Sulforaphane Target Protein Prediction: A Bioinformatics Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Prediction of Protein Targets
2.2. RNA Sequence Analysis
2.3. Functional Enrichment Analysis
2.4. Docking Analysis
2.5. Molecular Dynamics (MD) Simulation
3. Results
3.1. Predicted Targets of Sulforaphane
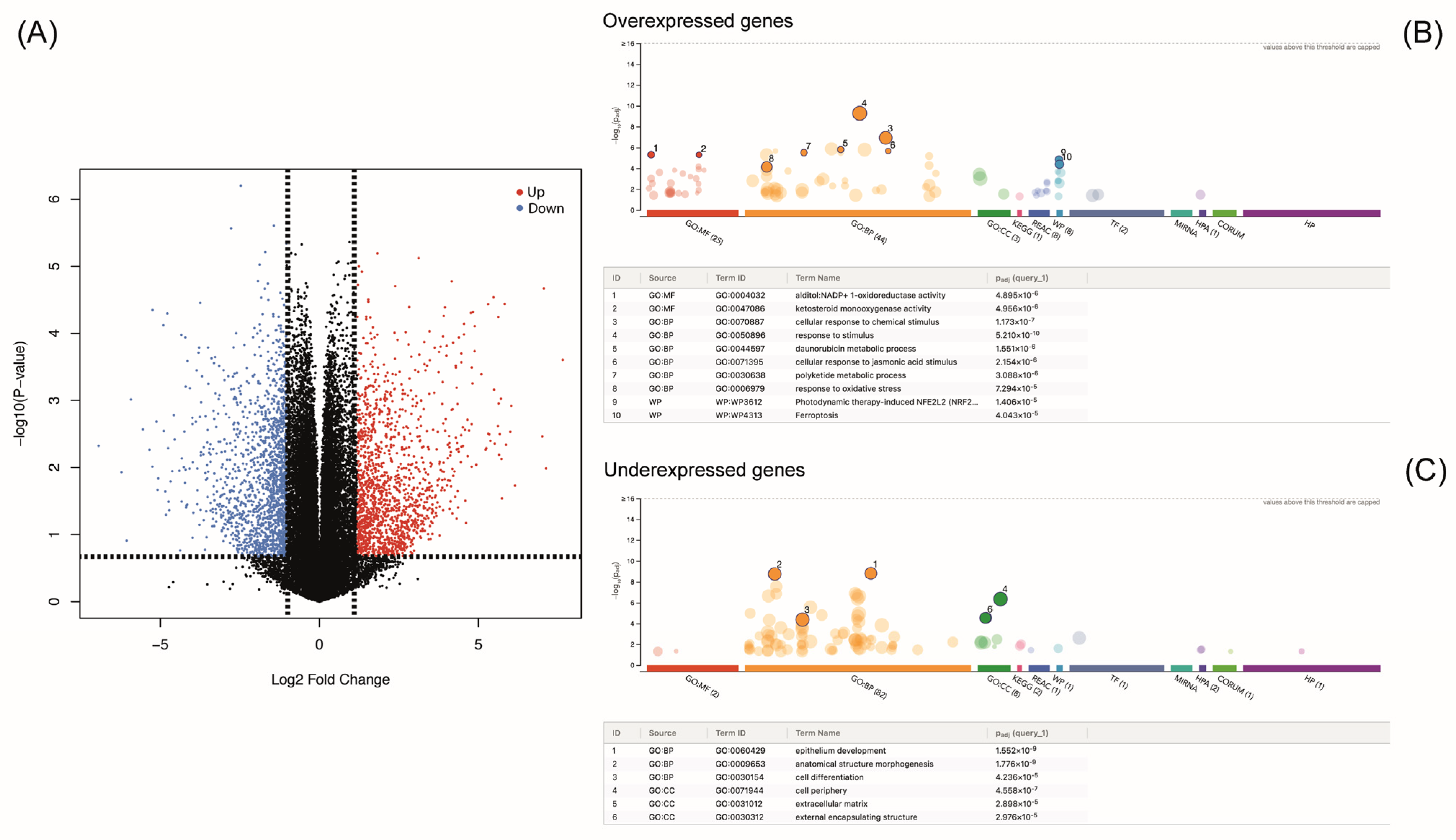
3.2. Changes in Transcriptomic Profile Caused by Sulforaphane
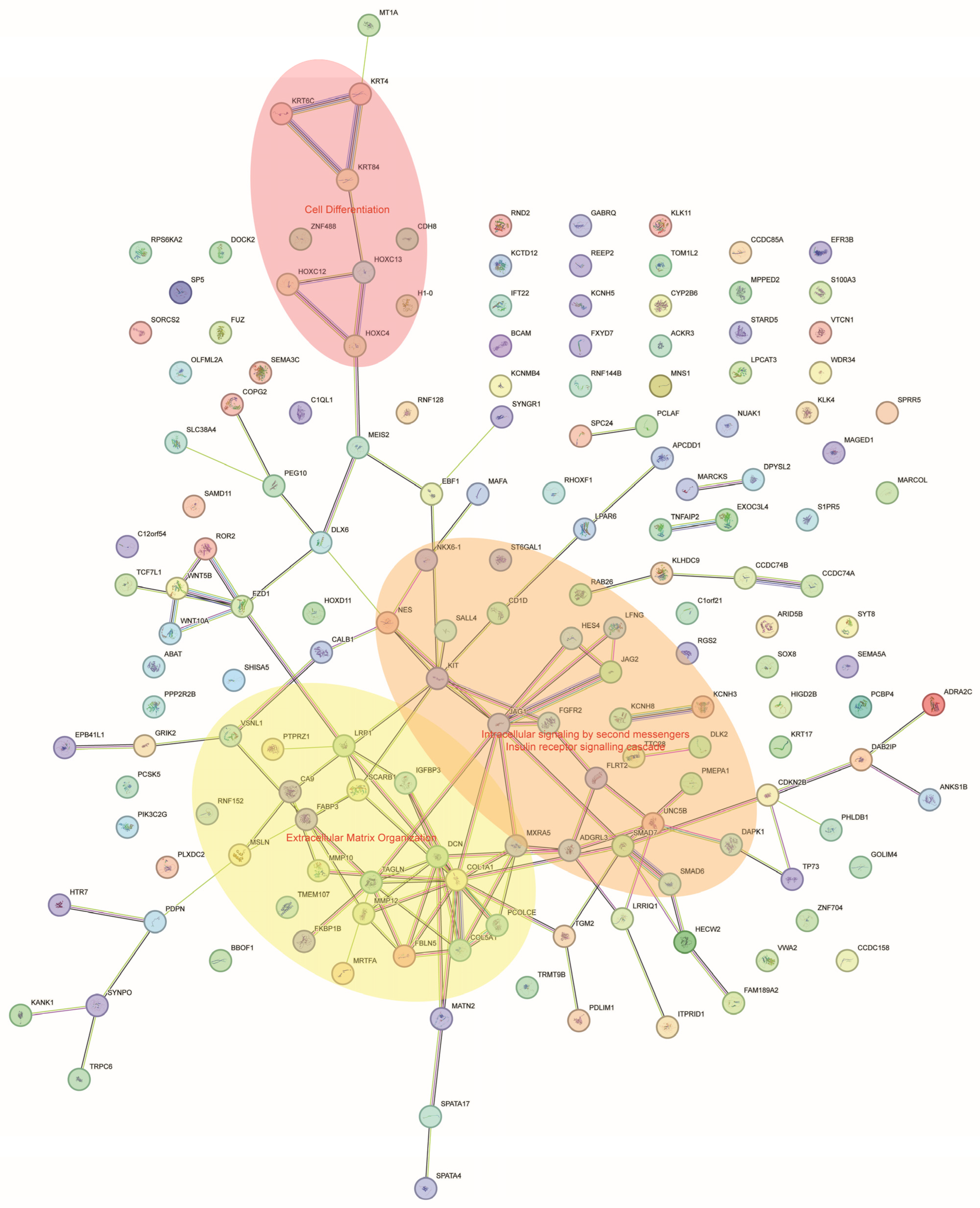
3.3. Functional Enrichment Analysis
3.4. Docking Analysis Predicts That Sulforaphane Interacts with NAMPT
4. Discussion
5. Limitations of the Study
6. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vanduchova, A.; Anzenbacher, P.; Anzenbacherova, E. Isothiocyanate from Broccoli, Sulforaphane, and Its Properties. J. Med. Food 2019, 22, 121–126. [Google Scholar] [CrossRef]
- Okada, M.; Yamamoto, A.; Aizawa, S.; Taga, A.; Terashima, H.; Kodama, S. HPLC Separation of Sulforaphane Enantiomers in Broccoli and Its Sprouts by Transformation into Diastereoisomers Using Derivatization with (S)-Leucine. J. Agric. Food Chem. 2017, 65, 244–250. [Google Scholar] [CrossRef]
- Abdull Razis, A.F.; Bagatta, M.; De Nicola, G.R.; Iori, R.; Ioannides, C. Induction of Epoxide Hydrolase and Glucuronosyl Transferase by Isothiocyanates and Intact Glucosinolates in Precision-Cut Rat Liver Slices: Importance of Side-Chain Substituent and Chirality. Arch. Toxicol. 2011, 85, 919–927. [Google Scholar] [CrossRef]
- Abdull Razis, A.F.; Iori, R.; Ioannides, C. The Natural Chemopreventive Phytochemical R-Sulforaphane Is a Far More Potent Inducer of the Carcinogen-Detoxifying Enzyme Systems in Rat Liver and Lung than the S-Isomer. Int. J. Cancer 2011, 128, 2775–2782. [Google Scholar] [CrossRef]
- Fahey, J.W.; Holtzclaw, W.D.; Wehage, S.L.; Wade, K.L.; Stephenson, K.K.; Talalay, P. Sulforaphane Bioavailability from Glucoraphanin-Rich Broccoli: Control by Active Endogenous Myrosinase. PLoS ONE 2015, 10, e0140963. [Google Scholar] [CrossRef]
- Conaway, C.C.; Getahun, S.M.; Liebes, L.L.; Pusateri, D.J.; Topham, D.K.W.; Botero-Omary, M.; Chung, F.-L. Disposition of Glucosinolates and Sulforaphane in Humans After Ingestion of Steamed and Fresh Broccoli. Nutr. Cancer 2000, 38, 168–178. [Google Scholar] [CrossRef]
