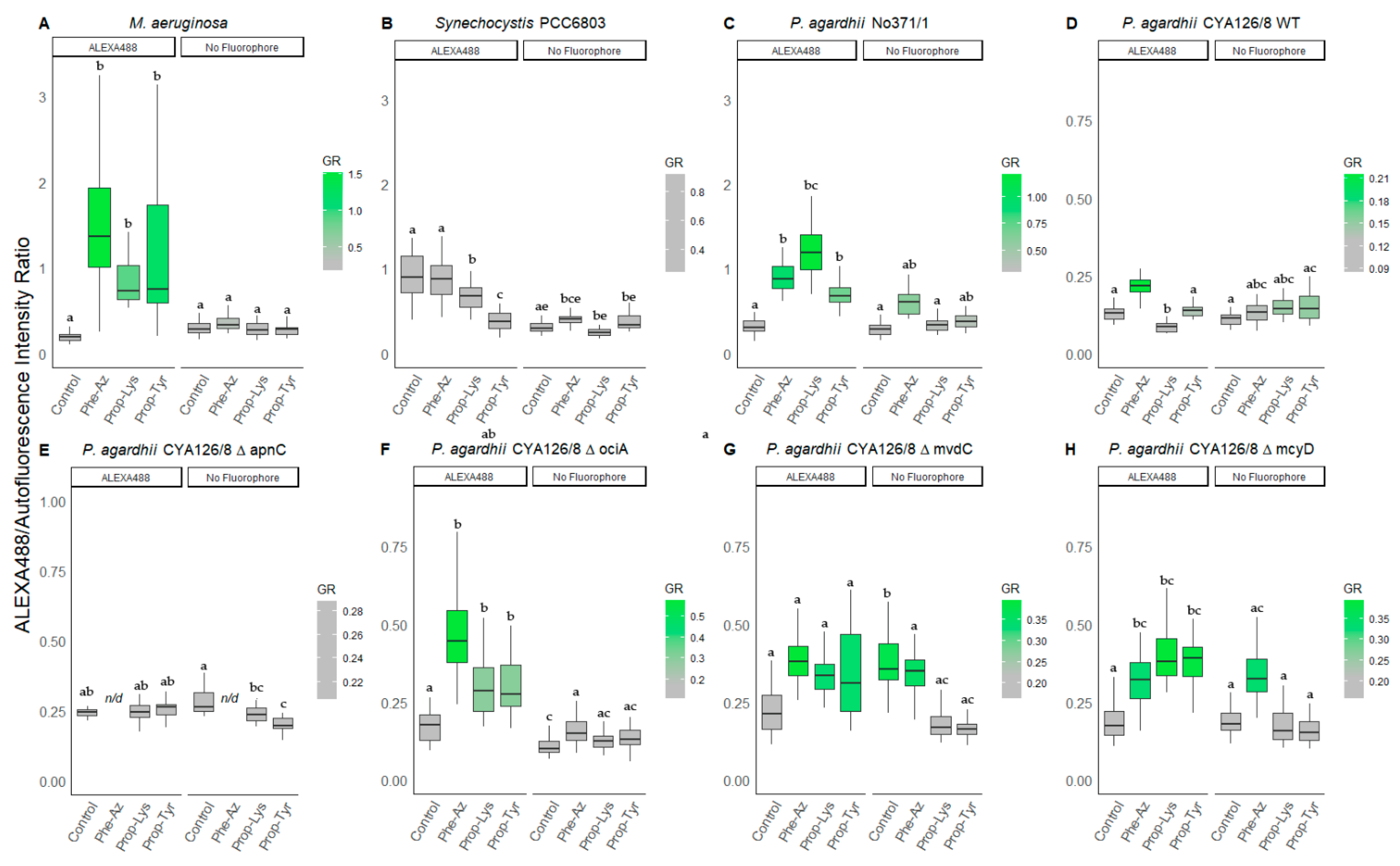
3.1. Growth Rate
3.2. Modified Peptides
3.2.1. Peptide Labeling Intensity
3.2.2. Peptide Intensity/Autofluorescence Ratio
3.3. Lipids
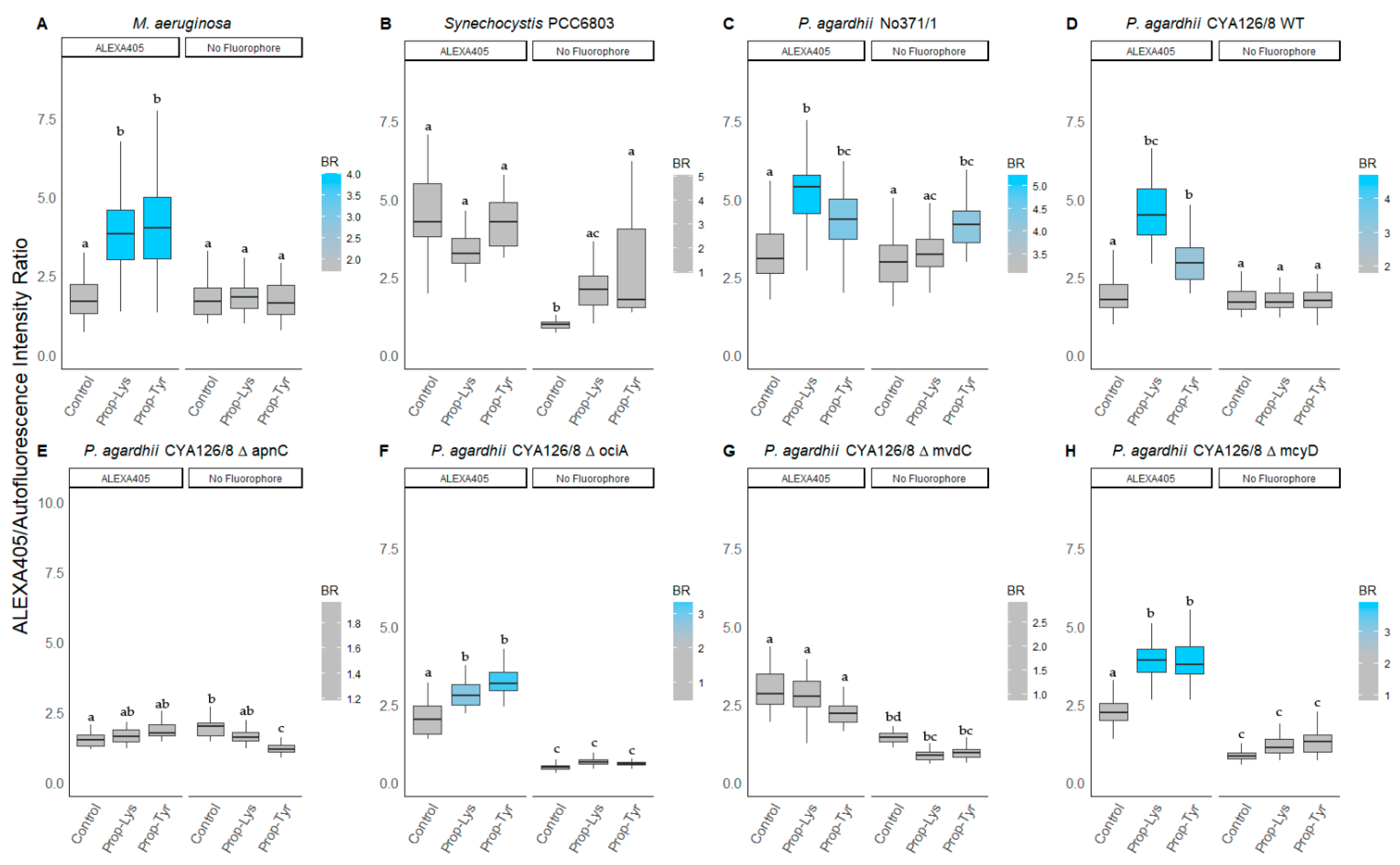
3.3.1. BODIPY Labeling
3.3.2. BODIPY/Autofluorescence Ratio
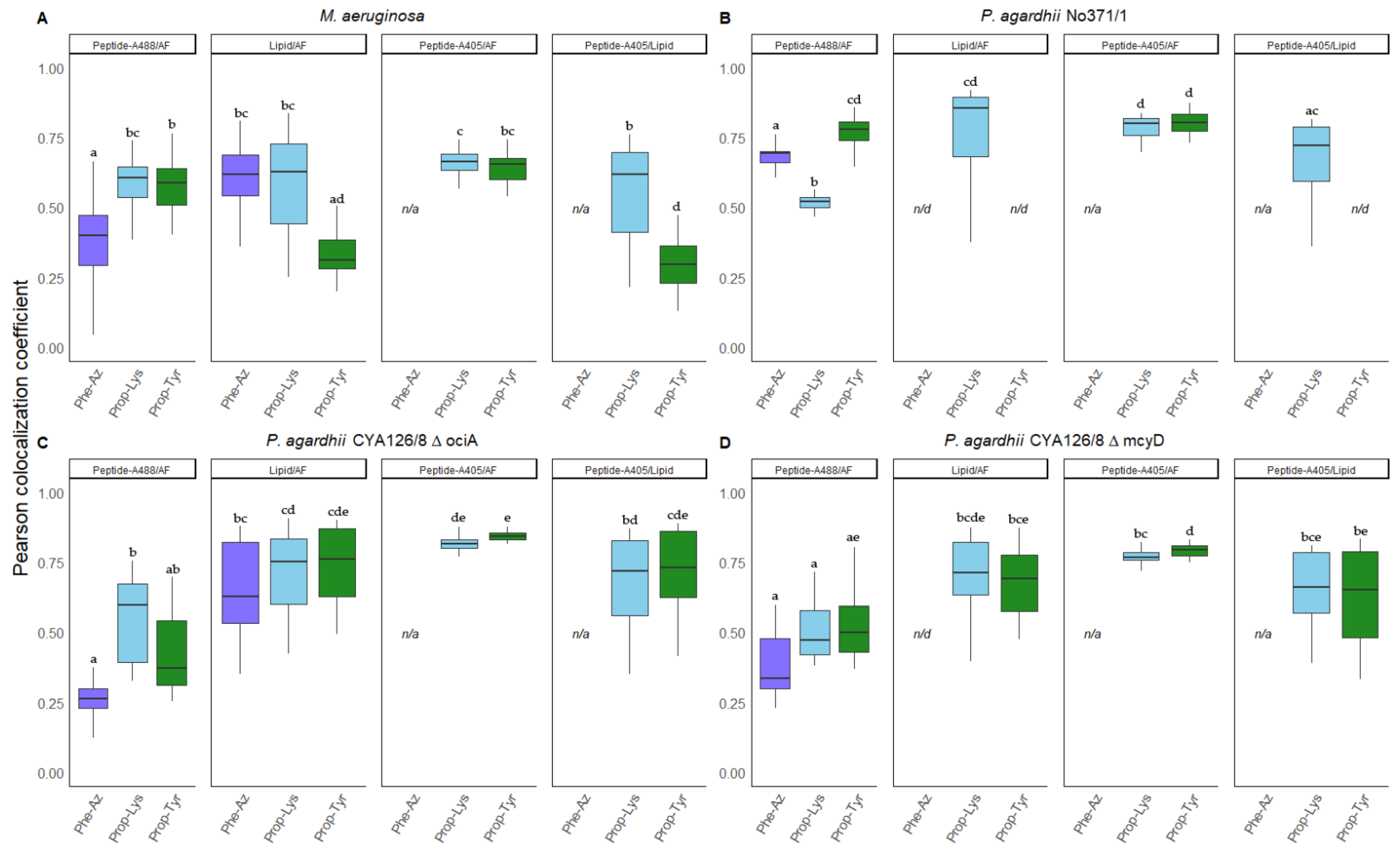
3.4. Colocalization of Peptides vs. Autofluorescence and Lipids
4. Discussions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Süssmuth, R.D.; Mainz, A. Nonribosomal Peptide Synthesis-Principles and Prospects. Angew. Chem. Int. Engl. 2017, 56, 3770–3821. [Google Scholar] [CrossRef] [PubMed]
- Entfellner, E.; Frei, M.; Christiansen, G.; Deng, L.; Blom, J.; Kurmayer, R. Evolution of Anabaenopeptin Peptide Structural Variability in the Cyanobacterium Planktothrix. Front. Microbiol. 2017, 8, 219. [Google Scholar] [CrossRef]
- Pearson, L.A.; Hisbergues, M.; Börner, T.; Dittmann, E.; Neilan, B.A. Inactivation of an ABC transporter gene, mcyH, results in loss of microcystin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Appl. Environ. Microbiol. 2004, 70, 6370–6378. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, G.; Philmus, B.; Hemscheidt, T.; Kurmayer, R. Genetic variation of adenylation domains of the anabaenopeptin synthesis operon and evolution of substrate promiscuity. J. Bacteriol. 2011, 193, 3822–3831. [Google Scholar] [CrossRef]
- Rohrlack, T.; Hyenstrand, P. Fate of intracellular microcystins in the cyanobacterium Microcystis aeruginosa (Chroococcales, Cyanophyceae). Phycologia 2007, 46, 277–283. [Google Scholar] [CrossRef]
- Jüttner, F.; Lüthi, H. Topology and enhanced toxicity of bound microcystins in Microcystis PCC 7806. Toxicon 2008, 51, 388–397. [Google Scholar] [CrossRef]
- Zilliges, Y.; Kehr, J.-C.; Meissner, S.; Ishida, K.; Mikkat, S.; Hagemann, M.; Kaplan, A.; Börner, T.; Dittmann, E. The cyanobacterial hepatotoxin microcystin binds to proteins and increases the fitness of Microcystis under oxidative stress conditions. PLoS ONE 2011, 6, e17615. [Google Scholar] [CrossRef]
