Does Exposure to Burning and Heated Tobacco Affect the Abundance of Perio-Pathogenic Species in the Subgingival Biofilm?
Abstract
1. Introduction
2. Materials and Methods
2.1. Inclusion Criteria
2.2. Exclusion Criteria
2.3. Drop-Outs
2.4. Anamnesis and Clinical Examination
2.5. The Microbiological Diagnostic Procedure
2.5.1. Microbiological Sampling
2.5.2. Isolation of Bacterial DNA
2.5.3. Next-Generation Sequencing (NGS)
2.5.4. Statistical Analysis
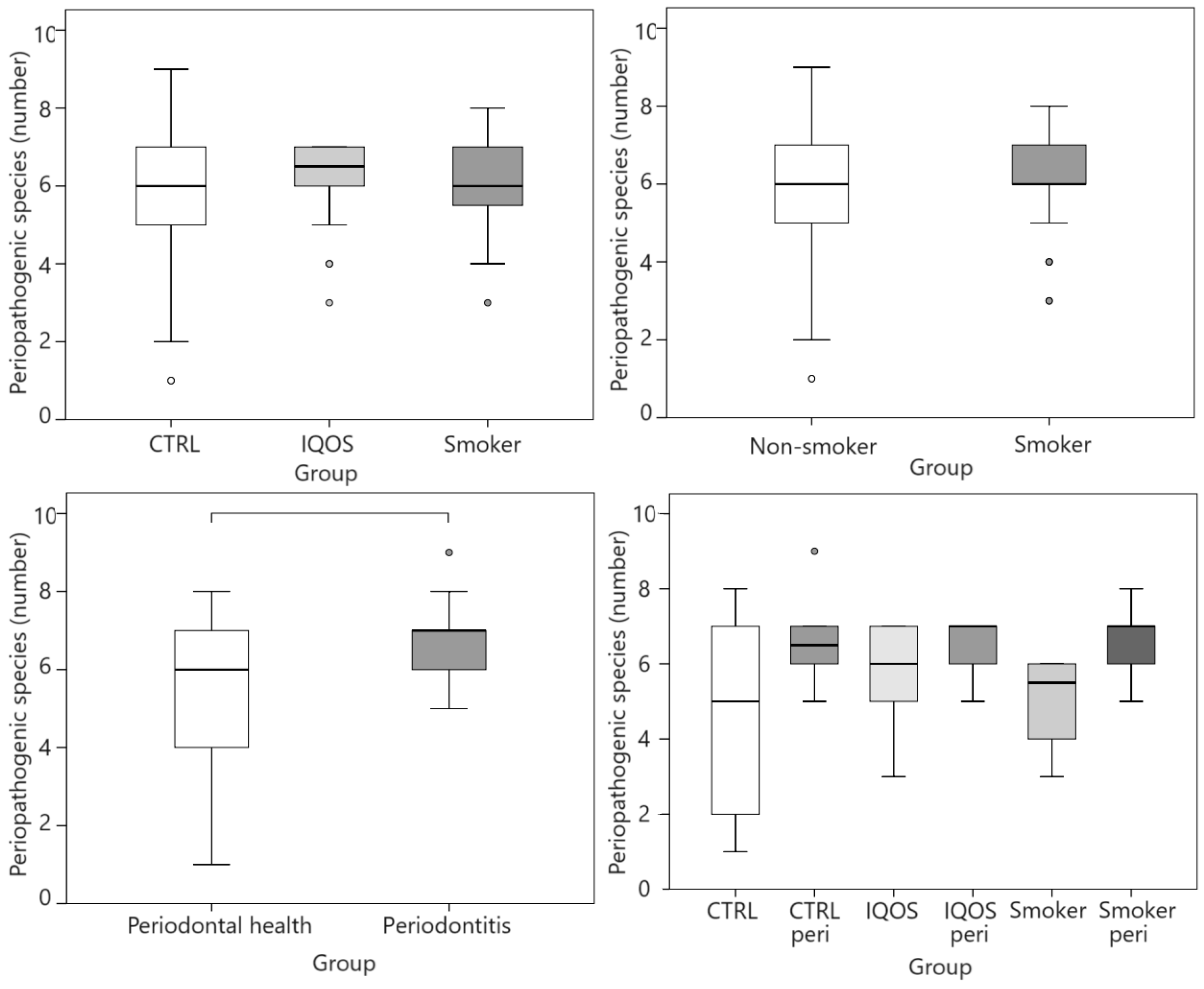
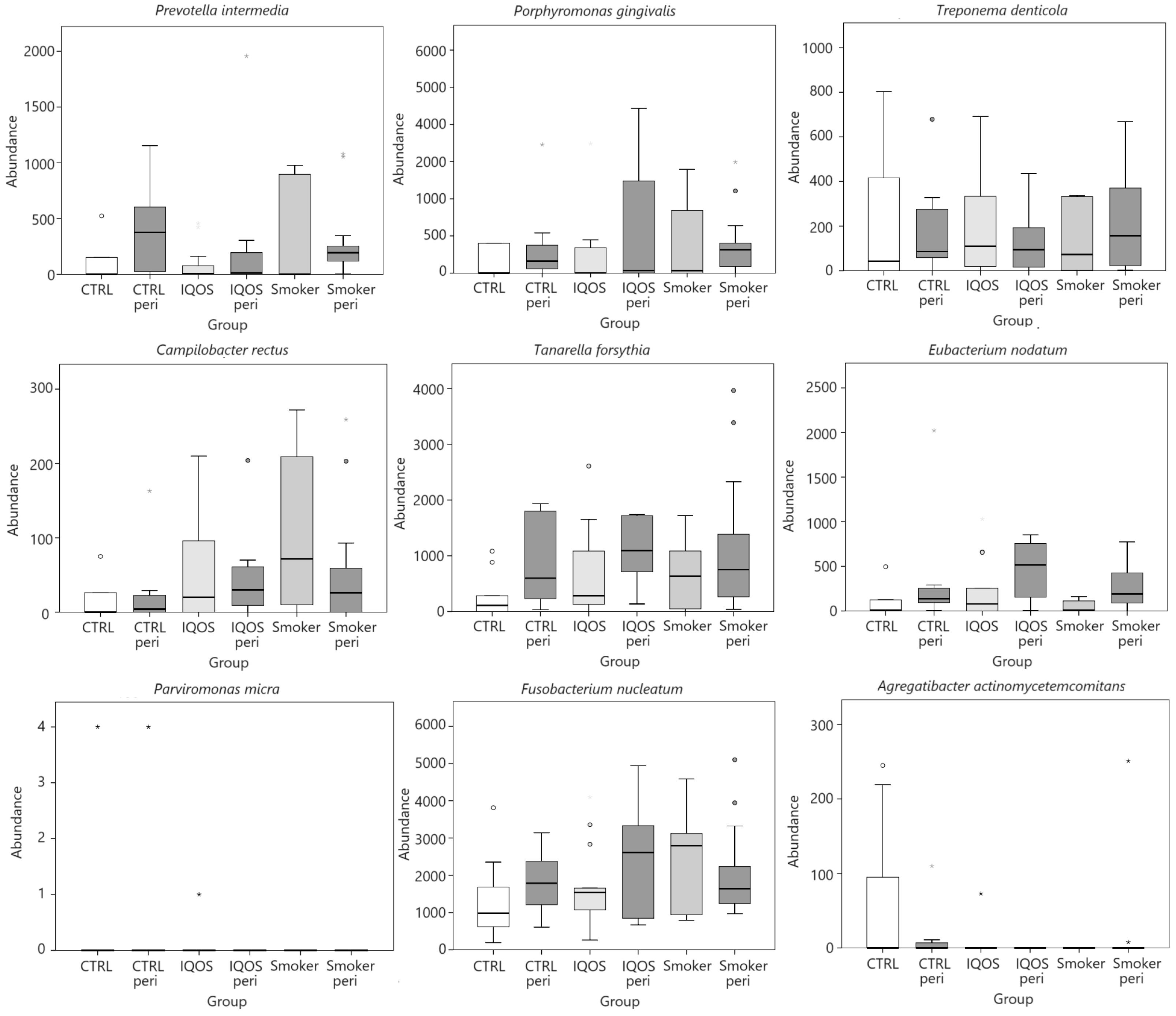
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pihlstrom, B.L.; Michalowicz, B.S.; Johnson, N.W. Periodontal diseases. Lancet 2005, 366, 1809–1820. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Li, Y.; Cao, Y.; Xue, J.; Zhou, X. The oral microbiome diversity and its relation to human diseases. Folia Microbiol. 2015, 60, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Sedghi, L.M.; Bacino, M.; Kapila, Y.L. Periodontal disease: The good, the bad, and the unknown. Front. Cell. Infect. Microbiol. 2021, 11, 766944. [Google Scholar] [CrossRef] [PubMed]