- Vermeulen, M.; Klöpping-Ketelaars, I.W.A.A.; van den Berg, R.; Vaes, W.H.J. Bioavailability and Kinetics of Sulforaphane in Humans after Consumption of Cooked versus Raw Broccoli. J. Agric. Food Chem. 2008, 56, 10505–10509. [Google Scholar] [CrossRef]
- Otoo, R.A.; Allen, A.R. Sulforaphane’s Multifaceted Potential: From Neuroprotection to Anticancer Action. Molecules 2023, 28, 6902. [Google Scholar] [CrossRef] [PubMed]
- Atwell, L.L.; Hsu, A.; Wong, C.P.; Stevens, J.F.; Bella, D.; Yu, T.-W.; Pereira, C.B.; Löhr, C.V.; Christensen, J.M.; Dashwood, R.H.; et al. Absorption and Chemopreventive Targets of Sulforaphane in Humans Following Consumption of Broccoli Sprouts or a Myrosinase-Treated Broccoli Sprout Extract. Mol. Nutr. Food Res. 2015, 59, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Elkashty, O.A.; Tran, S.D. Sulforaphane as a Promising Natural Molecule for Cancer Prevention and Treatment. Curr. Med. Sci. 2021, 41, 250–269. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wu, J.; Chen, D.; Jin, J.; Wu, Y.; Chen, Z. Effects of Sulforaphane in the Central Nervous System. Eur. J. Pharmacol. 2019, 853, 153–168. [Google Scholar] [CrossRef]
- Santín-Márquez, R.; Alarcón-Aguilar, A.; López-Diazguerrero, N.E.; Chondrogianni, N.; Königsberg, M. Sulforaphane—Role in Aging and Neurodegeneration. GeroScience 2019, 41, 655–670. [Google Scholar] [CrossRef]
- Wei, L.-Y.; Zhang, J.-K.; Zheng, L.; Chen, Y. The Functional Role of Sulforaphane in Intestinal Inflammation: A Review. Food Funct. 2022, 13, 514–529. [Google Scholar] [CrossRef]
- Lu, Y.; Sun, Y.; Liu, Z.; Lu, Y.; Zhu, X.; Lan, B.; Mi, Z.; Dang, L.; Li, N.; Zhan, W.; et al. Activation of NRF2 Ameliorates Oxidative Stress and Cystogenesis in Autosomal Dominant Polycystic Kidney Disease. Sci. Transl. Med. 2020, 12, eaba3613. [Google Scholar] [CrossRef]
- Wang, Y.; Petrikova, E.; Gross, W.; Sticht, C.; Gretz, N.; Herr, I.; Karakhanova, S. Sulforaphane Promotes Dendritic Cell Stimulatory Capacity Through Modulation of Regulatory Molecules, JAK/STAT3- and MicroRNA-Signaling. Front. Immunol. 2020, 11, 589818. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Q.; Liu, J.; Zhang, Z.; Ma, X.; Zhang, Y.; Zhu, J.; Thring, R.W.; Wu, M.; Gao, Y.; et al. Sulforaphane Alleviates High Fat Diet-Induced Insulin Resistance via AMPK/Nrf2/GPx4 Axis. Biomed. Pharmacother. 2022, 152, 113273. [Google Scholar] [CrossRef] [PubMed]