- Meissner, S.; Fastner, J.; Dittmann, E. Microcystin production revisited: Conjugate formation makes a major contribution. Environ. Microbiol. 2013, 15, 1810–1820. [Google Scholar] [CrossRef] [PubMed]
- Spassova, M.; Mellor, I.R.; Petrov, A.G.; Beattie, K.A.; Codd, G.A.; Vais, H.; Usherwood, P.N. Pores formed in lipid bilayers and in native membranes by nodularin, a cyanobacterial toxin. Eur. Biophys. J. 1995, 24, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Van De Meene, A.M.L.; Hohmann-Marriott, M.F.; Vermaas, W.F.J.; Roberson, R.W. The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch. Microbiol. 2006, 184, 259–270. [Google Scholar] [CrossRef]
- Young, F.M.; Thomson, C.; Metcalf, J.S.; Lucocq, J.M.; Codd, G.A. Immunogold localisation of microcystins in cryosectioned cells of Microcystis. J. Struct. Biol. 2005, 151, 208–214. [Google Scholar] [CrossRef]
- Kurmayer, R.; Entfellner, E.; Weisse, T.; Offterdinger, M.; Rentmeister, A.; Deng, L. Chemically labeled toxins or bioactive peptides show a heterogeneous intracellular distribution and low spatial overlap with autofluorescence in bloom-forming cyanobacteria. Sci. Rep. 2020, 10, 2781. [Google Scholar] [CrossRef]
- Presolski, S.I.; Hong, V.P.; Finn, M.G. Copper-Catalyzed Azide-Alkyne Click Chemistry for Bioconjugation. Curr. Protoc. Chem. Biol. 2011, 3, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Jewett, J.C.; Bertozzi, C.R. Cu-free click cycloaddition reactions in chemical biology. Chem. Soc. Rev. 2010, 39, 1272–1279. [Google Scholar] [CrossRef]
- Mikalsen, B.; Boison, G.; Skulberg, O.M.; Fastner, J.; Davies, W.; Gabrielsen, T.M.; Rudi, K.; Jakobsen, K.S. Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. J. Bacteriol. 2003, 185, 2774–2785. [Google Scholar] [CrossRef]
- Kurmayer, R.; Dittmann, E.; Fastner, J.; Chorus, I. Diversity of microcystin genes within a population of the toxic cyanobacterium Microcystis spp. in Lake Wannsee (Berlin, Germany). Microb. Ecol. 2002, 43, 107–118. [Google Scholar] [CrossRef]
- Philmus, B.; Christiansen, G.; Yoshida, W.Y.; Hemscheidt, T.K. Post-translational modification in microviridin biosynthesis. Chembiochem 2008, 9, 3066–3073. [Google Scholar] [CrossRef]