- Paster, B.J.; Olsen, I.; Aas, J.A.; Dewhirst, F.E. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000 2006, 42, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Wade, W.G. The oral microbiome in health and disease. Pharmacol. Res. 2013, 69, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, M.; Polizzi, A.; Santonocito, S.; Romano, A.; Lombardi, T.; Isola, G. Impact of oral microbiome in periodontal health and periodontitis: A critical review on prevention and treatment. Int. J. Mol. Sci. 2022, 23, 5142. [Google Scholar] [CrossRef] [PubMed]
- Lang, N.P.; Mombelli, A.; Attstrom, R. Dental plaque and calculus. In Textbook of Clinical Periodontology and Implant Dentistry, 5th ed.; Lindhe, J.A., Lang, N.P.B., Karring, T.C., Eds.; Wiley Blackwell: London, UK, 2008; pp. 184–196. [Google Scholar]
- Hajishengallis, G.; Lamont, R.J. Beyond the red complex and into more complexity: The polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral. Microbiol. 2012, 27, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Haffajee, A.D. Periodontal infections. In Textbook of Clinical Periodontology and Implant Dentistry, 5th ed.; Lindhe, J.A., Lang, N.P.B., Karring, T.C., Eds.; Wiley Blackwell: London, UK, 2008; pp. 208–209, 216–218, 232. [Google Scholar]
