Abstract
In Cone Beam Computed Tomography (CBCT) images, accurate tooth segmentation is crucial for oral health, providing essential guidance for dental procedures such as implant placement and difficult tooth extractions (impactions). However, due to the lack of a substantial amount of dental data and the complexity of tooth morphology in CBCT images, the task of tooth segmentation faces significant challenges. This may lead to issues such as overfitting and training instability in existing algorithms, resulting in poor model generalization. Ultimately, this may impact the accuracy of segmentation results and could even provide incorrect diagnostic and treatment information. In response to these challenges, we introduce PPA-SAM, an innovative dual-encoder segmentation network that merges the currently popular Segment Anything Model (SAM) with the 3D medical segmentation network, VNet. Through the use of adapters, we achieve parameter reuse and fine-tuning, enhancing the model’s adaptability to specific CBCT datasets. Simultaneously, we utilize a three-layer convolutional network as both a discriminator and a generator for adversarial training. The PPA-SAM model seamlessly integrates the high-precision segmentation performance of convolutional networks with the outstanding generalization capabilities of SAM models, achieving more accurate and robust three-dimensional tooth segmentation in CBCT images. Evaluation of a small CBCT dataset demonstrates that PPA-SAM outperforms other networks in terms of accuracy and robustness, providing a reliable and efficient solution for three-dimensional tooth segmentation in CBCT images. This research has a positive impact on the management of dentofacial conditions from oral implantology to orthognathic surgery, offering dependable technological support for future oral diagnostics and treatment planning.
1. Introduction
In the field of oral medicine, precise tooth segmentation is crucial for diagnosing oral diseases, formulating effective treatment plans, and restoring patients’ oral structures [1,2]. Medical imaging techniques such as X-rays, Cone Beam Computed Tomography (CBCT), Magnetic Resonance Imaging (MRI), etc., provide rich information about oral structures. By accurately segmenting tooth models from CBCT images, oral healthcare professionals can achieve more precise diagnoses of oral diseases, formulate personalized treatment plans effectively, and identify surface lesions and damages on teeth. This enables doctors to pinpoint and address specific areas with greater accuracy. For the restoration and reconstruction of oral structures, precise tooth segmentation provides crucial guidance for planning procedures such as dental implant placement and other deno-alveolar surgeries that involve cutting bone with minimal damage to roots. The bone cutting could be with burs or piezotome to remove impacted teeth or when performing corticotomies or osteotomies to remove bone blocks for grafting or accelerate orthodontic treatment, ensuring coordination with surrounding oral tissues. In addition, in orthognathic surgery, segmentation is equally crucial to prevent damage to tooth roots during osteotomy. For instance, in cases where the widening of the maxilla or the pushback of the premaxilla is planned, or when vertical movement of a section of the upper or lower jaw is required to correct an open bite. Artificial Intelligence has the potential to detect the severity of malocclusion and score it using occlusal indices or IOFTN [3]. Subsequently, this information can be utilized during orthognathic surgery to generate accurate segmentation and fabricate custom wafers [4]. Therefore, precise tooth segmentation not only enhances the accuracy of diagnostics in oral medicine but also promotes the precision of treatments, contributing to the effective restoration of oral structures.
Before the advent of machine learning, researchers predominantly relied on the level-set method for the segmentation of teeth CBCT images [5,6]. However, the level-set method typically requires an initial contour, which can impact the final segmentation results. Selecting a suitable initial contour may demand expertise or other prior information. Due to the potential existence of complex structures such as root canals and periodontal ligaments in CBCT images of teeth, the presence of these structures may make it challenging for the level set method to accurately segment the contours of each structure. With the advent of machine learning and neural networks, researchers have proposed various novel medical image segmentation networks, significantly reducing the time required for manual annotation by radiologists [7]. These networks not only diminish the subjective errors introduced by manual annotation but also enhance the stability and accuracy of segmentation results. Consequently, some researchers have applied deep learning methods to 3D tooth segmentation tasks [8,9,10,11,12]. This implies that doctors can rely more on these automated tools to quickly obtain precise segmentation results, providing more reliable support for patient diagnosis and treatment. Traditional convolutional networks often require extensive datasets to support them and are primarily designed for specific segmentation tasks, with weaker generalization performance.
Recently, the Segment Anything Model (SAM) [13] has emerged, showcasing outstanding zero-shot segmentation performance in various everyday image segmentation tasks. This model, proposed by Meta AI, has pushed the boundaries of segmentation and significantly advanced the development of foundational models in computer vision. However, due to SAM’s relatively limited training samples on medical image data, it exhibits a certain gap when applied to medical image segmentation tasks compared to existing models with precise segmentation and excellent performance. In some tasks, SAM even fails to correctly identify medical target images [14,15]. To harness SAM’s potential in the field of medical image segmentation, some researchers have made improvements to the SAM model. They froze the original parameters of the SAM model and introduced several optimizers in the image encoder. Fine-tuning these optimizer parameters allows the model to better adapt to specific medical image segmentation tasks [16]. However, these approaches are limited to processing individual slices in medical images, neglecting the interconnections between different slices. In comparison to three-dimensional holistic segmentation, the accuracy achieved by segmenting and subsequently stitching together individual slices is lower.
To address these issues, this paper proposes an effective plug-and-play three-dimensional tooth CBCT image segmentation architecture named VAG-SAM. The overall architecture of the network is a generative adversarial network composed of a discriminator and a generator. The generator consists of VNet and a SAM encoder with optimizers, while the discriminator is a three-layer convolutional network. VAG-SAM combines the SAM model with traditional convolutional models, fully integrating high-precision segmentation capability and outstanding generalization ability, demonstrating excellent performance in small-sample three-dimensional tooth CBCT image segmentation tasks.
The main contributions of this paper are as follows:
- We propose PPA-SAM, an innovative fusion method of the Visual Grand Model and traditional convolutional networks. By cleverly integrating the improved SAM encoder into VNet, forming a dual-encoder structure, it significantly enhances the multi-angle feature extraction capability.
- While retaining the reusable pre-trained weights of the SAM encoder, we introduce optimizers for parameter fine-tuning, making the segmentation network more suitable for three-dimensional tooth segmentation tasks while preserving the advantages of the large model. The combination of GAN (Generative Adversarial Networks) and SAM improves the network’s generalization performance, and the utilization of VNet enhances the network’s feature extraction capability, demonstrating excellent performance in small-sample tasks.
- We conducted a comprehensive performance evaluation on a small-sample 3D CBCT tooth dataset, comparing it with other advanced networks and considering different scales of training sample sizes. In this series of experiments, PPA-SAM exhibited outstanding segmentation performance, providing a solid foundation for addressing three-dimensional tooth segmentation tasks.