- Kurmayer, R.; Christiansen, G. The Genetic Basis of Toxin Production in Cyanobacteria. Freshw. Rev. 2009, 2, 31–50. [Google Scholar] [CrossRef]
- Ishida, K.; Christiansen, G.; Yoshida, W.Y.; Kurmayer, R.; Welker, M.; Valls, N.; Bonjoch, J.; Hertweck, C.; Börner, T.; Hemscheidt, T.; et al. Biosynthesis and structure of aeruginoside 126A and 126B, cyanobacterial peptide glycosides bearing a 2-carboxy-6-hydroxyoctahydroindole moiety. Chem. Biol. 2007, 14, 565–576. [Google Scholar] [CrossRef]
- Christiansen, G.; Fastner, J.; Erhard, M.; Börner, T.; Dittmann, E. Microcystin biosynthesis in Planktothrix: Genes, evolution, and manipulation. J. Bacteriol. 2003, 185, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Rippka, R. Recongnition and Identification of Cyanobacteria. Methods Enzymol. 1988, 167, 3–27. [Google Scholar]
- Christiansen, G.; Dittmann, E.; Via Ordorika, L.; Rippka, R.; Herdman, M.; Börner, T. Nonribosomal peptide synthetase genes occur in most cyanobacterial genera as evidenced by their distribution in axenic strains of the PCC. Arch. Microbiol. 2001, 176, 452–458. [Google Scholar] [CrossRef]
- Kohl, J.-G.; Nicklisch, A. Ökophysiologie der Algen: Wachstum u. Ressourcennutzung; Akad.-Verl.: Berlin, Germany, 1988; ISBN 9783055003158. [Google Scholar]
- Cooper, M.S.; D’Amico, L.A.; Henry, C.A. Chapter 11 Confocal Microscopic Analysis of Morphogenetic Movements; Elsevier: Amsterdam, The Netherlands, 1998; pp. 179–204. ISBN 9780125441612. [Google Scholar]
- Brennan, L.; Blanco Fernández, A.; Mostaert, A.S.; Owende, P. Enhancement of BODIPY505/515 lipid fluorescence method for applications in biofuel-directed microalgae production. J. Microbiol. Methods 2012, 90, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Peramuna, A.; Summers, M.L. Composition and occurrence of lipid droplets in the cyanobacterium Nostoc punctiforme. Arch. Microbiol. 2014, 196, 881–890. [Google Scholar] [CrossRef]
- Kosol, S.; Schmidt, J.; Kurmayer, R. Variation in peptide net production and growth among strains of the toxic cyanobacterium Planktothrix spp. Eur. J. Phycol. 2009, 44, 49–62. [Google Scholar] [CrossRef]