- Lei, P.; Tian, S.; Teng, C.; Huang, L.; Liu, X.; Wang, J.; Zhang, Y.; Li, B.; Shan, Y. Sulforaphane Improves Lipid Metabolism by Enhancing Mitochondrial Function and Biogenesis In Vivo and In Vitro. Mol. Nutr. Food Res. 2019, 63, 1800795. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Shao, R.; Wang, N.; Zhou, N.; Du, K.; Shi, J.; Wang, Y.; Zhao, Z.; Ye, X.; Zhang, X.; et al. Sulforaphane Activates a Lysosome-Dependent Transcriptional Program to Mitigate Oxidative Stress. Autophagy 2021, 17, 872–887. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, S.; Kasai, S.; Yamazaki, H.; Tatara, Y.; Mimura, J.; Engler, M.J.; Tanji, K.; Nikaido, Y.; Inoue, T.; Suganuma, H.; et al. Sulforaphane Increase Mitochondrial Biogenesis-Related Gene Expression in the Hippocampus and Suppresses Age-Related Cognitive Decline in Mice. Int. J. Mol. Sci. 2022, 23, 8433. [Google Scholar] [CrossRef] [PubMed]
- Yagishita, Y.; Fahey, J.W.; Dinkova-Kostova, A.T.; Kensler, T.W. Broccoli or Sulforaphane: Is It the Source or Dose That Matters? Molecules 2019, 24, 3593. [Google Scholar] [CrossRef] [PubMed]
- Issa, N.T.; Stathias, V.; Schürer, S.; Dakshanamurthy, S. Machine and Deep Learning Approaches for Cancer Drug Repurposing. Semin. Cancer Biol. 2021, 68, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Du, Y.; Li, L.; Wei, D.-Q. Bioinformatics Approaches for Anti-Cancer Drug Discovery. Curr. Drug Targets 2019, 21, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Patel, L.; Shukla, T.; Huang, X.; Ussery, D.W.; Wang, S. Machine Learning Methods in Drug Discovery. Molecules 2020, 25, 5277. [Google Scholar] [CrossRef] [PubMed]
- Zielinski, J.M.; Luke, J.J.; Guglietta, S.; Krieg, C. High Throughput Multi-Omics Approaches for Clinical Trial Evaluation and Drug Discovery. Front. Immunol. 2021, 12, 590742. [Google Scholar] [CrossRef]
- Cheng, F.; Kovács, I.A.; Barabási, A.-L. Network-Based Prediction of Drug Combinations. Nat. Commun. 2019, 10, 1197. [Google Scholar] [CrossRef]
- Lagunas-Rangel, F.A. Prediction of Resveratrol Target Proteins: A Bioinformatics Analysis. J. Biomol. Struct. Dyn. 2023, 1–10. [Google Scholar] [CrossRef]
