Microbial Mediated Synthesis of Silver Nanoparticles by Lactobacillus Plantarum TA4 and its Antibacterial and Antioxidant Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Bacterial Culture and Biomass Extraction
2.3. Determination of Maximum Tolerable Concentration (MTC) against Ag+
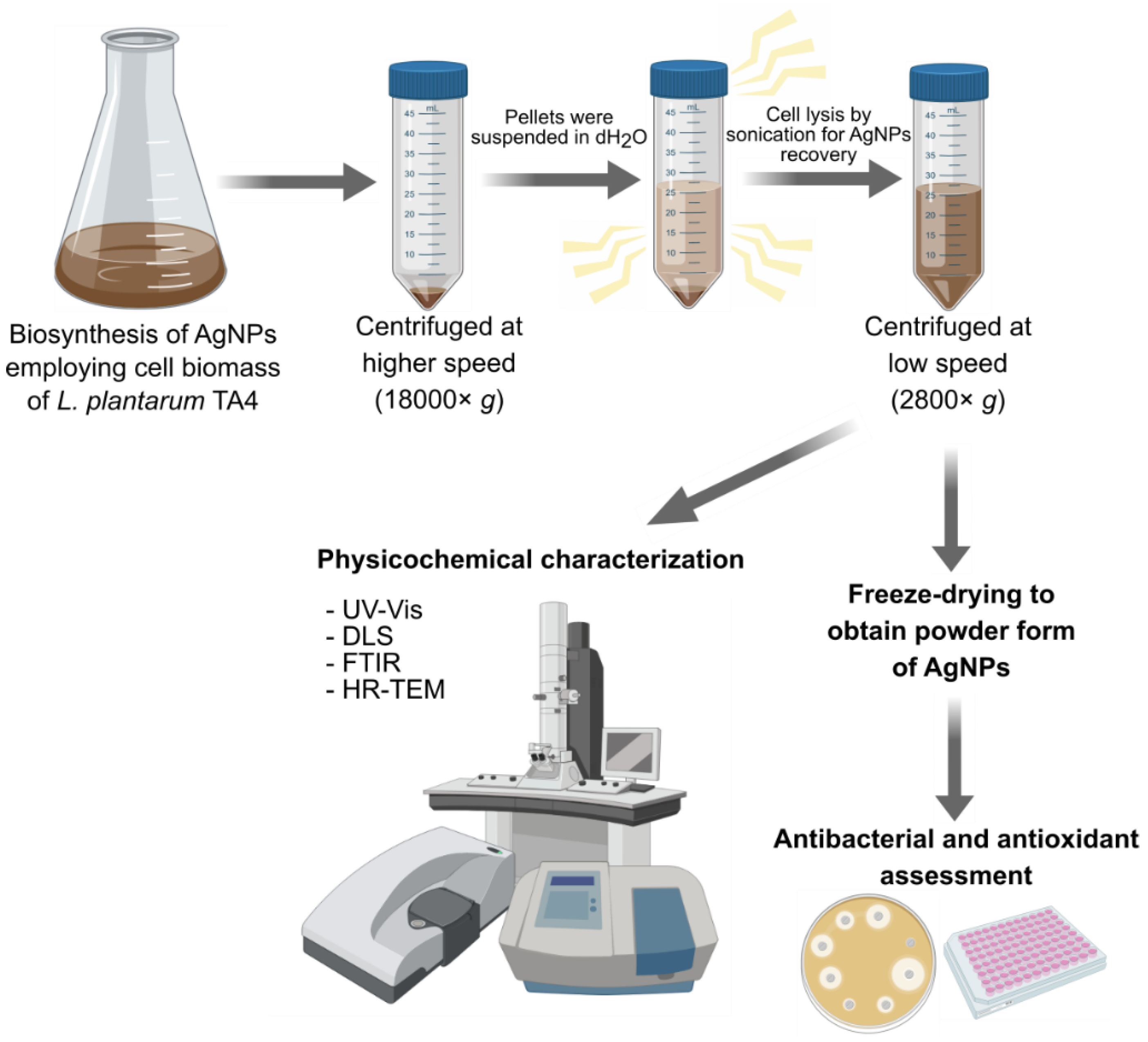
2.4. Biosynthesis of Silver Nanoparticles (AgNPs) Using L. plantarum TA4
2.5. Characterization of Biosynthesized AgNPs
2.6. Scanning Electron Microscope (SEM) and Energy Dispersive X-ray (EDX) of L. plantarum TA4 Exposed to Ag+
2.7. Antibacterial Assay of Biosynthesized AgNPs
2.8. Antioxidant Activity of Biosynthesized AgNPs
2.9. Statistical Analysis
3. Results and Discussion
3.1. Maximum Tolerable Concentration (MTC) of L. plantarum TA4 against Ag+
3.2. Biosynthesis of AgNPs Using L. plantarum TA4 Biomass
3.3. Dynamic Light Scattering (DLS) Analysis
3.4. FTIR Analysis
3.5. SEM-EDX and HR-TEM Analysis
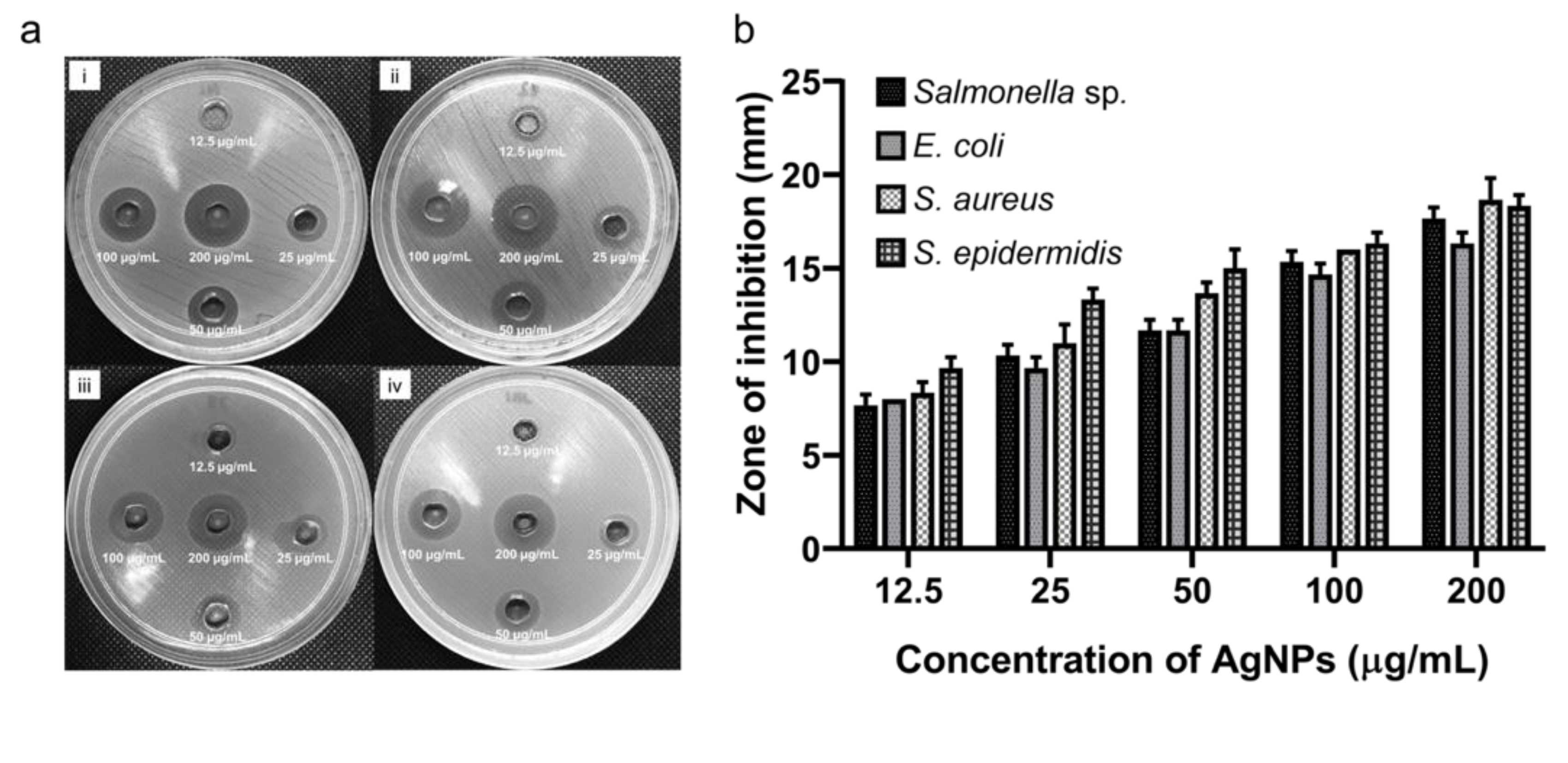
3.6. Antibacterial Activity of Biosynthesized AgNPs
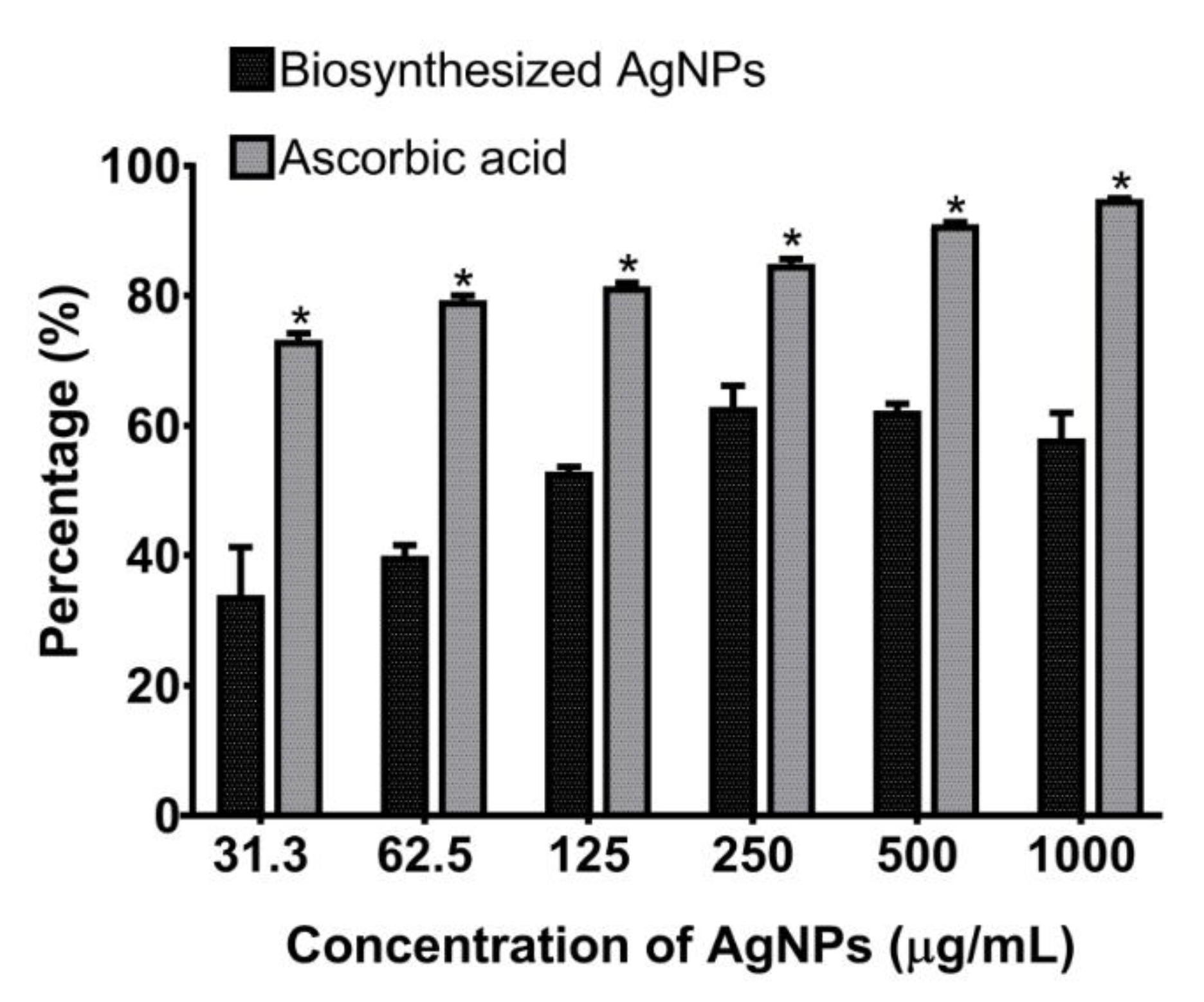
3.7. Antioxidant Activity of Biosynthesized AgNPs
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Firdhouse, M.J.; Lalitha, P. Biosynthesis of Silver Nanoparticles and Its Applications. J. Nanotechnol. 2015, 2015, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Jun, B.-H. Silver Nanoparticles: Synthesis and Application for Nanomedicine. Int. J. Mol. Sci. 2019, 20, 865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Abdelgawad, A.M.; El-Naggar, M.E.; Rojas, O.J. Antibacterial activity of silver nanoparticles synthesized In-situ by solution spraying onto cellulose. Carbohydr. Polym. 2016, 147, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Paladini, F.; Pollini, M. Antimicrobial Silver Nanoparticles for Wound Healing Application: Progress and Future Trends. Materials 2019, 12, 2540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simbine, E.O.; Rodrigues, L.; Da, C.; Lapa-Guimarães, J.; Kamimura, E.S.; Corassin, C.H.; Oliveira, C.A.F.D. Application of silver nanoparticles in food packages: A review. Food Sci. Technol. 2019, 39, 793–802. [Google Scholar] [CrossRef] [Green Version]
- Dakal, T.C.; Kumar, A.; Majumdar, R.S.; Yadav, V. Mechanistic Basis of Antimicrobial Actions of Silver Nanoparticles. Front. Microbiol. 2016, 7, 1831. [Google Scholar] [CrossRef] [Green Version]
- Durán, N.; Marcato, P.D.; Durán, M.; Yadav, A.; Gade, A.; Rai, M. Mechanistic aspects in the biogenic synthesis of extracellular metal nanoparticles by peptides, bacteria, fungi, and plants. Appl. Microbiol. Biotechnol. 2011, 90, 1609–1624. [Google Scholar] [CrossRef]