- Costes, S.V.; Daelemans, D.; Cho, E.H.; Dobbin, Z.; Pavlakis, G.; Lockett, S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 2004, 86. [Google Scholar] [CrossRef]
- Dunn, K.W.; Kamocka, M.M.; McDonald, J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011, 300, C723–C742. [Google Scholar] [CrossRef] [PubMed]
- Fastner, J.; Erhard, M.; Carmichael, W.W.; Sun, F.; Chorus, I. Characterization and diversity of microcystins in natural blooms and strains of the genera Microcystis and Planktothrix from German freshwaters. Arch. Hydrobiol. 1999, 145, 147–163. [Google Scholar] [CrossRef]
- Welker, M.; Fastner, J.; Erhard, M.; Döhren, H. von. Applications of MALDI-TOF MS analysis in cyanotoxin research. Environ. Toxicol. 2002, 17, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Spoof, L.; Błaszczyk, A.; Meriluoto, J.; Cegłowska, M.; Mazur-Marzec, H. Structures and Activity of New Anabaenopeptins Produced by Baltic Sea Cyanobacteria. Mar. Drugs 2015, 14, 8. [Google Scholar] [CrossRef]
- Okumura, H.S.; Philmus, B.; Portmann, C.; Hemscheidt, T.K. Homotyrosine-containing cyanopeptolins 880 and 960 and anabaenopeptins 908 and 915 from Planktothrix agardhii CYA 126/8. J. Nat. Prod. 2009, 72, 172–176. [Google Scholar] [CrossRef]
- Lichstein, H.C.; Soule, M.H. Studies of the effect of sodium azide on microbic growth and respiration I. The action of sodium azide on microbic growth. J. Bacteriol. 1943, 239–251. [Google Scholar]
- Rozycki, M.; Bartha, R. Problems associated with the use of azide as an inhibitor of microbial activity in soil. Appl. Environ. Microbiol. 1981, 41, 833–836. [Google Scholar] [CrossRef] [PubMed]
- Cabrol, L.; Quéméneur, M.; Misson, B. Inhibitory effects of sodium azide on microbial growth in experimental resuspension of marine sediment. J. Microbiol. Methods 2017, 133, 62–65. [Google Scholar] [CrossRef] [PubMed]
- Zotina, T.; Köster, O.; Jüttner, F. Photoheterotrophy and light-dependent uptake of organic and organic nitrogenous compounds by Planktothrix rubescens under low irradiance. Freshw. Biol. 2003, 48, 1859–1872. [Google Scholar] [CrossRef]