- Caton, J.G.; Armitage, G.; Berglundh, T.; Chapple, I.L.C.; Jepsen, S.; Kornman, K.S.; Mealey, B.L.; Papapanou, P.N.; Sanz, M.; Tonetti, M.S. A new classification scheme for periodontal and peri-implant diseases and conditions—Introduction and key changes from the 1999 classification. J. Clin. Periodontol. 2018, 45 (Suppl. 20), S1–S8. [Google Scholar] [CrossRef]
- Başaran, R.; Güven, N.M.; Eke, B.C. An overview of iQOS® as a new heat-not-burn tobacco product and its potential effects on human health and the environment. Turk. J. Pharm. Sci. 2019, 16, 371–374. [Google Scholar] [CrossRef]
- World Health Organization. Heated Tobacco Products (HTPs) Information Sheet; World Health Organization: Geneva, Switzerland, 2020; Available online: https://www.who.int/publications/i/item/WHO-HEP-HPR-2020.2 (accessed on 30 December 2023).
- European Respiratory Society. ERS Position Paper on Heated Tobacco Products; European Respiratory Society: Lausanne, Switzerland, 2018; Available online: https://www.ersnet.org/news-and-features/news/ers-position-paper-on-heated-tobacco-products/ (accessed on 30 December 2023).