2. Related Work
2.1. Visual Fundamental Models
Convolutional Neural Networks (CNNs) play a pivotal role in computer vision, demonstrating remarkable success in tasks such as image classification, object detection, and semantic segmentation. Recently, U-shaped segmentation networks, as exemplified by the U-Net architecture proposed by Ronneberger et al. [17], consist primarily of an encoder and a decoder, forming a U-shaped structure. The encoder is responsible for feature extraction, while the decoder maps features back to the segmented result of the original input image. U-Net finds broad applications in fields such as medical image segmentation and satellite image analysis. Subsequently, numerous networks based on U-Net improvements have emerged [18,19,20]. For example, UNet++ [19] adopts a nested structure and dense connections to better capture multi-scale and hierarchical features, thereby enhancing the expressiveness of U-Net. It is widely applied in tasks such as medical image segmentation and natural image segmentation.
Simultaneously, the Transformer, as a model capable of capturing global information and pixel relationships in images, has garnered significant attention from researchers for its integration into the field of image processing. TransUNet [18], based on the attention mechanism of the Transformer, has the ability to capture global contextual information, thereby contributing to a better understanding of the overall structure of images. Dosovitskiy et al. [21] introduced the Vision Transformer (ViT), which utilizes a self-attention mechanism to process images. Prior to inputting images into ViT, they are segmented into blocks and then mapped into a sequence of vectors suitable for processing by a Transformer. This approach enables ViT to excel in handling images of different sizes and effectively capturing long-range contextual information.
The success of ViT underscores the significance of self-attention mechanisms in the field of computer vision. However, traditional U-shaped segmentation networks and Transformer-based networks often require extensive training datasets, leading to relatively poor generalization performance. With the continuous development of techniques such as adversarial training [22,23,24] and pre-training [19,25], the robustness and generalization capabilities of deep learning models have significantly improved.
Adversarial training enhances the robustness of neural networks against adversarial attacks by introducing adversarial examples during the training process, thereby preventing adversarial inputs. This method is widely applied in various domains such as image classification, speech recognition, and natural language processing. It makes models more resilient to targeted perturbations, preventing the model from being misled. Pretraining involves training a model on a large-scale dataset to learn generic feature representations, thereby enhancing its performance on related tasks. Widely applied in the fields of image processing and natural language processing, this approach strengthens the model’s generalization capabilities. CLIP [26], through pre-training on large-scale image-text pairs, successfully learned the connections between images and text, demonstrating strong generalization capabilities. Generative Pre-trained Transformer (GPT) is a language model based on the Transformer architecture. Through self-supervised learning pretraining on extensive text data, it acquires universal language representations. GPT excels in various natural language processing tasks, including text generation, text classification, and language understanding, attracting significant attention due to its versatility and generalization capabilities. OpenAI’s DALL·E 2 [27] adopted a pre-training mechanism similar to GPT [28], achieving significant improvements in generating diversity, image quality, and understanding text descriptions, expanding the model’s application range and performance. However, these large-scale models often encounter substantial data pressure and computational demands during training, requiring a significant amount of high-quality data to ensure the model learns diverse language representations adequately.
2.2. Fine-Tuning Models
In the field of machine learning, fine-tuning models have been widely employed in transfer learning tasks. Fine-tuning refers to the process of further training a neural network model that has already undergone pre-training, using data specific to a particular task. The aim is to adapt the model more effectively to the new task, thereby enhancing its performance.
Adapters are lightweight structures introduced into the model, allowing specific task fine-tuning on a pre-trained model without modifying the overall architecture. Adapters provide a means of preserving generality while facilitating flexible fine-tuning for specific tasks. For example, the GPT [28] model proposed by OpenAI is a pre-trained generative model based on the Transformer architecture. Widely employed in natural language processing tasks such as text generation, text classification, and dialogue generation, GPT demonstrates powerful language understanding and generation capabilities. During the pre-training phase, GPT learns general language representations through unsupervised learning on massive text data. Subsequently, it adapts to specific downstream tasks through parameter fine-tuning. ViT-Adapter [29] is an approach that introduces adapter layers into the Visual Transformer (ViT) model, allowing for customized task fine-tuning on pre-trained models to enhance performance on specific visual tasks. SAM [13] is a universal image segmentation model proposed by Kirillov et al. Pre-trained on over 11 million images and utilizing over a billion masks, SAM has demonstrated outstanding performance in image segmentation tasks. This success highlights the importance of large-scale data and powerful pre-training in the field of deep learning. These research achievements collectively drive the development of computer vision, providing more powerful and flexible solutions for various application scenarios. SAM segments targets through single points or borders for target hints, and its excellent segmentation performance is attributed to the fact that its pre-training data mostly have clear and regular boundaries.
However, in medical images, the boundaries of tissues and content are often fuzzy, rendering SAM unable to accurately identify target content through local information alone. To enhance SAM’s adaptability to downstream tasks, some researchers have employed parameter fine-tuning techniques to improve its performance. MedSAM [30] utilizes parameter-efficient fine-tuning (PEFT) to fine-tune the pre-trained SAM model, demonstrating excellent performance in medical image segmentation. SAM-Med 2D [14] introduces adapters into the encoder for fine-tuning, making it well-suited for the medical domain. The Medical SAM Adapter [16] retains most parameters of the SAM model and applies popular adapters from NLP techniques to medical image segmentation, showcasing surprisingly good performance.
Despite their success, fine-tuning models encounter challenges, including the need for large-scale annotated data, high computational costs, and the requirement for adaptability in new domains. Fine-tuning may also face constraints due to conceptual differences during domain transfer, necessitating careful adjustments to accommodate varying tasks and data distributions.
2.3. Medical Image Segmentation
Medical image segmentation holds significant importance in clinical diagnosis, treatment planning, and disease research [31]. Accurate segmentation results can assist doctors in precisely locating and quantifying lesion areas, providing crucial diagnostic information, and supporting disease diagnosis and treatment. For instance, MRI brain image segmentation is employed to localize and quantify brain structures such as the cerebral cortex, hippocampus, and basal ganglia. This is crucial for studying the impact of brain disorders like epilepsy, Alzheimer’s disease, and brain tumors, as well as for planning surgical procedures. In CT lung imaging, medical image segmentation assists in locating and quantifying pulmonary structures, thereby supporting the diagnosis and treatment of diseases such as lung cancer and chronic obstructive pulmonary disease (COPD). In dermatology, image segmentation techniques aid doctors in identifying and analyzing the boundaries of skin lesions, facilitating diagnosis and treatment planning, especially in the early detection of skin cancer.