- Lagunas-Rangel, F.A. Bioinformatic Analysis of SIRT7 Sequence and Structure. J. Biomol. Struct. Dyn. 2023, 41, 8081–8091. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated Data and New Features for Efficient Prediction of Protein Targets of Small Molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Nickel, J.; Gohlke, B.-O.; Erehman, J.; Banerjee, P.; Rong, W.W.; Goede, A.; Dunkel, M.; Preissner, R. SuperPred: Update on Drug Classification and Target Prediction. Nucleic Acids Res. 2014, 42, W26–W31. [Google Scholar] [CrossRef]
- Yao, Z.-J.; Dong, J.; Che, Y.-J.; Zhu, M.-F.; Wen, M.; Wang, N.-N.; Wang, S.; Lu, A.-P.; Cao, D.-S. TargetNet: A Web Service for Predicting Potential Drug–Target Interaction Profiling via Multi-Target SAR Models. J. Comput. Aided Mol. Des. 2016, 30, 413–424. [Google Scholar] [CrossRef]
- Edgar, R. Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilo, J. G:Profiler—A Web-Based Toolset for Functional Profiling of Gene Lists from Large-Scale Experiments. Nucleic Acids Res. 2007, 35, W193–W200. [Google Scholar] [CrossRef] [PubMed]
- von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A Database of Predicted Functional Associations between Proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein–Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, R.; Nakamura, S.; Nakazawa, T.; Minoura, K.; Yoshida, T.; Nishi, Y.; Kobayashi, Y.; Ohkubo, T. Structure and Reaction Mechanism of Human Nicotinamide Phosphoribosyltransferase. J. Biochem. 2010, 147, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC 15—Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a Protein-Small Molecule Docking Web Service Based on EADock DSS. Nucleic Acids Res. 2011, 39, W270–W277. [Google Scholar] [CrossRef]
- Yao, H.; Liu, M.; Wang, L.; Zu, Y.; Wu, C.; Li, C.; Zhang, R.; Lu, H.; Li, F.; Xi, S.; et al. Discovery of Small-Molecule Activators of Nicotinamide Phosphoribosyltransferase (NAMPT) and Their Preclinical Neuroprotective Activity. Cell Res. 2022, 32, 570–584. [Google Scholar] [CrossRef]
- Case, D.; Aktulga, H.; Belfon, K.; Ben-Shalom, I.Y.; Berryman, J.T.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E.; Cisneros, G.A.; Cruzeiro, V.W.D.; et al. Amber 2022. 2022. Available online: https://ambermd.org/doc12/Amber20.pdf (accessed on 2 January 2024).