- Xu, C.; Guo, Y.; Qiao, L.; Ma, L.; Cheng, Y.; Roman, A. Biogenic Synthesis of Novel Functionalized Selenium Nanoparticles by Lactobacillus casei ATCC 393 and Its Protective Effects on Intestinal Barrier Dysfunction Caused by Enterotoxigenic Escherichia coli K88. Front. Microbiol. 2018, 9, 1129. [Google Scholar] [CrossRef] [Green Version]
- Markus, J.; Mathiyalagan, R.; Kim, Y.J.; Abbai, R.; Singh, P.; Ahn, S.; Perez, Z.E.J.; Hurh, J.; Yang, D.C. Intracellular synthesis of gold nanoparticles with antioxidant activity by probiotic Lactobacillus kimchicus DCY51T isolated from Korean kimchi. Enzym. Microb. Technol. 2016, 95, 85–93. [Google Scholar] [CrossRef]
- Mohd Yusof, H.; Mohamad, R.; Zaidan, U.H.; Rahman, N.A. Sustainable microbial cell nanofactory for zinc oxide nanoparticles production by zinc-tolerant probiotic Lactobacillus plantarum strain TA4. Microb. Cell Factories 2020, 19, 10. [Google Scholar] [CrossRef]
- Sintubin, L.; De Windt, W.; Dick, J.; Mast, J.; van der Ha, D.; Verstraete, W.; Boon, N. Lactic acid bacteria as reducing and capping agent for the fast and efficient production of silver nanoparticles. Appl. Microbiol. Biotechnol. 2009, 84, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Mrvčić, J.; Stanzer, D.; Šolić, E.; Stehlik-Tomas, V. Interaction of lactic acid bacteria with metal ions: Opportunities for improving food safety and quality. World J. Microbiol. Biotechnol. 2012, 28, 2771–2782. [Google Scholar] [CrossRef] [PubMed]
- Król, A.; Railean-Plugaru, V.; Pomastowski, P.; Złoch, M.; Buszewski, B. Mechanism study of intracellular zinc oxide nanocomposites formation. Colloids Surf. A Physicochem. Eng. Asp. 2018, 553, 349–358. [Google Scholar] [CrossRef]
- Jain, N.; Bhargava, A.; Tarafdar, J.C.; Singh, S.K.; Panwar, J. A biomimetic approach towards synthesis of zinc oxide nanoparticles. Appl. Microbiol. Biotechnol. 2013, 97, 859–869. [Google Scholar] [CrossRef]
- Bhargava, A.; Jain, N.; Khan, M.A.; Pareek, V.; Dilip, R.V.; Panwar, J. Utilizing metal tolerance potential of soil fungus for efficient synthesis of gold nanoparticles with superior catalytic activity for degradation of rhodamine B. J. Environ. Manag. 2016, 183, 22–32. [Google Scholar] [CrossRef]
- George Kerry, R.; Patra, J.K.; Gouda, S.; Park, Y.; Shin, H.-S.; Das, G. Benefaction of probiotics for human health: A review. J. Food Drug Anal. 2018, 26, 927–939. [Google Scholar] [CrossRef] [Green Version]