- Ganesan, S.M.; Dabdoub, S.M.; Nagaraja, H.N.; Scott, M.L.; Pamulapati, S.; Berman, M.L.; Shields, P.G.; Wewers, M.E.; Kumar, P.S. Adverse effects of electronic cigarettes on the disease-naive oral microbiome. Sci. Adv. 2020, 6, eaaz0108. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Luo, Y.; Jiang, X.; Zhang, H.; Zhu, F.; Hu, S.; Hou, H.; Hu, Q.; Pang, Y. Chemical analysis and simulated pyrolysis of tobacco heating system 2.2 compared to conventional cigarettes. Nicotine Tob. Res. 2019, 21, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Glantz, S.A. Heated tobacco products: The example of IQOS. Tob. Control 2018, 27 (Suppl. 1), s1–s6. [Google Scholar] [CrossRef] [PubMed]
- Gee, J.; Prasad, K.; Slayford, S.; Gray, A.; Nother, K.; Cunningham, A.; Mavropoulou, E.; Proctor, C. Assessment of tobacco heating product THP1.0. Part 8: Study to determine puffing topography, mouth level exposure and consumption among Japanese users. Regul. Toxicol. Pharmacol. 2018, 93, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Phillip Morris Int., Stamford, Connecticut. About Us, Our Products. Available online: https://www.pmi.com/markets/croatia/hr/about-us/our-products (accessed on 24 December 2023).
- Tishchenko, O.V.; Kryvenko, L.S.; Gargina, V.V. Influence of smoking heating up tobacco products and e-cigarettes on the microbiota of dental plaque. Pol. Merkur. Lekarski. 2022, 50, 16–20. [Google Scholar] [PubMed]
- Karasneh, J.A.; Al Habashneh, R.A.; Marzouka, N.A.; Thornhill, M.H. Effect of cigarette smoking on subgingival bacteria in healthy subjects and patients with chronic periodontitis. BMC Oral Health 2017, 17, 64. [Google Scholar] [CrossRef]
- Shchipkova, A.Y.; Nagaraja, H.N.; Kumar, P.S. Subgingival microbial profiles of smokers with periodontitis. J. Dent. Res. 2010, 89, 1247–1253. [Google Scholar] [CrossRef]
- Yu, G.; Phillips, S.; Gail, M.H.; Goedert, J.J.; Humphrys, M.S.; Ravel, J.; Ren, Y.; Caporaso, N.E. The effect of cigarette smoking on the oral and nasal microbiota. Microbiome 2017, 5, 3. [Google Scholar] [CrossRef]
- Shiloah, J.; Patters, M.R.; Waring, M.B. The prevalence of pathogenic periodontal microflora in healthy young adult smokers. J. Periodontol. 2000, 71, 562–567. [Google Scholar] [CrossRef]
- Kozak, M.; Pawlik, A. The role of the oral microbiome in the development of diseases. Int. J. Mol. Sci. 2023, 24, 5231. [Google Scholar] [CrossRef]
- Zijian, C.; Josephine, M.; Kulveer, M.; Paul, E.; Deirdre, A.D. Periodontal disease and periodontal bacteria as triggers for rheumatoid arthritis. Best. Pract. Res. Clin. Rheumatol. 2017, 31, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Visentin, D.; Gobin, I.; Maglica, Ž. Periodontal pathogens and their links to neuroinflammation and neurodegeneration. Microorganisms 2023, 11, 1832. [Google Scholar] [CrossRef] [PubMed]