However, medical images often exhibit complex structures and diversity. Magnetic Resonance Imaging (MRI) utilizes magnetic fields and harmless radio waves to generate high-contrast images with detailed anatomical information. Medical image segmentation in MRI faces several challenges, including the integration of multi-channel information, handling high-resolution complex structures, coping with intense contrast variations, addressing the need for local and global consistency, and sensitivity to noise and artifacts. Computed Tomography (CT) images, obtained through X-ray imaging, provide detailed cross-sectional images of internal body structures. Automatically segmenting medical images in CT also faces several challenges, including artifact interference, radiation dose noise, voxel resolution differences, and a wide dynamic range. These issues make segmenting complex structures and small features from the images more challenging.
Compared to 2D images, 3D images can provide more spatial information, better representing the three-dimensional structures and morphological features of organs and lesions. Some researchers have proposed segmentation networks based on 3D convolutions [32,33,34,35], applying convolution operations directly to 3D data. With the emergence of SAM models based on large datasets, continuous improvements and adaptations of segmentation models for medical image tasks have been proposed [14,15,16].
2.4. CBCT Tooth Segmentation
Segmenting tooth models from CBCT images holds significant importance for the subsequent diagnosis and treatment of dentists [1,2]. For example, accurate tooth models assist dentists in detailed assessments of the morphology, density, and structure of the alveolar bone, aiding in the diagnosis of various bone disorders such as fractures, cysts, or inflammation. In dental implant surgery, CBCT tooth segmentation provides crucial information for planning the position, angle, and depth of implants. By precisely segmenting tooth and alveolar bone structures, doctors can evaluate the suitability of implants and determine the optimal surgical approach. Before the advent of deep learning, people commonly used variants of level-set methods [5,6] and incorporated prior knowledge for tooth segmentation. With the emergence of deep learning, automated tooth segmentation from CBCT using neural networks has been explored [10,11,12]. However, accurate tooth segmentation from CBCT images remains a challenging task due to the following reasons: (1) The shape and size differences between different teeth in CBCT images, as well as their intersections with surrounding tissues, make the segmentation task complex. (2) Acquiring a large amount of three-dimensional CBCT image data is relatively difficult, leading to overfitting and insufficient generalization ability of deep learning models in small sample situations. (3) CBCT images provide rich three-dimensional information, but traditional two-dimensional segmentation methods may not fully leverage this information, making it challenging to effectively process and integrate three-dimensional information.
3. Methodology
3.1. Architecture Overview
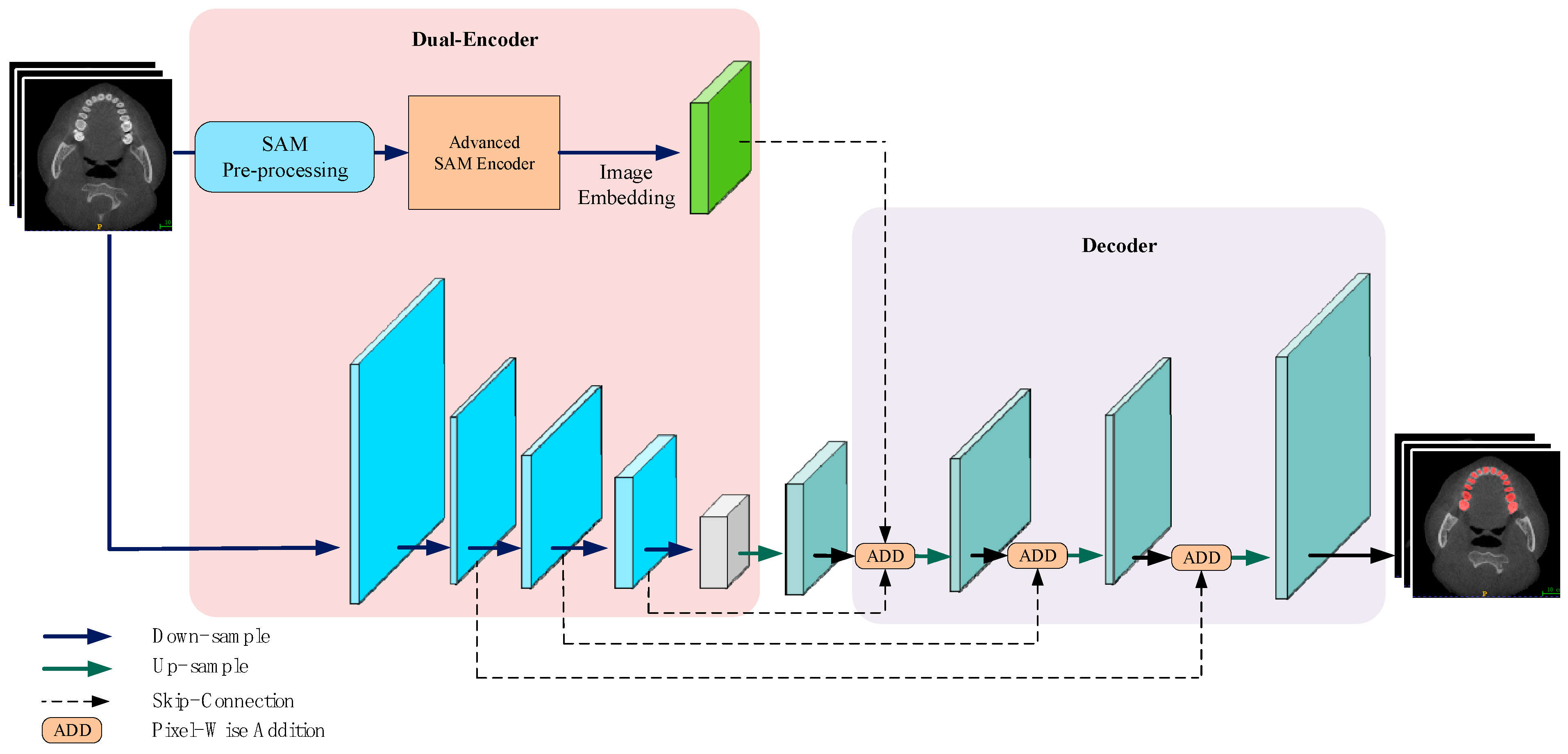
The proposed PPA-SAM, as illustrated in Figure 1, is an overall generative adversarial network architecture consisting of a generator and a discriminator. They undergo adversarial training to enhance the quality of the output labels. The generator integrates the strengths of VNet and SAM, comprising two parallel encoders: the VNet encoder and the enhanced SAM encoder, where the enhanced SAM encoder retains the original parameters for reuse. The outputs of these two encoders are concatenated and passed to the decoding layer, resulting in the prediction labels. The discriminator consists of three layers of convolutional networks designed to judge the authenticity of input samples. The discriminator’s output is utilized for backward propagation to both the generator and discriminator, enhancing the overall performance.