- Rajesh, S.; Dharanishanthi, V.; Kanna, A.V. Antibacterial mechanism of biogenic silver nanoparticles of Lactobacillus acidophilus. J. Exp. Nanosci. 2015, 10, 1143–1152. [Google Scholar] [CrossRef]
- Chaudhari, P.R.; Masurkar, S.A.; Shidore, V.B.; Kamble, S.P. Antimicrobial activity of extracellularly synthesized silver nanoparticles using Lactobacillus species obtained from VIZYLAC capsule. J. Appl. Pharm. Sci. 2012, 2, 25. [Google Scholar]
- Dakhil, A.S. Biosynthesis of silver nanoparticle (AgNPs) using Lactobacillus and their effects on oxidative stress biomarkers in rats. J. King Saud Univ.-Sci. 2017, 29, 462–467. [Google Scholar] [CrossRef]
- Viorica, R.-P.; Pawel, P.; Kinga, M.; Michal, Z.; Katarzyna, R.; Boguslaw, B. Lactococcus lactis as a safe and inexpensive source of bioactive silver composites. Appl. Microbiol. Biotechnol. 2017, 101, 7141–7153. [Google Scholar] [CrossRef] [Green Version]
- Das, V.L.; Thomas, R.; Varghese, R.T.; Soniya, E.V.; Mathew, J.; Radhakrishnan, E.K. Extracellular synthesis of silver nanoparticles by the Bacillus strain CS 11 isolated from industrialized area. 3 Biotech 2014, 4, 121–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Presentato, A.; Piacenza, E.; Anikovskiy, M.; Cappelletti, M.; Zannoni, D.; Turner, R.J. Biosynthesis of selenium-nanoparticles and -nanorods as a product of selenite bioconversion by the aerobic bacterium Rhodococcus aetherivorans BCP1. New Biotechnol. 2018, 41, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Janardhanan, A.; Varghese, R.T.; Soniya, E.V.; Mathew, J.; Radhakrishnan, E.K. Antibacterial properties of silver nanoparticles synthesized by marine Ochrobactrum sp. Braz. J. Microbiol. 2014, 45, 1221–1227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safawo, T.; Sandeep, B.; Pola, S.; Tadesse, A. Synthesis and characterization of zinc oxide nanoparticles using tuber extract of anchote (Coccinia abyssinica (Lam.) Cong.) for antimicrobial and antioxidant activity assessment. OpenNano 2018, 3, 56–63. [Google Scholar] [CrossRef]
- Mohd Yusof, H.; Mohamad, R.; Zaidan, U.H.; Abdul Rahman, N. Microbial synthesis of zinc oxide nanoparticles and their potential application as an antimicrobial agent and a feed supplement in animal industry: A review. J. Anim. Sci. Biotechnol. 2019, 10, 1–22. [Google Scholar] [CrossRef]