- Young, F.M.; Morrison, L.F.; James, J.; Codd, G.A. Quantification and localization of microcystins in colonies of a laboratory strain of Microcystis (Cyanobacteria) using immunological methods. Eur. J. Phycol. 2008, 43, 217–225. [Google Scholar] [CrossRef][Green Version]
- Gerbersdorf, S.U. An advanced technique for immuno-labelling of microcystins in cryosectioned cells of Microcystis aeruginosa PCC 7806 (cyanobacteria): Implementations of an experiment with varying light scenarios and culture densities. Toxicon 2006, 47, 218–228. [Google Scholar] [CrossRef]
- Mandakovic, D.; Trigo, C.; Andrade, D.; Riquelme, B.; Gómez-Lillo, G.; Soto-Liebe, K.; Díez, B.; Vásquez, M. CyDiv, a Conserved and Novel Filamentous Cyanobacterial Cell Division Protein Involved in Septum Localization. Front. Microbiol. 2016, 7, 94. [Google Scholar] [CrossRef] [PubMed]


| Peptide | Original Mass [M+H]+ | Exchange AA | Non-Natural AA | Predicted Mass [M+H]+ | Detected Mass [M+H]+ | Fragmentation MSn [M+H]+ | Proportion of Modified MC or AP (%) | Elution Time (min) |
|---|---|---|---|---|---|---|---|---|
| M. aeruginosa | ||||||||
| D-Asp-MC-YR | 1031.5 | 181.4 (Tyr) | 206.1 (Phe-Az) | 1056.4 | 1056.5 | nd 1 | 1 | 38.1 |
| MC-YR | 1045.5 | “ | “ | 1070.4 | 1070.5 | 599.3 2 | 2 | 38.5 |
| D-Asp-MC-YR | 1031.5 | “ | 228.3 (Prop-Lys) | 1078.5 | 1078.4 | nd 1 | <1 | 32.6 |
| MC-YR | 1045.5 | “ | “ | 1092.6 | 1092.5 | 599.3 2 | 2 | 33.7 |
| D-Asp-MC-YR | 1031.5 | “ | 219.2 (Prop-Tyr) | 1069.4 | 1069.5 | 599.3 2, 135.0 3 | 2 | 37.4 |
| MC-YR | 1045.5 | “ | “ | 1083.5 | 1083.5 | 949.4 4, 599.3 2, 135.0 3 | 45 | 38.1 |
| P. agardhii No371/1 | ||||||||
| AP-A | 844.4 | 181.4 (Tyr) | 206.1 (Phe-Az) | 869.3 (843.3 reduced) | 843.3 | 637.4 5, 619.4 6, 534.4 7, 460.3 8, 387.3 9 | 14 | 16.7 |
| “ | “ | “ | 228.3 (Prop-Lys) | 891.5 | 891.5 | 637.4 5, 619.4 6, 534.4 7, 460.3 8, 387.3 9 | 81 | 25.1 |
| “ | “ | “ | 219.2 (Prop-Tyr) | 882.4 | 882.4 | nd 1 | 4 | 31.9 |
| P. agardhii CYA126/8 WT and ΔociA, ΔmvdC, ΔmcyD mutants | ||||||||