- Kaliamoorthy, S.; Priya Sayeeram, S.; Gowdhaman, N.; Jayaraj, M.; Radhika, B.; Chellapandi, S.; Elumalai, A.; Archana, S.P.; Raju, K.; Palla, S.; et al. Association of periodontal red complex bacteria with the incidence of gastrointestinal cancers: A systematic review and meta-Analysis. Cureus 2024, 16, e59251. [Google Scholar] [CrossRef] [PubMed]
- Pardo, A.; Signoriello, A.; Signoretto, C.; Messina, E.; Carelli, M.; Tessari, M.; De Manna, N.D.; Rossetti, C.; Albanese, M.; Lombardo, G.; et al. Detection of periodontal pathogens in oral samples and cardiac specimens in patients undergoing aortic valve replacement: A pilot study. J. Clin. Med. 2021, 10, 3874. [Google Scholar] [CrossRef]
- Chang, J.T.; Anic, G.M.; Rostron, B.L.; Tanwar, M.; Chang, C.M. Cigarette smoking reduction and health risks: A systematic review and meta-analysis. Nicotine Tob. Res. 2021, 19, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Larsson, S.C.; Burgess, S. Appraising the causal role of smoking in multiple diseases: A systematic review and meta-analysis of Mendelian randomization studies. EBioMedicine 2022, 82, 104154. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Shi, G. Smoking and microbiome in oral, airway, gut and some systemic diseases. J. Transl. Med. 2019, 17, 225. [Google Scholar] [CrossRef] [PubMed]
- Aldakheel, F.M.; Alduraywish, S.A.; Jhugroo, P.; Jhugroo, C.; Divakar, D.D. Quantification of pathogenic bacteria in the subgingival oral biofilm samples collected from cigarette-smokers, individuals using electronic nicotine delivery systems and non-smokers with and without periodontitis. Arch Oral Biol. 2020, 117, 104793. [Google Scholar] [CrossRef] [PubMed]
- Mišković, I.; Kuiš, D.; Špalj, S.; Pupovac, A.; Prpić, J. Periodontal health status in adults exposed to tobacco heating system aerosol and cigarette smoke vs. non-smokers: A cross-sectional study. Dent. J. 2024, 12, 26. [Google Scholar] [CrossRef]
- Ristrl, R. Sample Size Calculator Version 1.061. Available online: https://homepage.univie.ac.at/robin.ristl/samplesize.php (accessed on 2 November 2023).
- Mintzker, Y.; Blum, D.; Adler, L. Replacing PICO in non-interventional studies. BMJ Evid. Based. Med. 2023, 28, 284. [Google Scholar] [CrossRef]
- Palmer, R.A.; Soory, M.; Lang, P.N.B. Modifying factors. In Textbook of Clinical Periodontology and Implant Dentistry, 5th ed.; Lindhe, J.A., Lang, N.P.B., Karring, T.C., Eds.; Wiley Blackwell: London, UK, 2008; pp. 307–314. [Google Scholar]