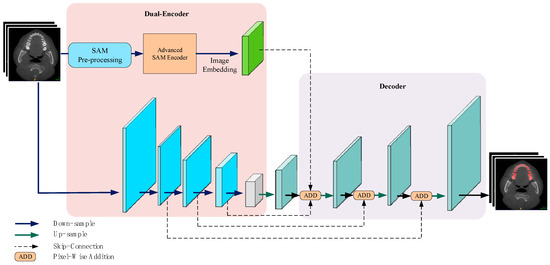
Figure 1.
PPA-SAM integrates the enhanced SAM encoder into the VNet network, creating a dual-encoder structure. The outputs from both the VNet encoder and the enhanced SAM encoder are combined and input into the decoder, allowing PPA-SAM to achieve three-dimensional segmentation of teeth.
3.2. VNet Structure
VNet is a convolutional neural network structure designed for three-dimensional image segmentation. It has been widely applied in the field of medical image segmentation, especially showing significant advantages in segmentation tasks involving three-dimensional medical images such as CT (computed tomography) and MRI (magnetic resonance imaging). In the medical domain, precise and reliable image segmentation is crucial as it directly relates to accurate diagnosis and treatment decisions by medical professionals. VNet introduces skip connections, allowing end-to-end information transfer from input to output, which is crucial for handling complex data structures like medical images. In the encoder, features are progressively extracted and compressed through multiple convolutional blocks, concurrently reducing the size of feature maps. In the decoder section, upsampling and deconvolution operations restore the feature space, enabling the network to reconstruct and retain detailed image information. Skip connections are established between each layer of the decoder and its corresponding encoder layer, creating a close relationship between the encoder and decoder, allowing low-level features to be directly transmitted to the decoder, thereby enhancing the network’s ability to integrate multi-scale feature information. Additionally, VNet utilizes residual connections, a mechanism that helps the network better learn complex medical image features, particularly in accurately understanding organ boundaries and local features, leading to superior performance in medical image segmentation tasks.
3.3. Dual-Encoder Structure
The dual-encoder structure comprises the VNet encoder and an improved version of the SAM encoder. The enhanced SAM encoder, based on the Transformer architecture, aids in capturing global information and sequence relationships. VNet is more suitable for processing three-dimensional data in medical images and possesses good local feature extraction capabilities. This dual-encoder structure enables the model to benefit simultaneously from the strengths of both Transformer and convolutional neural networks, enriching feature representation across multiple levels, including global, local, and sequential information. This enhances segmentation accuracy in tasks such as medical image segmentation and improves the model’s generalization ability to adapt to a broader range of datasets and scenarios.
The improved SAM encoder is designed based on the prototype of the Vision Transformer (ViT). During fine-tuning, we freeze the original parameters of ViT and introduce adapters to better adapt to medical image tasks. To reduce the computational burden on the model and ensure adaptability after image feature extraction, we first linearly process the image size to fit the inputs of the improved SAM encoder and VNet, which are (1024 × 1024 × 1024) and (256 × 256 × 256), respectively. When inputting into the improved SAM encoder, the process starts with Patch Embedding, dividing the input image into fixed-size blocks and transforming each block into a deep feature vector. Patch Embedding uses a 2D convolution with a kernel size of 16, a stride of 16, and an output dimension of 768, resulting in 64 × 64 768-dimensional vectors. Next, we add positional encoding, which provides the model with positional information for each pixel or patch in the image, enabling the model to understand the spatial arrangement of objects in the image more accurately for segmentation tasks. Subsequently, 12 Transformer blocks are applied sequentially, enhancing the understanding and extraction of details in various parts of the image. Finally, we obtain a high-dimensional image embedding. To adapt to the output size of the VNet encoder, we use convolution for dimension reduction and concatenate the feature blocks obtained from both encoders for subsequent data reconstruction and prediction in the VNet decoder.