- Zonaro, E.; Piacenza, E.; Presentato, A.; Monti, F.; Dell’Anna, R.; Lampis, S.; Vallini, G. Ochrobactrum sp. MPV1 from a dump of roasted pyrites can be exploited as bacterial catalyst for the biogenesis of selenium and tellurium nanoparticles. Microb. Cell Factories 2017, 16, 215. [Google Scholar] [CrossRef] [Green Version]
- Giller, K.E.; Witter, E.; McGrath, S.P. Heavy metals and soil microbes. Soil Biol. Biochem. 2009, 41, 2031–2037. [Google Scholar] [CrossRef]
- Klaus, T.; Joerger, R.; Olsson, E.; Granqvist, C.-G. Silver-based crystalline nanoparticles, microbially fabricated. Proc. Natl. Acad. Sci. USA 1999, 96, 13611–13614. [Google Scholar] [CrossRef] [Green Version]
- Iravani, S. Bacteria in Nanoparticle Synthesis: Current Status and Future Prospects. Int. Sch. Res. Not. 2014, 2014, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Kamika, I.; Momba, M.N.B. Assessing the resistance and bioremediation ability of selected bacterial and protozoan species to heavy metals in metal-rich industrial wastewater. BMC Microbiol. 2013, 13, 28. [Google Scholar] [CrossRef] [Green Version]
- Mohanta, Y.K.; Panda, S.K.; Bastia, A.K.; Mohanta, T.K. Biosynthesis of Silver Nanoparticles from Protium serratum and Investigation of their Potential Impacts on Food Safety and Control. Front. Microbiol. 2017, 8, 826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibrahim, E.; Fouad, H.; Zhang, M.; Zhang, Y.; Qiu, W.; Yan, C.; Li, B.; Mo, J.; Chen, J. Biosynthesis of silver nanoparticles using endophytic bacteria and their role in inhibition of rice pathogenic bacteria and plant growth promotion. RSC Adv. 2019, 9, 29293–29299. [Google Scholar] [CrossRef] [Green Version]
- El-Shanshoury, A.E.-R.R.; ElSilk, S.E.; Ebeid, M.E. Extracellular Biosynthesis of Silver Nanoparticles Using Escherichia coli ATCC 8739, Bacillus subtilis ATCC 6633, and Streptococcus thermophilus ESh1 and Their Antimicrobial Activities. ISRN Nanotechnol. 2011, 2011, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Nanda, A.; Saravanan, M. Biosynthesis of silver nanoparticles from Staphylococcus aureus and its antimicrobial activity against MRSA and MRSE. Nanomed. Nanotechnol. Biol. Med. 2009, 5, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Garmasheva, I.; Kovalenko, N.; Voychuk, S.; Ostapchuk, A.; Livins’ka, O.; Oleschenko, L. Lactobacillus species mediated synthesis of silver nanoparticles and their antibacterial activity against opportunistic pathogens in vitro. BioImpacts 2016, 6, 219–223. [Google Scholar] [CrossRef] [Green Version]