- Tonetti, M.S.; Sanz, M. Implementation of the new classification of periodontal diseases: Decision-making algorithms for clinical practice and education. J. Clin. Periodontol. 2019, 46, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Bašić, K.; Peroš, K.; Bošnjak, Z.; Šutej, I. Subgingival microbiota profile in association with cigarette smoking in young adults: A cross-sectional study. Dent. J. 2021, 9, 150. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- Bhar, A. The application of next generation sequencing technology in medical diagnostics: A perspective. Proc. Indian Natl. Sci. Acad. Part A Phys. Sci. 2022, 88, 592–600. [Google Scholar] [CrossRef]
- Gonzalez-Garay, M.L. The road from next-generation sequencing to personalized medicine. Per. Med. 2014, 11, 523–544. [Google Scholar] [CrossRef] [PubMed]
- ArRejaie, A.S.; Al-Aali, K.A.; Alrabiah, M.; Vohra, F.; Mokeem, S.A.; Basunbul, G.; Alrahlah, A.; Abduljabbar, T. Proinflammatory cytokine levels and peri-implant parameters among cigarette smokers, individuals vaping electronic cigarettes, and non-smokers. J. Periodontol. 2019, 90, 367–374. [Google Scholar] [CrossRef]
- Bizzarro, S.; Loos, B.G.; Laine, M.L.; Crielaard, W.; Zaura, E. Subgingival microbiome in smokers and non-smokers in periodontitis: An exploratory study using traditional targeted techniques and a next-generation sequencing. J. Clin. Periodontol. 2013, 40, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Jervøe-Storm, P.M.; Alahdab, H.; Koltzscher, M.; Fimmers, R.; Jepsen, S. Comparison of curet and paper point sampling of subgingival bacteria as analyzed by real-time polymerase chain reaction. J. Periodontol. 2007, 78, 909–917. [Google Scholar] [CrossRef]
- Pérez-Chaparro, P.J.; Duarte, P.M.; Pannuti, C.M.; Figueiredo, L.C.; Mestnik, M.J.; Gonçalves, C.P.; Faveri, M.; Feres, M. Evaluation of human and microbial DNA content in subgingival plaque samples collected by paper points or curette. J. Microbiol. Methods 2015, 111, 19–20. [Google Scholar] [CrossRef]
- Singh, M.; Sahota, J.K.; Singh, P.; Kour, H. Association of red complex bacteria with periodontal disease: A clinico microbiological study. Apollo Med. 2021, 18, 224–227. [Google Scholar] [CrossRef]
- Abdulkareem, A.A.; Al-Taweel, F.B.; Al-Sharqi, A.J.B.; Gul, S.S.; Sha, A.; Chapple, I.L.C. Current concepts in the pathogenesis of periodontitis: From symbiosis to dysbiosis. J. Oral Microbiol. 2023, 15, 2197779. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Pahumunto, N.; Naorungroj, S.; Teanpaisan, R. Association between periodontal pathogens and severity of periodontal diseases among adolescents in Kunming City: China. J. Health Sci. Med. Res. 2024, 42, 20231029. [Google Scholar] [CrossRef]