3.4. Adapter
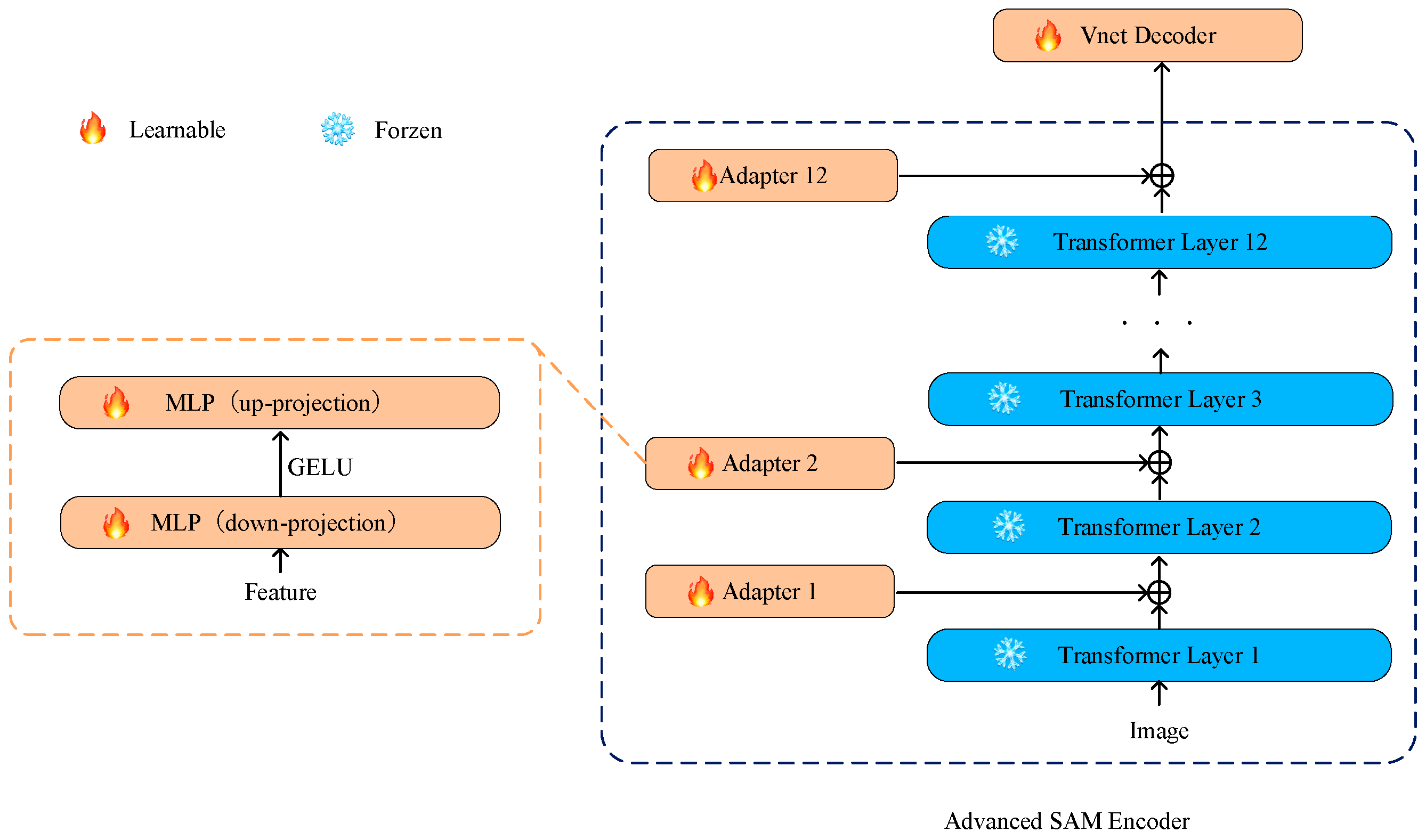
We adopted an effective strategy by introducing simple yet efficient adapters between each Transformer layer on the basis of freezing the parameters of the SAM encoder. This design allows fine-tuning for specific tasks while preserving the general features learned in the pretraining model to prevent excessive adjustments that might compromise the model’s general knowledge obtained from large-scale pretraining tasks. This adaptation helps the model better adapt to downstream tasks like medical image segmentation [36]. Additionally, this design helps prevent catastrophic forgetting, making the model more robust on new medical image segmentation tasks. The structure of the adapter is illustrated in Figure 2.
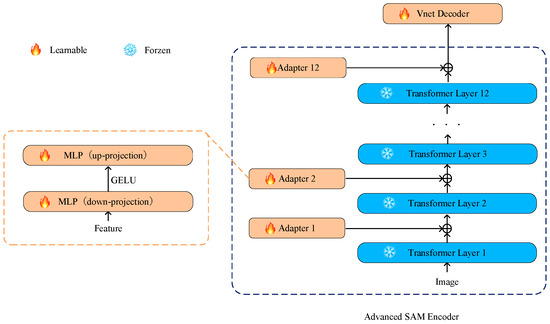
Figure 2.
The adapters are placed at the connections between each Transformer layer in the SAM encoder. The enhanced SAM encoder allows only the parameters of the optimizer to undergo iterative updates, while other parameters remain frozen.
We drew inspiration from SAM-Adapter [35] and designed the adapter as a bottleneck model, comprising a down-projecting MLP layer, the GELU activation function, and an up-projecting MLP layer. The formula is as follows:
Specifically, Fi represents the output of each Transformer layer, and MLPdown is responsible for down-projecting these outputs, compressing the input data to a lower dimension. The GELU function, introduced as the activation function, adds non-linearity and aids the model in learning more complex feature representations. Subsequently, the MLPup layer restores the data from the lower dimension to the original shape, achieving the up-projection of the adapter. This bottleneck structure design introduces more flexibility into the adapter, allowing the model to maintain computational efficiency while learning task-specific adaptability.
3.5. Discriminator
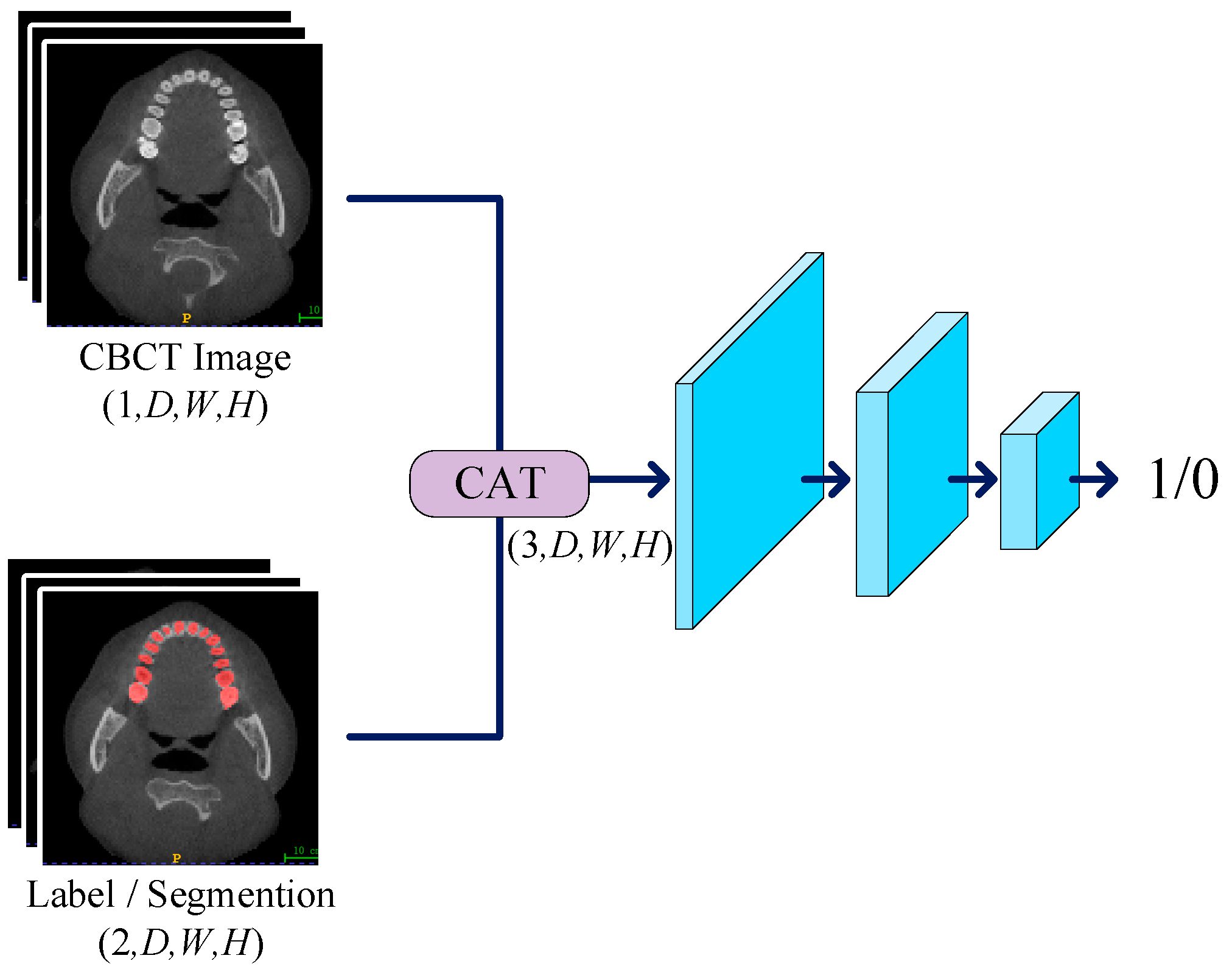
We devised a three-layer convolutional network to serve as the discriminator, working in conjunction with the generator for adversarial training. The primary role of the discriminator is to discern the authenticity of the input labels, and its network structure is illustrated in Figure 3. In the discriminator, we set the kernel size of the first two convolutional layers to 3, with a stride of 2 and padding of 1, resulting in a halved data shape after each pass through these layers. For the third convolutional layer, the kernel size is set to 1, with a stride of 1 and no padding to maintain the shape. The LeakyReLU activation function is employed in the hidden layers with a negative slope set to 0.05. When inputting data into the discriminator, CT images are concatenated with labels as conditions. This concatenation strategy enhances the discriminator’s sensitivity, allowing it to more accurately assess the authenticity of input labels. During downsampling, we progressively reduce the spatial resolution of the input data while retaining essential feature information. Finally, we utilize the Sigmoid function as the activation function, producing outputs within the range of 0 to 1. The discriminator’s output can be interpreted as the authenticity of the input labels, providing a basis for subsequent updates to the discriminator and generator parameters.