- Saravanan, M.; Nanda, A.; Kingsley, S.J. Lactobacillus delbrueckii mediated synthesis of silver nanoparticles and their evaluation of antibacterial efficacy against MDR clinical pathogens. In Proceedings of the International Conference on Nanoscience, Engineering and Technology (ICONSET 2011), Chennai, India, 28–30 November 2011; pp. 386–390. [Google Scholar]
- Matei, A.; Matei, S.; Matei, G.-M.; Cogălniceanu, G.; Cornea, C.P. Biosynthesis of silver nanoparticles mediated by culture filtrate of lactic acid bacteria, characterization and antifungal activity. EuroBiotech J. 2020, 4, 97–103. [Google Scholar] [CrossRef]
- Dhoondia, Z.H.; Chakraborty, H. Lactobacillus Mediated Synthesis of Silver Oxide Nanoparticles. Nanomater. Nanotechnol. 2012, 2, 15. [Google Scholar] [CrossRef] [Green Version]
- Anil Kumar, S.; Abyaneh, M.K.; Gosavi, S.W.; Kulkarni, S.K.; Pasricha, R.; Ahmad, A.; Khan, M.I. Nitrate reductase-mediated synthesis of silver nanoparticles from AgNO3. Biotechnol. Lett. 2007, 29, 439–445. [Google Scholar] [CrossRef]
- Samuel, M.S.; Jose, S.; Selvarajan, E.; Mathimani, T.; Pugazhendhi, A. Biosynthesized silver nanoparticles using Bacillus amyloliquefaciens; Application for cytotoxicity effect on A549 cell line and photocatalytic degradation of p-nitrophenol. J. Photochem. Photobiol. B Biol. 2020, 202, 111642. [Google Scholar] [CrossRef]
- Wang, Y.; Shu, X.; Hou, J.; Lu, W.; Zhao, W.; Huang, S.; Wu, L. Selenium Nanoparticle Synthesized by Proteus mirabilis YC801: An Efficacious Pathway for Selenite Biotransformation and Detoxification. Int. J. Mol. Sci. 2018, 19, 3809. [Google Scholar] [CrossRef] [Green Version]
- Chapot-Chartier, M.-P.; Kulakauskas, S. Cell wall structure and function in lactic acid bacteria. Microb. Cell Factories 2014, 13, S9. [Google Scholar] [CrossRef] [Green Version]
- Omran, B.A.; Nassar, H.N.; Younis, S.A.; Fatthallah, N.A.; Hamdy, A.; El-Shatoury, E.H.; El-Gendy, N.S. Physiochemical properties of Trichoderma longibrachiatum DSMZ 16517-synthesized silver nanoparticles for the mitigation of halotolerant sulphate-reducing bacteria. J. Appl. Microbiol. 2019, 126, 138–154. [Google Scholar] [CrossRef]
- Tomaszewska, E.; Soliwoda, K.; Kadziola, K.; Tkacz-Szczesna, B.; Celichowski, G.; Cichomski, M.; Szmaja, W.; Grobelny, J. Detection Limits of DLS and UV-Vis Spectroscopy in Characterization of Polydisperse Nanoparticles Colloids. J. Nanomater. 2013, 2013, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Hamouda, R.A.; Hussein, M.H.; Abo-elmagd, R.A.; Bawazir, S.S. Synthesis and biological characterization of silver nanoparticles derived from the cyanobacterium Oscillatoria limnetica. Sci. Rep. 2019, 9, 13071. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Martin, G.; Pescuma, M.; Pérez-Corona, T.; Mozzi, F.; Madrid, Y. Determination of size and mass-and number-based concentration of biogenic SeNPs synthesized by lactic acid bacteria by using a multimethod approach. Anal. Chim. Acta 2017, 992, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Nair, B.; Pradeep, T. Coalescence of Nanoclusters and Formation of Submicron Crystallites Assisted by Lactobacillus Strains. Cryst. Growth Des. 2002, 2, 293–298. [Google Scholar] [CrossRef]
- Butler, K.S.; Peeler, D.J.; Casey, B.J.; Dair, B.J.; Elespuru, R.K. Silver nanoparticles: Correlating nanoparticle size and cellular uptake with genotoxicity. Mutagenesis 2015, 30, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Gosens, I.; Post, J.A.; de la Fonteyne, L.J.J.; Jansen, E.H.J.M.; Geus, J.W.; Cassee, F.R.; de Jong, W.H. Impact of agglomeration state of nano- and submicron sized gold particles on pulmonary inflammation. Part. Fibre Toxicol. 2010, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Bilal, M.; Rasheed, T.; Iqbal, H.M.N.; Li, C.; Hu, H.; Zhang, X. Development of silver nanoparticles loaded chitosan-alginate constructs with biomedical potentialities. Int. J. Biol. Macromol. 2017, 105, 393–400. [Google Scholar] [CrossRef]
- Priyadarshini, S.; Gopinath, V.; Meera Priyadharsshini, N.; MubarakAli, D.; Velusamy, P. Synthesis of anisotropic silver nanoparticles using novel strain, Bacillus flexus and its biomedical application. Colloids Surf. B Biointerfaces 2013, 102, 232–237. [Google Scholar] [CrossRef]
- Yan, X.; He, B.; Liu, L.; Qu, G.; Shi, J.; Hu, L.; Jiang, G. Antibacterial mechanism of silver nanoparticles in: Pseudomonas aeruginosa: Proteomics approach. Metallomics 2018, 10, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Seil, J.T.; Webster, T.J. Antimicrobial applications of nanotechnology: Methods and literature. Int. J. Nanomed. 2012, 7, 2767–2781. [Google Scholar]
- Raffi, M.; Hussain, F.; Bhatti, T.M.; Akhter, J.I.; Hameed, A.; Hasan, M.M. Antibacterial characterization of silver nanoparticles against E.coli ATCC-15224. J. Mater. Sci. Technol. 2008, 24, 192–196. [Google Scholar]
- Quinteros, M.A.; Cano Aristizábal, V.; Dalmasso, P.R.; Paraje, M.G.; Páez, P.L. Oxidative stress generation of silver nanoparticles in three bacterial genera and its relationship with the antimicrobial activity. Toxicol. Vitr. 2016, 36, 216–223. [Google Scholar] [CrossRef]