- Ponder, J.W.; Case, D.A. Force Fields for Protein Simulations. In Advances in Protein Chemistry; Academic Press: Cambridge, MA, USA, 2003; pp. 27–85. [Google Scholar]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, Efficient Generation of High-Quality Atomic Charges. AM1-BCC Model: II. Parameterization and Validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.Py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Healy, Z.R.; Liu, H.; Holtzclaw, W.D.; Talalay, P. Inactivation of Tautomerase Activity of Macrophage Migration Inhibitory Factor by Sulforaphane: A Potential Biomarker for Anti-Inflammatory Intervention. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1516–1523. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, H.; Dong, N.; Su, X.; Duan, M.; Wei, Y.; Wei, J.; Liu, G.; Peng, Q.; Zhao, Y. Sulforaphane Induces S-Phase Arrest and Apoptosis via P53-Dependent Manner in Gastric Cancer Cells. Sci. Rep. 2021, 11, 2504. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.-K.; Mi, Y.-H.; Xiong, X.-Y.; Wang, X.-H. Sulforaphane Ameliorates the Intestinal Injury in Necrotizing Enterocolitis by Regulating the PI3K/Akt/GSK-3β Signaling Pathway. Can. J. Gastroenterol. Hepatol. 2022, 2022, 6529842. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Jiang, H.; Li, S.; Han, B.; Liu, Y.; Yang, D.; Li, J.; Yang, Q.; Wu, P.; Zhang, Z. Sulforaphane Prevents Chromium-Induced Lung Injury in Rats via Activation of the Akt/GSK-3β/Fyn Pathway. Environ. Pollut. 2020, 259, 113812. [Google Scholar] [CrossRef]
- Shang, G.; Tang, X.; Gao, P.; Guo, F.; Liu, H.; Zhao, Z.; Chen, Q.; Jiang, T.; Zhang, N.; Li, H. Sulforaphane Attenuation of Experimental Diabetic Nephropathy Involves GSK-3 Beta/Fyn/Nrf2 Signaling Pathway. J. Nutr. Biochem. 2015, 26, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zou, Y.; Zhuang, X.; Chen, S.; Lin, Y.; Li, W.; Lin, J.; Lin, Z. Sulforaphane Suppresses EMT and Metastasis in Human Lung Cancer through MiR-616-5p-Mediated GSK3β/β-Catenin Signaling Pathways. Acta Pharmacol. Sin. 2017, 38, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Piberger, A.L.; Keil, C.; Platz, S.; Rohn, S.; Hartwig, A. Sulforaphane Inhibits Damage-Induced Poly (ADP-Ribosyl)Ation via Direct Interaction of Its Cellular Metabolites with PARP-1. Mol. Nutr. Food Res. 2015, 59, 2231–2242. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tang, Y. Sulforaphane Ameliorates Amyloid-β-Induced Inflammatory Injury by Suppressing the PARP1/SIRT1 Pathway in Retinal Pigment Epithelial Cells. Bioengineered 2022, 13, 7079–7089. [Google Scholar] [CrossRef]
- Çakır, I.; Lining Pan, P.; Hadley, C.K.; El-Gamal, A.; Fadel, A.; Elsayegh, D.; Mohamed, O.; Rizk, N.M.; Ghamari-Langroudi, M. Sulforaphane Reduces Obesity by Reversing Leptin Resistance. Elife 2022, 11, e67368. [Google Scholar] [CrossRef] [PubMed]
- Schepici, G.; Bramanti, P.; Mazzon, E. Efficacy of Sulforaphane in Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 8637. [Google Scholar] [CrossRef]
- Russo, M.; Spagnuolo, C.; Russo, G.L.; Skalicka-Woźniak, K.; Daglia, M.; Sobarzo-Sánchez, E.; Nabavi, S.F.; Nabavi, S.M. Nrf2 Targeting by Sulforaphane: A Potential Therapy for Cancer Treatment. Crit. Rev. Food Sci. Nutr. 2018, 58, 1391–1405. [Google Scholar] [CrossRef]
- Abassi, P.; Abassi, F.; Yari, F.; Hashemi, M.; Nafisi, S. Study on the Interaction of Sulforaphane with Human and Bovine Serum Albumins. J. Photochem. Photobiol. B Biol. 2013, 122, 61–67. [Google Scholar] [CrossRef]
- Rorke, E.A.; Adhikary, G.; Szmacinski, H.; Lakowicz, J.R.; Weber, D.J.; Godoy-Ruiz, R.; Puranik, P.; Keillor, J.W.; Gates, E.W.J.; Eckert, R.L. Sulforaphane Covalently Interacts with the Transglutaminase 2 Cancer Maintenance Protein to Alter Its Structure and Suppress Its Activity. Mol. Carcinog. 2022, 61, 19–32. [Google Scholar] [CrossRef]
- Massudi, H.; Grant, R.; Guillemin, G.J.; Braidy, N. NAD + Metabolism and Oxidative Stress: The Golden Nucleotide on a Crown of Thorns. Redox Rep. 2012, 17, 28–46. [Google Scholar] [CrossRef]