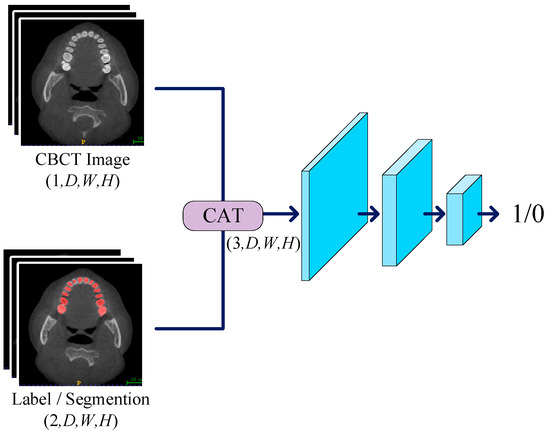
Figure 3.
The CT image is concatenated with constraint conditions and labels before being input into the discriminator. The discriminator sequentially extracts feature information layer by layer, ultimately producing a value between 0 and 1, which is used for parameter updates.
3.6. Generative Adversarial Structure
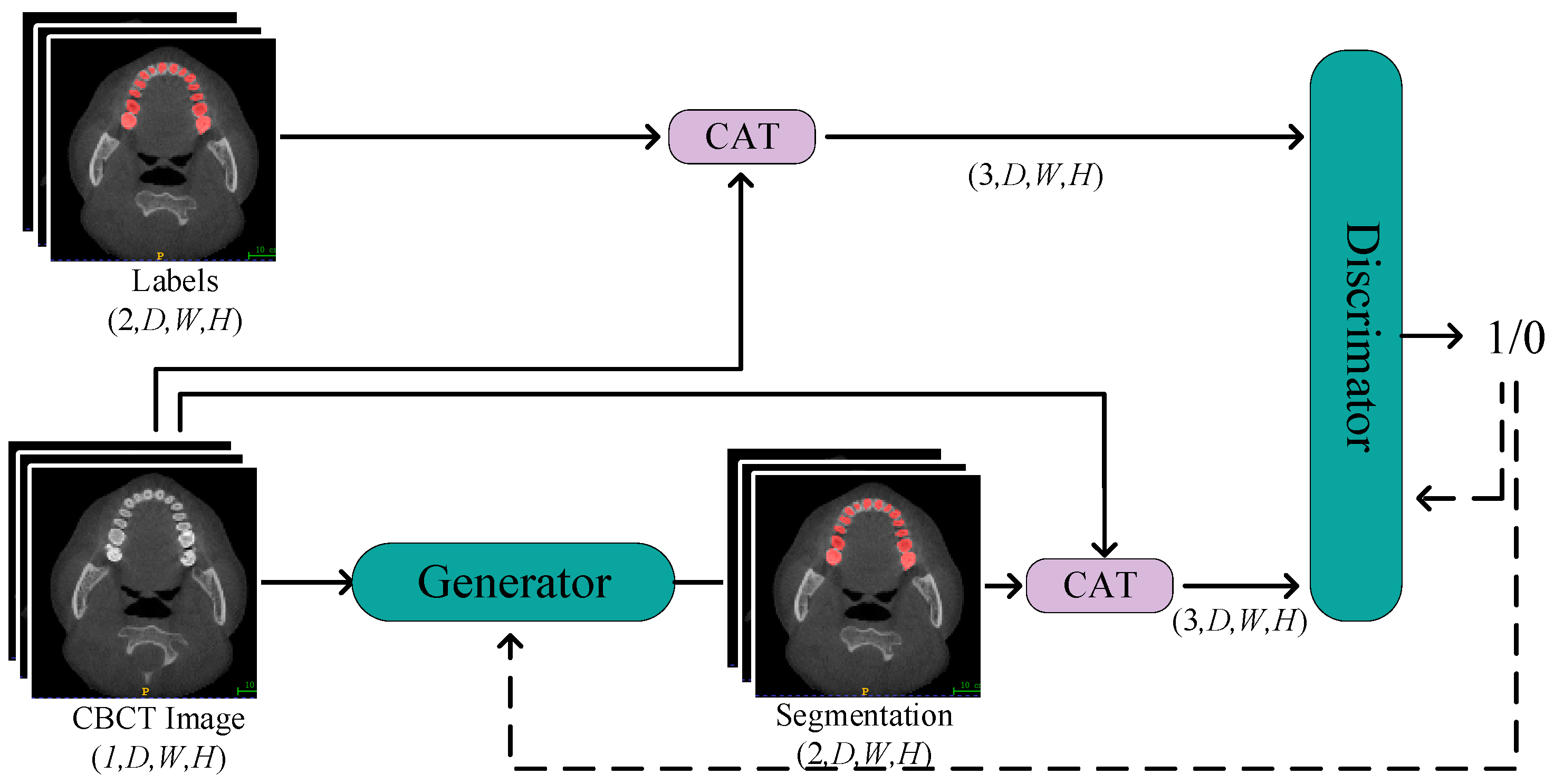
We employed a segmentation network with a dual-encoder as the generator, working in tandem with the discriminator to constitute adversarial training. The training process is depicted in Figure 4.