- Morones, J.R.; Elechiguerra, J.L.; Camacho, A.; Holt, K.; Kouri, J.B.; Ramírez, J.T.; Yacaman, M.J. The bactericidal effect of silver nanoparticles. Nanotechnology 2005, 16, 2346–2353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qing, Y.; Cheng, L.; Li, R.; Liu, G.; Zhang, Y.; Tang, X.; Wang, J.; Liu, H.; Qin, Y. Potential antibacterial mechanism of silver nanoparticles and the optimization of orthopedic implants by advanced modification technologies. Int. J. Nanomed. 2018, 12, 3311. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.; Rong, K.; Li, J.; Yang, H.; Chen, R. Size-dependent antibacterial activities of silver nanoparticles against oral anaerobic pathogenic bacteria. J. Mater. Sci. Mater. Med. 2013, 24, 1465–1471. [Google Scholar] [CrossRef]
- Dong, Y.; Zhu, H.; Shen, Y.; Zhang, W.; Zhang, L. Antibacterial activity of silver nanoparticles of different particle size against Vibrio Natriegens. PLoS ONE 2019, 14, e0222322. [Google Scholar] [CrossRef] [Green Version]
- Das, D.; Nath, B.C.; Phukon, P.; Kalita, A.; Dolui, S.K. Synthesis of ZnO nanoparticles and evaluation of antioxidant and cytotoxic activity. Colloids Surf. B Biointerfaces 2013, 111, 556–560. [Google Scholar] [CrossRef]
- Valgimigli, L.; Baschieri, A.; Amorati, R. Antioxidant activity of nanomaterials. J. Mater. Chem. B 2018, 6, 2036–2051. [Google Scholar] [CrossRef]
- Das, G.; Patra, J.K.; Debnath, T.; Ansari, A.; Shin, H.-S. Investigation of antioxidant, antibacterial, antidiabetic, and cytotoxicity potential of silver nanoparticles synthesized using the outer peel extract of Ananas comosus (L.). PLoS ONE 2019, 14, e0220950. [Google Scholar] [CrossRef] [Green Version]
- Kumar, B.; Smita, K.; Seqqat, R.; Benalcazar, K.; Grijalva, M.; Cumbal, L. In vitro evaluation of silver nanoparticles cytotoxicity on Hepatic cancer (Hep-G2) cell line and their antioxidant activity: Green approach for fabrication and application. J. Photochem. Photobiol. B Biol. 2016, 159, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, A.B.; Moniri, M.; Azizi, S.; Rahim, R.A.; Ariff, A.B.; Saad, W.Z.; Namvar, F.; Navaderi, M.; Mohamad, R. Biosynthesis of ZnO nanoparticles by a new Pichia kudriavzevii yeast strain and evaluation of their antimicrobial and antioxidant activities. Molecules 2017, 22, 872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khorrami, S.; Zarrabi, A.; Khaleghi, M.; Danaei, M.; Mozafari, M. Selective cytotoxicity of green synthesized silver nanoparticles against the MCF-7 tumor cell line and their enhanced antioxidant and antimicrobial properties. Int. J. Nanomed. 