| AP-908 | 909.2 | 174.2 (Arg) | 206.1 (Phe-Az) | 941.1 (915.1 reduced) | 915.1 | 709.2 10, 691.2 11, 387.2 9, 500.2 12, 277.1 13 | 3–5 | 18.5 |
| AP-915 | 916.2 | 181.4 (Tyr) | 219.2 (Prop-Tyr) | 954.2 | 954.2 | 709.2 10, 691.2 11, 387.1 9, 500.1 12, 277.1 13 | 3 | 33.5 |
| ALEXA488 1 | No Fluorophore 1 | ||||||
|---|---|---|---|---|---|---|---|
| n | Phe-Az | Prop-Lys | Prop-Tyr | Phe-Az | Prop-Lys | Prop-Tyr | |
| M. aeruginosa Hofbauer | 43 | 4.6 ± 2.2 a | 4.4 ± 3.7 a | 5.3 ± 2.6 a | 1.2 ± 0.3 b | 1.0 ± 0.3 b | 1.0 ± 0.3 b |
| 1.1–12.5 | 1.6–19.4 | 1.1–13.0 | 0.6–1.7 | 0.5–1.6 | 0.5–1.6 | ||
| Synechocystis PCC6803 | 20 | 1.0 ± 0.8 ab | 0.5 ± 0.6 a | 0.3 ± 0.1 a | 1.5 ± 0.6 b | 2.1 ± 0.8 bc | 2.4 ± 1.8 bc |
| 0.3–3.2 | 0.2–2.7 | 0.1–0.4 | 0.8–2.6 | 0.8–4.0 | 0.9–6.9 | ||
| P. agardhii No371/1 | 16 | 1.3 ± 0.6 a | 4.0 ± 1.2 b | 1.9 ± 0.4 c | 0.6 ± 0.7 a | 0.8 ± 0.2 a | 0.9 ± 0.2 a |
| 0.5–3.3 | 2.1–7.5 | 1.3–2.7 | 0.1–2.2 | 0.5–1.2 | 0.6–1.2 | ||
| P. agardhii CYA126/8 WT | 38 | 2.0 ± 0.5 b | 1.3 ± 0.3 ac | 1.6 ± 0.2 ad | 1.2 ± 0.2 a | 1.2 ± 0.3 a | 1.4 ± 0.5 a |
| 0.5–2.5 | 0.8–1.9 | 0.8–1.9 | 0.8–1.7 | 0.8–1.7 | 0.8–2.3 | ||
| P. agardhii CYA126/8 ΔapnC | 20 | n/d | 1.0 ± 0.1a | 1.3 ± 0.4 b | n/d | 0.9 ± 0.2 ac | 0.7 ± 0.2 c |
| 0.8–1.3 | 1.0–2.6 | 0.7–1.3 | 0.5–1.1 | ||||
| P. agardhii CYA126/8 ΔociA | 38 | 1.8 ± 0.5 a | 2.5 ± 0.7 a | 2.4 ± 0.6 a | 1.0 ± 0.2 b | 1.1 ± 0.5 b | 1.0 ± 0.3 b |
| 1.0–3.1 | 1.5–3.6 | 1.4–3.6 | 0.6–1.3 | 0.6–2.5 | 0.2–2.0 | ||
| P. agardhii CYA126/8 ΔmvdC | 40 | 0.9 ± 0.3 a | 1.7 ± 0.3 ab | 1.5 ± 0.7 a | 0.6 ± 0.2 c | 0.6 ± 0.2 c | 0.7 ± 0.1 c |
| 0.6–1.9 | 1.2–2.4 | 0.6–2.5 | 0.3–0.9 | 0.4–1.1 | 0.5–0.8 | ||
| P. agardhii CYA126/8 ΔmcyD | 39 | 0.9 ± 0.3 b | 1.7 ± 0.3 a | 1.6 ± 0.3 a | 1.0 ± 0.3 b | 0.9 ± 0.3 b | 0.9 ± 0.2 b |
| 0.5–1.9 | 1.3–2.3 | 1.0–2.2 | 0.6–1.7 | 0.5–1.5 | 0.6–1.5 | ||
| ALEXA405 1 | No Fluorophore 1 | ||||
|---|---|---|---|---|---|
| (n) | Prop-Lys | Prop-Tyr | Prop-Lys | Prop-Tyr | |
| M. aeruginosa Hofbauer | 50 | 1.9 ± 0.6 a | 2.4 ± 0.9 a | 1.1 ± 0.4 bc | 1.0 ± 0.4 b |