- Chigasaki, O.; Aoyama, N.; Sasaki, Y.; Takeuchi, Y.; Mizutani, K.; Ikeda, Y.; Gokyu, M.; Umeda, M.; Izumi, Y.; Iwata, T.; et al. Porphyromonas gingivalis, the most influential pathogen in red-complex bacteria: A cross-sectional study on the relationship between bacterial count and clinical periodontal status in Japan. J. Periodontol. 2021, 92, 1719–1729. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A. Persistence of Tannerella forsythia and Fusobacterium nucleatum in dental plaque: A strategic alliance. Curr. Oral Health Rep. 2020, 7, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhou, X.; Cheng, L.; Li, M. The impact of smoking on subgingival microflora: From periodontal health to disease. Front. Microbiol. 2020, 11, 506311. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Chavakis, T.; Lambris, J.D. Current understanding of periodontal disease pathogenesis and targets for host-modulation therapy. Periodontology 2000 2020, 84, 14–34. [Google Scholar] [CrossRef] [PubMed]
- Van Assche, N.; Van Essche, M.; Pauwels, M.; Teughels, W.; Quirynen, M. Do periodontopathogens disappear after full-mouth tooth extraction? J. Clin. Periodontol. 2009, 36, 1043–1047. [Google Scholar] [CrossRef] [PubMed]
- Veseli, E.; Staka, G.; Tovani-Palone, M.R. Evaluation of red-complex bacteria loads in complete denture patients: A pilot study. BDJ Open 2023, 9, 7. [Google Scholar] [CrossRef]
- Nath, S.G.; Raveendran, R. Microbial dysbiosis in periodontitis. J. Indian Soc. Periodontol. 2013, 17, 543–545. [Google Scholar] [CrossRef]
- Zhang, J.; Yu, J.; Dou, J.; Hu, P.; Guo, Q. The impact of smoking on subgingival plaque and the development of periodontitis: A literature review. Front. Oral Health 2021, 2, 751099. [Google Scholar] [CrossRef] [PubMed]
- Hanioka, T.; Morita, M.; Yamamoto, T.; Inagaki, K.; Wang, P.L.; Ito, H.; Morozumi, T.; Takeshita, T.; Suzuki, N.; Shigeishi, H.; et al. Smoking and periodontal microorganisms. Jpn. Dent. Sci. Rev. 2019, 55, 88–94. [Google Scholar] [CrossRef] [PubMed]
| Presence of Periodontitis | |||
|---|---|---|---|
| Exposure to Tobacco | Non-Periodontitis | Periodontitis | Total |
| CTRL (N (%)) | 10 (55.6%) | 8 (44.4%) | 18 (100%) |
| IQOS (N (%)) | 13 (59.1%) | 9 (40.9%) | 22 (100%) |
| S (N (%)) | 6 (31.6%) | 13 (68.4%) | 19 (100%) |
| Total (N (%)) | 29 (49.2%) | 30 (50.8%) | 59 (100%) |
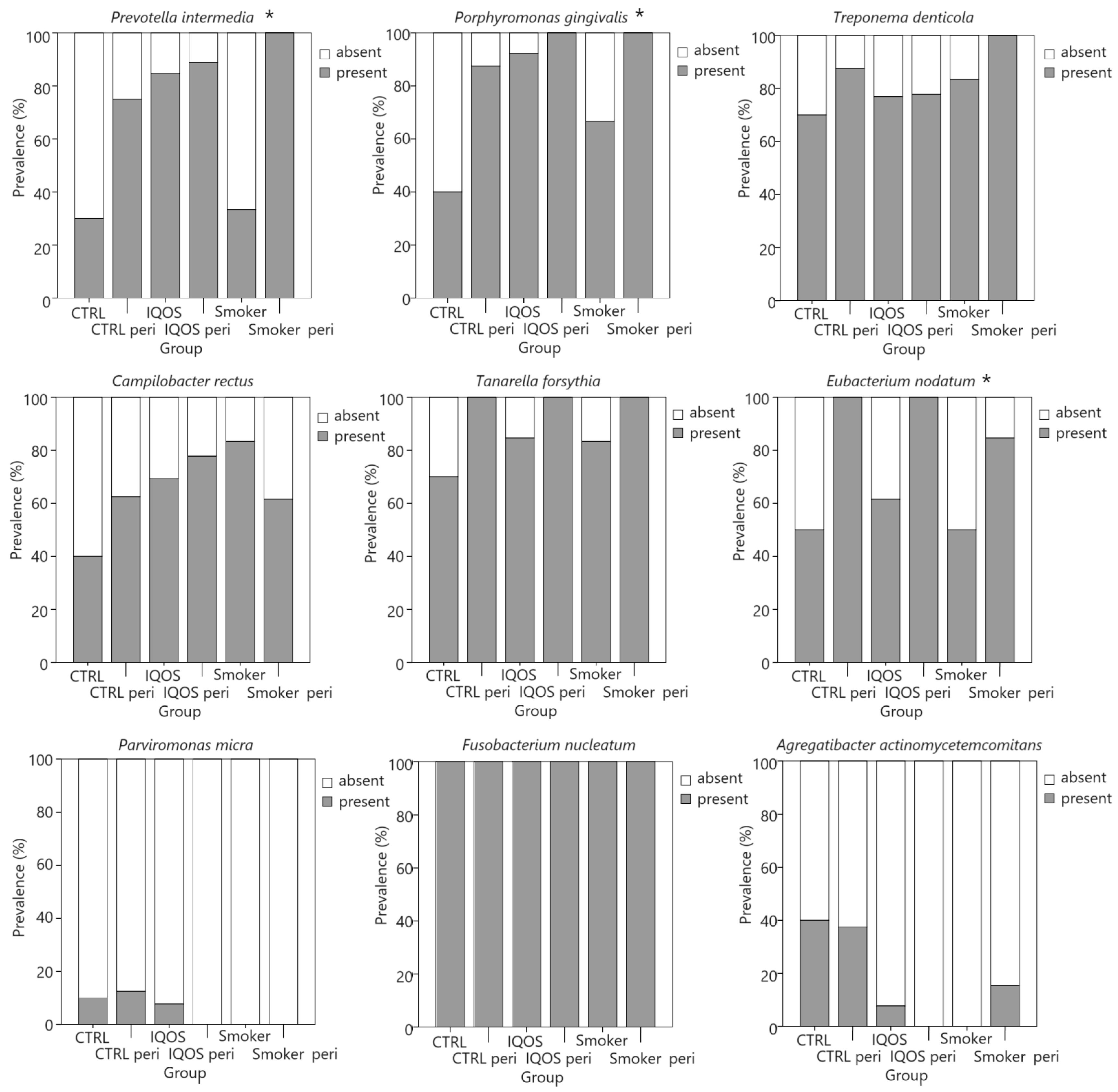
| Bacteria | S (N (%)) | IQOS (N (%)) | CTRL (N (%)) | p * | V |
|---|---|---|---|---|---|