2018, 13, 8013–8024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sytu, M.R.C.; Camacho, D.H. Green Synthesis of Silver Nanoparticles (AgNPs) from Lenzites betulina and the Potential Synergistic Effect of AgNP and Capping Biomolecules in Enhancing Antioxidant Activity. Bionanoscience 2018, 8, 835–844. [Google Scholar] [CrossRef]





| Species | Route | Size (nm) | Reference |
|---|---|---|---|
| Lactobacillus sp. | Supernatant | 30–100 | [19] |
| Lactobacillus acidophilus | Cell biomass | 30 | [35] |
| Lactobacillus plantarum | Cell biomass | 33.4 | [35] |
| L. acidophilus | Supernatant | 4–50 | [17] |
| Lactobacillus delbrueckii | Supernatant | 54–113 | [36] |
| Lactobacillus sp. | Supernatant | 14 | [37] |
| Lactobacillus mindensis | Supernatant | 20 | [38] |
| Spectra Band (cm−1) | Functional Group and Vibrational Assignment | |
|---|---|---|
| L. plantarum TA4 | Biosynthesized AgNPs | |
| 3254 | 3197 | Hydroxyl group (-OH)/amide group (N–H stretching vibration) |
| 2924 | C–H stretching vibration | |
| 1636 | 1634 | Amide I (C=O stretching vibration) |
| 1151 | 1518 | Amide II |
| 1456 | 1450 | C–H bending vibration |
| 1224 | 1209 | C–N stretching vibration, PO2- asymmetric stretching |
| 1078 | 1030 | C–O stretching vibration |
| 994 | 961 | C=C bending vibration |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohd Yusof, H.; Abdul Rahman, N.; Mohamad, R.; Zaidan, U.H. Microbial Mediated Synthesis of Silver Nanoparticles by Lactobacillus Plantarum TA4 and its Antibacterial and Antioxidant Activity. Appl. Sci. 2020, 10, 6973. https://doi.org/10.3390/app10196973
Mohd Yusof H, Abdul Rahman N, Mohamad R, Zaidan UH. Microbial Mediated Synthesis of Silver Nanoparticles by Lactobacillus Plantarum TA4 and its Antibacterial and Antioxidant Activity. Applied Sciences. 2020; 10(19):6973. https://doi.org/10.3390/app10196973
Chicago/Turabian StyleMohd Yusof, Hidayat, Nor’Aini Abdul Rahman, Rosfarizan Mohamad, and Uswatun Hasanah Zaidan. 2020. "Microbial Mediated Synthesis of Silver Nanoparticles by Lactobacillus Plantarum TA4 and its Antibacterial and Antioxidant Activity" Applied Sciences 10, no. 19: 6973. https://doi.org/10.3390/app10196973
APA StyleMohd Yusof, H., Abdul Rahman, N., Mohamad, R., & Zaidan, U. H. (2020). Microbial Mediated Synthesis of Silver Nanoparticles by Lactobacillus Plantarum TA4 and its Antibacterial and Antioxidant Activity. Applied Sciences, 10(19), 6973. https://doi.org/10.3390/app10196973