| 0.8–3.5 | 0.5–3.7 | 0.4–2.1 | 0.3–1.9 | ||
| Synechocystis PCC6803 | 14 | 0.9 ± 0.6 a | 0.9 ± 0.3 a | 5.5 ± 2.4 b | 5.6 ± 5.3 b |
| 0.3–2.1 | 0.4–1.5 | 2.1–9.7 | 1.6–19.8 | ||
| P. agardhii No371/1 | 39 | 1.2 ± 0.2 a | 0.9 ± 0.3 a | 0.8 ± 0.2 ab | 1.0 ± 0.2 a |
| 0.7–1.7 | 0.4–1.5 | 0.5–1.1 | 0.6–1.4 | ||
| P. agardhii CYA126/8 WT | 38 | 2.3 ± 0.5 c | 1.6 ± 0.3 a | 0.9 ± 0.2 b | 1.0 ± 0.2 b |
| 1.3–3.6 | 0.9–2.2 | 0.6–1.5 | 0.7–1.5 | ||
| P. agardhii CYA126/8 ΔapnC | 20 | 1.0 ± 0.2 a | 1.4 ± 0.2 b | 0.9 ± 0.1 a | 0.6 ± 0.1 d |
| 0.7–1.3 | 1.0–1.9 | 0.8–1.1 | 0.5–0.8 | ||
| P. agardhii CYA126/8 ΔociA | 39 | 1.4 ± 0.3 ac | 1.6 ± 0.3 a | 0.7 ± 0.2 bc | 1.0 ± 0.3 b |
| 0.9–2.0 | 1.2–2.4 | 0.7–2.2 | 0.4–1.7 | ||
| P. agardhii CYA126/8 ΔmvdC | 40 | 1.1 ± 0.3 a | 1.0 ± 0.2 a | 0.8 ± 0.2 c | 1.0 ± 0.2 a |
| 0.6–1.7 | 0.6–1.4 | 0.5–1.3 | 0.8–1.3 | ||
| P. agardhii CYA126/8 ΔmcyD | 38 | 1.5 ± 0.2 a | 1.3 ± 0.3 a | 1.2 ± 0.3 abd | 1.5 ± 0.3 ab |
| 1.0–1.9 | 0.8–1.8 | 0.9–2.0 | 0.8–2.2 | ||
| BODIPY 505/515 | No Fluorophore | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| (n) | Phe-Az | Prop-Lys | Prop-Tyr | Control | Phe-Az | Prop-Lys | Prop-Tyr | Control | |
| M. aeruginosa Hofbauer | 50 | 2.9 ± 0.6 b | 2.0 ± 0.8 a | 3.0 ± 1.1 b | 1.9 ± 1.0 a | 1.0 ± 0.3 c | 1.0 ± 0.5 c | 1.0 ± 0.3 c | 1.0 ± 0.4 c |
| 1.8–4.7 | 0.8–3.6 | 1.5–5.3 | 0.9–5.0 | 0.5–1.8 | 0.4–2.5 | 0.5–1.7 | 0.6–1.8 | ||
| Synechocystis PCC6803 | 14 | 3.7 ± 1.2 ab | 5.2 ± 3.5 b | 3.3 ± 3.1 a | 1.3 ± 0.6 ac | 1.0 ± 0.4 c | 1.0 ± 0.4 c | 1.0 ± 0.8 c | 1.0 ± 0.6 c |
| 2.1–6.8 | 2.7–13.6 | 1.2–10.5 | 0.6–2.5 | 0.5–1.7 | 0.4–1.9 | 0.4–2.9 | 0.5–2.9 | ||
| P. agardhii No371/1 | 24 | 1.4 ± 1.3 b | 1.7 ± 0.6 a | 1.2 ± 0.6 b | 1.6 ± 0.3 a | 1.0 ± 1.1 b | 1.0 ± 0.2 b | 1.0 ± 0.2 b | 1.0 ± 0.4 b |
| 0.2–4.7 | 0.9–3.4 | 0.3–2.7 | 1.2–2.4 | 0.2–3.6 | 0.6–1.4 | 0.6–1.3 | 0.4–2.0 | ||
| P. agardhii CYA126/8 WT | 38 | 0.9 ± 0.2 b | 0.9 ± 0.2 b | 0.9 ± 0.2 b | 1.3 ± 0.2 a | 1.0 ± 0.2 b | 1.0 ± 0.2 b | 1.0 ± 0.3 b | 1.0 ± 0.3 b |
| 0.5–1.2 | 0.4–1.2 | 0.6–1.1 | 0.7–1.7 | 0.7–1.4 | 0.6–1.4 | 0.6–1.7 | 0.2–1.8 | ||