- Garten, A.; Schuster, S.; Penke, M.; Gorski, T.; de Giorgis, T.; Kiess, W. Physiological and Pathophysiological Roles of NAMPT and NAD Metabolism. Nat. Rev. Endocrinol. 2015, 11, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.M.; Park, C.W.; Kim, S.W.; Nam, Y.J.; Yu, J.H.; Shin, J.H.; Yun, C.H.; Im, S.-H.; Kim, K.-T.; Sung, Y.C.; et al. NAMPT Suppresses Glucose Deprivation-Induced Oxidative Stress by Increasing NADPH Levels in Breast Cancer. Oncogene 2016, 35, 3544–3554. [Google Scholar] [CrossRef] [PubMed]
- Oka, S.; Byun, J.; Huang, C.; Imai, N.; Ralda, G.; Zhai, P.; Xu, X.; Kashyap, S.; Warren, J.S.; Alan Maschek, J.; et al. Nampt Potentiates Antioxidant Defense in Diabetic Cardiomyopathy. Circ. Res. 2021, 129, 114–130. [Google Scholar] [CrossRef] [PubMed]
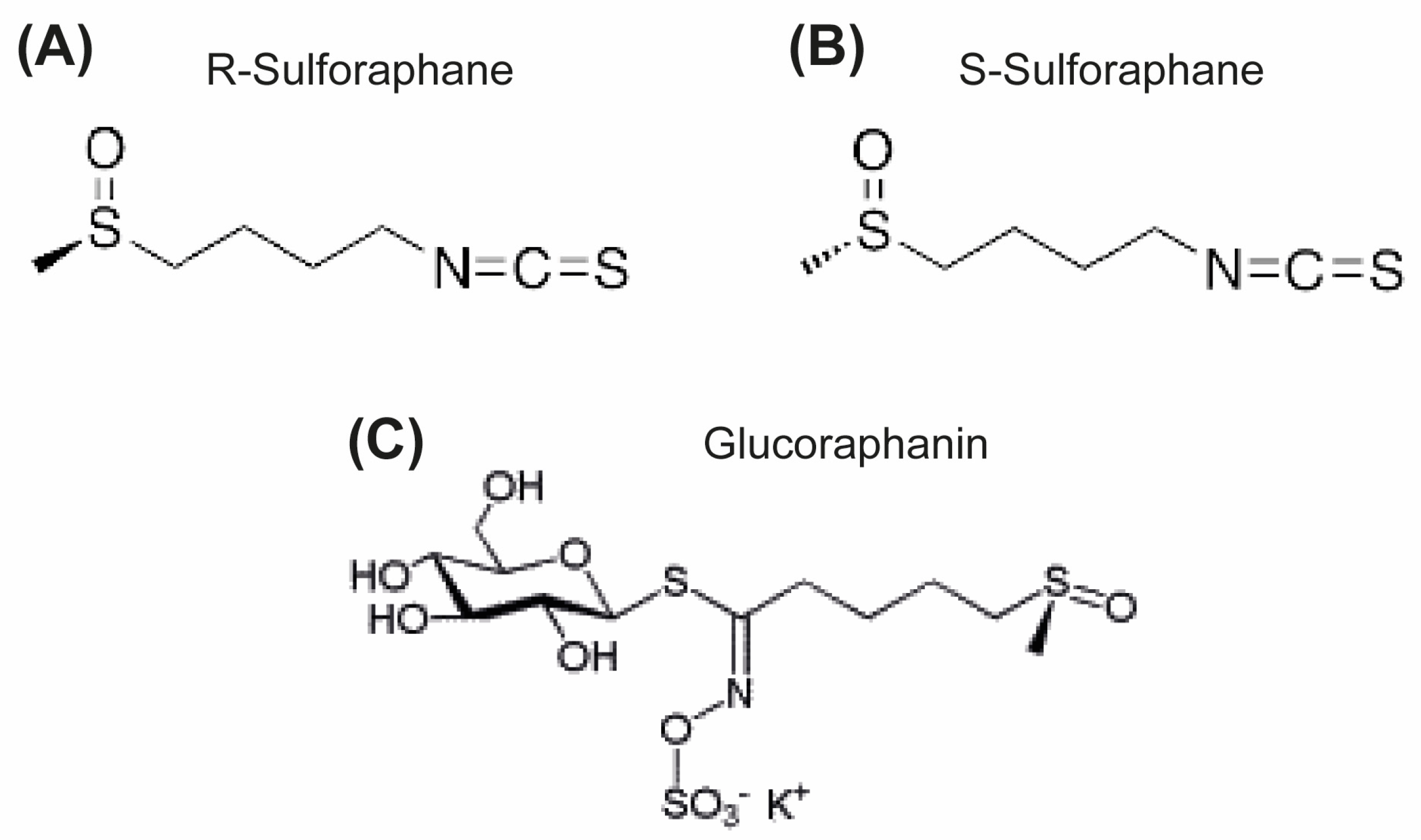
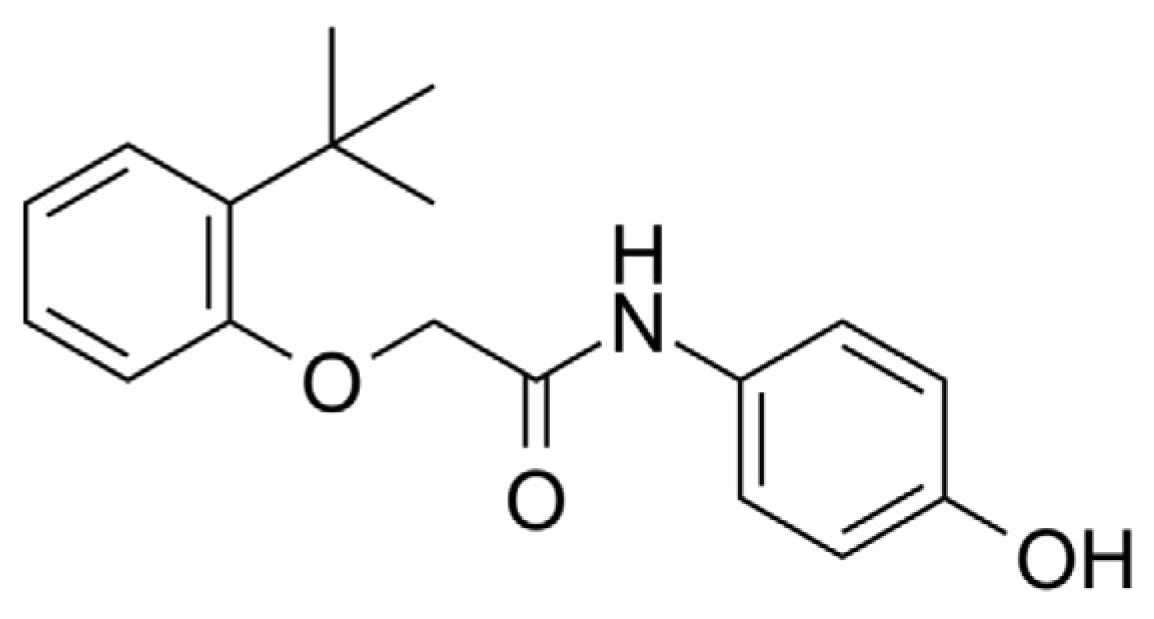


| Target | Gene | Uniprot ID | ChEMBL ID | Target Class |
|---|---|---|---|---|
| Macrophage migration inhibitory factor | MIF | P14174 | CHEMBL2085 | Tautomerase |
| Cytochrome P450 19A1 | CYP19A1 | P11511 | CHEMBL1978 | Aromatase |
| Nicotinamide phosphoribosyltransferase | NAMPT | P43490 | CHEMBL1744525 | Transferase |
| Poly [ADP-ribose] polymerase-1 | PARP1 | P09874 | CHEMBL3105 | Transferase |
| P2X purinoceptor 7 | P2RX7 | Q99572 | CHEMBL4805 | Ligand-gated ion channel |
| Glycogen synthase kinase-3 β | GSK3B | P49841 | CHEMBL262 | Kinase |
| Cyclin-dependent kinase 2 | CDK2 | P24941 | CHEMBL301 | Kinase |
| Cathepsin L | CTSL | P07711 | CHEMBL3837 | Protease |
| Adenosine A1 receptor | ADORA1 | P30542 | CHEMBL226 | Family A G protein-coupled receptor |
| Adenosine A2a receptor | ADORA2A | P29274 | CHEMBL251 | Family A G protein-coupled receptor |
| Monoamine oxidase A | MAOA | P21397 | CHEMBL1951 | Oxidoreductase |
| Upregulated | ||||||
|---|---|---|---|---|---|---|
| Gene | logFC | AveExpr | t | p Value | adj.P.Val | B |
| LINC00536 | 7.64755127 | −3.0149304 | 21.9961465 | 0.00024888 | 0.03788923 | 0.36312489 |
| IL1R2 | 7.05727373 | −3.3119119 | 38.9200915 | 2.15 × 10−5 | 0.0314058 | 1.11208622 |
| BBOX1-AS1 | 5.82578022 | −3.9274667 | 32.9561069 | 3.65 × 10−5 | 0.0314058 | 0.96501409 |
| PLEKHS1 | 5.70468219 | −3.9959986 | 13.8792101 | 0.00094197 | 0.04820704 | −0.1886544 |