| Prevotella intermedia | 15/19 (79%) | 19/22 (86%) | 9/18 (50%) | 0.028 | 0.348 |
| Porphyromonas gingvalis | 17/19 (90%) | 21/22 (96%) | 11/18 (61%) | 0.010 | 0.393 |
| Treponema denticola | 18/19 (95%) | 7/22 (77%) | 14/18 (78%) | 0.257 | 0.215 |
| Campylobacter rectus | 13/19 (68%) | 16/22 (73%) | 9/18 (50%) | 0.297 | 0.203 |
| Tannerella forsythia | 18/19 (95%) | 20/22 (91%) | 15/18 (83%) | 0.507 | 0.152 |
| Eubacterium nodatum | 14/19 (74%) | 17/22 (77%) | 13/18 (72%) | 0.930 | 0.050 |
| Parvimonas micra | 0 | 1/22 (5%) | 2/18 (11%) | 0.303 | 0.201 |
| Fusobacterium nucleatum | 19/19 (100%) | 22/22 (100%) | 18/18 (100%) | - | - |
| Aggregatibacter actinomycetemcommitans | 2/19 (11%) | 1/21 (5%) | 7/18 (39%) | 0.010 | 0.393 |
| Bacteria | Periodontitis | No Periodontitis | p * | V |
|---|---|---|---|---|
| Prevotella intermedia | 27/30 (90%) | 16/29 (55%) | 0.003 | 0.392 |
| Porphyromonas gingvalis | 29/30 (97%) | 20/29 (69%) | 0.005 | 0.369 |
| Treponema denticola | 27/30 (90%) | 22/29 (76%) | 0.148 | 0.188 |
| Campylobacter rectus | 20/30 (67%) | 18/29 (62%) | 0.712 | 0.048 |
| Tannerella forsythia | 30/30 (100%) | 23/29 (79%) | 0.001 | 0.342 |
| Eubacterium nodatum | 28/30 (93%) | 16/29 (55%) | 0.011 | 0.438 |
| Parvimonas micra | 1/30 (3%) | 2/29 (7%) | 0.533 | 0.081 |
| Fusobacterium nucleatum | 30/30 (100%) | 29/29 (100%) | - | - |
| Aggregatibacter actinomycetemcommitans | 5/30 (17%) | 5/29 (17%) | 0.958 | 0.008 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mišković, I.; Kuiš, D.; Špalj, S.; Pupovac, A.; Mohar-Vitezić, B.; Prpić, J. Does Exposure to Burning and Heated Tobacco Affect the Abundance of Perio-Pathogenic Species in the Subgingival Biofilm? Appl. Sci. 2024, 14, 4824. https://doi.org/10.3390/app14114824
Mišković I, Kuiš D, Špalj S, Pupovac A, Mohar-Vitezić B, Prpić J. Does Exposure to Burning and Heated Tobacco Affect the Abundance of Perio-Pathogenic Species in the Subgingival Biofilm? Applied Sciences. 2024; 14(11):4824. https://doi.org/10.3390/app14114824
Chicago/Turabian StyleMišković, Ivana, Davor Kuiš, Stjepan Špalj, Aleksandar Pupovac, Bojana Mohar-Vitezić, and Jelena Prpić. 2024. "Does Exposure to Burning and Heated Tobacco Affect the Abundance of Perio-Pathogenic Species in the Subgingival Biofilm?" Applied Sciences 14, no. 11: 4824. https://doi.org/10.3390/app14114824
APA StyleMišković, I., Kuiš, D., Špalj, S., Pupovac, A., Mohar-Vitezić, B., & Prpić, J. (2024). Does Exposure to Burning and Heated Tobacco Affect the Abundance of Perio-Pathogenic Species in the Subgingival Biofilm? Applied Sciences, 14(11), 4824. https://doi.org/10.3390/app14114824