| P. agardhii CYA126/8 ΔapnC | 40 | n/d | 1.3 ± 0.3 ab | 1.1 ± 0.2 a | 1.0 ± 0.2 b | n/d | 1.0 ± 0.2 a | 1.0 ± 0.3 a | 1.0 ± 0.2 a |
| 0.6–2.0 | 0.6–1.5 | 0.6–1.3 | 0.7–1.5 | 0.3–1.5 | 0.7–1.3 | ||||
| P. agardhii CYA126/8 ΔociA | 39 | 2.4 ± 1.1 a | 2.1 ± 0.3 a | 2.6 ± 0.5 a | 1.9 ± 0.5 a | 1.0 ± 0.2 b | 1.0 ± 0.4 b | 1.0 ± 0.3 b | 1.0 ± 0.3 b |
| 1.1–5.5 | 1.6–2.9 | 2.0–4.2 | 1.0–2.8 | 0.6–1.1 | 0.5–2.2 | 0.2–2.0 | 0.6–1.7 | ||
| P. agardhii CYA126/8 ΔmvdC | 40 | 2.6 ± 1.5 ab | 2.7 ± 0.7 a | 2.2 ± 0.5 a | 1.3 ± 0.6 b | 1.0 ± 0.3 b | 1.0 ± 0.3 b | 1.0 ± 0.2 b | 1.0 ± 0.2 b |
| 0.5–5.4 | 1.5–4.5 | 1.5–3.2 | 0.7–3.5 | 0.6–1.5 | 0.7–1.8 | 0.7–1.3 | 0.4–1.4 | ||
| P. agardhii CYA126/8 ΔmcyD | 6 | 0.9 ± 0.1 b | 2.6 ± 0.4 a | 3.5 ± 0.8 a | 2.1 ± 0.5 a | 1.0 ± 0.4 b | 1.0 ± 0.4 b | 1.0 ± 0.3 b | 1.0 ± 0.2 b |
| 0.8–1.1 | 1.5–3.4 | 1.6–4.5 | 1.3–3.6 | 0.7–1.9 | 0.6–1.8 | 0.7–1.7 | 0.6–1.5 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morón-Asensio, R.; Schuler, D.; Wiedlroither, A.; Offterdinger, M.; Kurmayer, R. Differential Labeling of Chemically Modified Peptides and Lipids among Cyanobacteria Planktothrix and Microcystis. Microorganisms 2021, 9, 1578. https://doi.org/10.3390/microorganisms9081578
Morón-Asensio R, Schuler D, Wiedlroither A, Offterdinger M, Kurmayer R. Differential Labeling of Chemically Modified Peptides and Lipids among Cyanobacteria Planktothrix and Microcystis. Microorganisms. 2021; 9(8):1578. https://doi.org/10.3390/microorganisms9081578
Chicago/Turabian StyleMorón-Asensio, Rubén, David Schuler, Anneliese Wiedlroither, Martin Offterdinger, and Rainer Kurmayer. 2021. "Differential Labeling of Chemically Modified Peptides and Lipids among Cyanobacteria Planktothrix and Microcystis" Microorganisms 9, no. 8: 1578. https://doi.org/10.3390/microorganisms9081578
APA StyleMorón-Asensio, R., Schuler, D., Wiedlroither, A., Offterdinger, M., & Kurmayer, R. (2021). Differential Labeling of Chemically Modified Peptides and Lipids among Cyanobacteria Planktothrix and Microcystis. Microorganisms, 9(8), 1578. https://doi.org/10.3390/microorganisms9081578