| AC105177.1 | 5.62076293 | −4.0337932 | 28.649169 | 5.71 × 10−5 | 0.0314058 | 0.88104941 |
| LAMC3 | 5.49154119 | −4.1016187 | 15.2901064 | 0.00069452 | 0.04291078 | −0.0618761 |
| TNIP3 | 5.4717553 | −4.1066281 | 35.425051 | 2.90 × 10−5 | 0.0314058 | 0.95355416 |
| AC020571.1 | 5.30127359 | −4.1922612 | 32.8798182 | 3.68 × 10−5 | 0.0314058 | 0.90642193 |
| AKR1C3 | 5.09234014 | 5.97048189 | 18.5451872 | 0.00040802 | 0.04045807 | 0.71525274 |
| LINC00565 | 4.88357902 | −4.402276 | 25.9281408 | 7.85 × 10−5 | 0.03315288 | 0.73875417 |
| Downregulated | ||||||
| Gene | logFC | AveExpr | t | p Value | adj.P.Val | B |
| DCN | −5.9219991 | −3.5794442 | −13.758792 | 0.00096589 | 0.04824652 | −0.1635847 |
| MARCOL | −5.2456221 | −3.9144047 | −30.897349 | 4.49 × 10−5 | 0.0314058 | 0.94001916 |
| NKX6-1 | −4.9009606 | −4.0843505 | −26.237668 | 7.56 × 10−5 | 0.03284137 | 0.82265699 |
| TRPC6 | −4.3352769 | −4.3671491 | −23.63015 | 0.00010555 | 0.03426141 | 0.69136095 |
| AC138866.2 | −4.2150679 | −4.4272963 | −23.013573 | 0.00011483 | 0.03426141 | 0.65751575 |
| LINC02208 | −4.2045923 | −4.4300338 | −14.309786 | 0.00062259 | 0.04291078 | 0.0378538 |
| AC016026.1 | −4.1467169 | −4.4601074 | −17.831801 | 0.0002586 | 0.03811126 | 0.43408431 |
| AC106738.1 | −4.0685678 | −4.5062475 | −13.933678 | 0.00066005 | 0.04291078 | 0.0053519 |
| AC008687.6 | −3.8557043 | −4.6052296 | −16.118801 | 0.00035632 | 0.03919762 | 0.28826772 |
| AL121761.2 | −3.8557043 | −4.6052296 | −16.118801 | 0.00035632 | 0.03919762 | 0.28826772 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lagunas-Rangel, F.A. Sulforaphane Target Protein Prediction: A Bioinformatics Analysis. Appl. Sci. 2024, 14, 1052. https://doi.org/10.3390/app14031052
Lagunas-Rangel FA. Sulforaphane Target Protein Prediction: A Bioinformatics Analysis. Applied Sciences. 2024; 14(3):1052. https://doi.org/10.3390/app14031052
Chicago/Turabian StyleLagunas-Rangel, Francisco Alejandro. 2024. "Sulforaphane Target Protein Prediction: A Bioinformatics Analysis" Applied Sciences 14, no. 3: 1052. https://doi.org/10.3390/app14031052