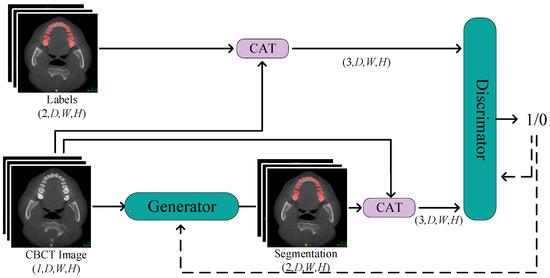
Figure 4.
The Process of Generative Adversarial Training. Through continuous alternating training of the discriminator and generator, the objective is to enhance the quality of generated labels from the generator and improve the precision of the discriminator’s judgments.
In this network structure, CBCT images are initially fed into the segmentation network of the generator. After a series of operations, including downsampling, upsampling, and skip connections, the generator produces predicted labels. To further enhance segmentation accuracy, we concatenate CBCT images with the predicted labels and input them as constraint conditions into the discriminator. By calculating the loss between the discriminator output and a matrix of all ones, and then performing backpropagation, we update the weights of the generator. This process helps the generator better fit the real labels, thus improving its performance in image segmentation tasks. In this way, one training update process for the generator is completed.
Subsequently, we freeze the parameters of the generator and proceed to train the discriminator. The discriminator receives input either with CBCT images and real labels or with CBCT images and concatenated predicted labels. It calculates the loss between the discriminator output and a matrix of all ones or all zeros. Through backpropagation, the weights of the discriminator are updated to enhance its ability to distinguish between real and fake labels. By iteratively updating the parameters of the generator and discriminator, the network becomes more adept at the CBCT tooth image segmentation task, improving its overall performance. This alternating training strategy effectively propels the learning process of the network, gradually optimizing it to achieve higher segmentation accuracy.
Overall, our approach fully leverages the collaborative training between the generator and discriminator. Through an effective network structure and alternating update strategy, it demonstrates improved performance in three-dimensional tooth segmentation tasks.
4. Experiments
4.1. Dataset
In this study, we utilized a tooth dataset provided by Hangzhou Dental Hospital for training, testing, and evaluation. Before the utilization of the dataset, informed consent was obtained from the hospital, granting permission to use their three-dimensional CBCT images for research purposes. Additionally, to ensure the privacy and anonymity of the patients, their data was thoroughly anonymized during the preprocessing phase. The dataset consists of 46 samples, each being a three-dimensional CBCT image with a shape size of (D × 536 × 536). Here, D represents the depth, ranging from 186 to 359. Each sample has been carefully annotated, providing detailed descriptions of the three-dimensional model of the teeth. In medical small-sample segmentation tasks, experimenting with varying quantities and sizes of training samples contributes to a comprehensive evaluation of model performance. This experimental design can reveal the model’s robustness to changes in data volume and its performance across diverse scenarios. By incrementally increasing the number of training samples, we can observe variations in the model’s performance under different sample conditions. To thoroughly validate the segmentation performance of PPA-GAN under small-sample scenarios, we conducted four sets of experiments based on different training sample sizes. In these four experiments, we randomly selected 5, 10, 15, and 20 samples as the training set, using the remaining samples as the test set. In each experiment set, training and testing were performed for each network, ensuring consistency in the training and test sets among different networks within the same experiment set to minimize the impact of other factors on the experiment results. Our goal is to gain insights into the segmentation effectiveness of PPA-GAN when dealing with a restricted training dataset and to compare its performance with current state-of-the-art segmentation models. These advanced models include 3DUnet, Swin Unetr, SAM, TransBTS, and the original VNet.
To ensure the smooth operation of the network and enhance its performance, we performed a series of operations on the data: (1) Prior to experimentation, we scaled the values of CBCT images to the range of 0–255, improving data stability and avoiding issues like gradient explosion or vanishing gradients. (2) We transformed the image shape to (256 × 256 × 256), reducing the computational burden on the network and adapting to the input requirements of the dual encoder. (3) Before inputting the images into the network, we performed data augmentation by flipping the images along the X and Y axes with a 15% probability. This increased the model’s generalization capability, enabling it to better adapt to previously unseen data, while also enhancing the diversity of the dataset.
4.2. Evaluation Metrics
In this study, we employ the Dice coefficient (Dice), the Hausdorff distance (HD95), the Average Surface Distance (ASD) and the Jaccard coefficient (Jaccard) metrics for comparative evaluation of experimental results and models.
The Dice is utilized to measure the similarity between two sets, commonly applied in medical image segmentation to assess the segmentation accuracy of predicted labels. Assuming the segmentation result is represented by S and the ground truth annotation is denoted as G, the Dice coefficient formula between S and G is expressed as follows in Equation (2):
The Hausdorff distance is defined as the maximum distance from one set to another. It is commonly used in image segmentation tasks to calculate the distance between the farthest points on the predicted image’s edge and the true image’s edge. A smaller value indicates a higher similarity between the two sets. Assuming the segmentation result is denoted as S and the ground truth annotation is denoted as G, the Hausdorff formula between S and G is expressed as shown in Equation (3):
where h(S, G) represents the distance from S to G, i.e., for a point s in S, find the nearest point g in G, calculate the Euclidean distance between them, and then take the maximum of these distances. Hausdorff corresponds to the most dissimilar regions between two sets. To avoid unreasonable distances caused by certain points being far from the main cluster, this paper calculates the distance of the farthest 95% of pixels as the indicator (HD95) to ensure numerical stability.
ASD denotes the average distance between the segmentation result’s edge and the ground truth’s edge. Assuming the segmentation result is denoted as S and the ground truth annotation is denoted as G, the ASD formula between S and G is expressed as follows in Equation (4):
where nS and nG represent the number of non-zero pixels in S and G respectively, and d(s, G) denotes the Euclidean distance from pixel s to the nearest non-zero pixel in G. Both the Hausdorff and ASD measure the distance between two sets. However, Hausdorff focuses on the distance between the most dissimilar pair of points, while ASD is based on the average distance among all points. Therefore, ASD can provide a more comprehensive assessment of the accuracy of the segmentation results.
The Jaccard coefficient is commonly used to assess the performance of segmentation algorithms, especially when comparing the similarity between segmentation results and ground truth. It calculates the intersection ratio between predicted and true values. The formula is represented as Equation (5).
A larger Jaccard distance indicates a higher overlap between two sets, implying a smaller difference between the segmentation result and the true annotation.
4.3. Loss Function
To enhance the robustness and precision of the model in small-sample segmentation tasks and reduce the occurrence of false positives and false negatives, we employed a combination of Dice loss and generative adversarial network (GAN) loss. The specific loss function is shown in Equation (6).
where Lossgan represents the loss function in generative adversarial training, as shown in Equation (7):
where E represents the expected value of the distribution function, where x represents the CBCT image after voxel preprocessing, y represents the normalized ground truth labels, and z represents the fake labels obtained by x after passing through the generator. Lossdice represents the Dice loss, formulated as shown in Equation (8):
4.4. Experimental Setting
In our research, we selected the Nvidia GeForce RTX 4090, produced by Nvidia Corporation located in California, United States, as the experimental platform for the three-dimensional tooth segmentation task, equipped with 24 GB of VRAM. During the training process, we chose AdamW as the optimizer for both the generator and discriminator, with beta_1 set to 0.9, beta_2 set to 0.999, and an initial learning rate set to 2 × 10−4. To efficiently adjust the learning rate, we employed a dynamic learning rate adjustment strategy using the Dice coefficient as the experimental metric. Specifically, when the optimal Dice coefficient does not decrease after 40 epochs, we halved the learning rate. This process continued until either there was no decrease for 80 consecutive epochs or training concluded after 250 epochs. This strategy helps maintain the convergence of the model during training and effectively prevents it from getting stuck in local optima.
5. Result
To comprehensively validate the segmentation performance of PPA-SAM, we chose VNet as a baseline and assessed the impact of introducing the enhanced SAM encoder and generative adversarial network on segmentation performance. Simultaneously, we conducted performance comparisons with commonly used models. Table 1 presents the segmentation performance of multiple models on the tooth dataset with varying numbers of training samples (5, 10, 15, 20).

Table 1.
Evaluating the performance of PPA-SAM and other models in the small-sample tooth segmentation task.
By incrementally increasing the number of training samples, we evaluated each model’s performance under small-sample conditions. The results indicate that the introduction of the enhanced SAM encoder and generative adversarial network indeed enhances segmentation performance. In the tooth segmentation task with a training sample size of 20, PPA-SAM exhibited outstanding performance, achieving a Dice coefficient of 94.63, a 95 Hausdorff distance of 0.845, ASD of 0.207, and Jaccard coefficient of 9018. In comparison to other advanced models, PPA-SAM demonstrated superior tooth segmentation results under small-sample training conditions. Conversely, Swin Unetr showed inferior performance in small-sample tasks, with metrics consistently lower than those of 3DUnet. For the SAM model, being a 2D segmentation model, we extracted individual slices from the three-dimensional tooth model, constructing datasets for training and testing. The testing results of the slices were subsequently concatenated and evaluated. Due to SAM’s limited pre-training on medical image datasets and its inability to fully leverage three-dimensional information, SAM’s performance in segmentation tasks was relatively subpar.
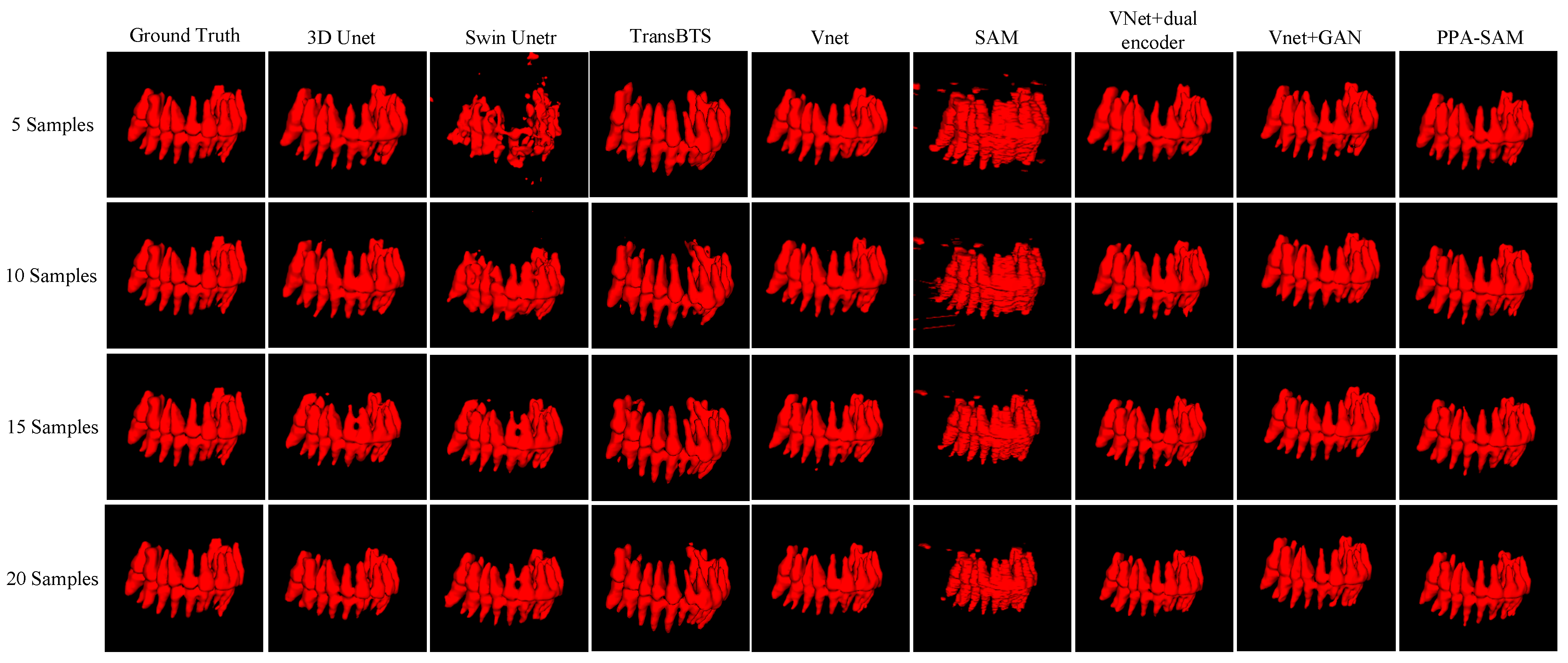
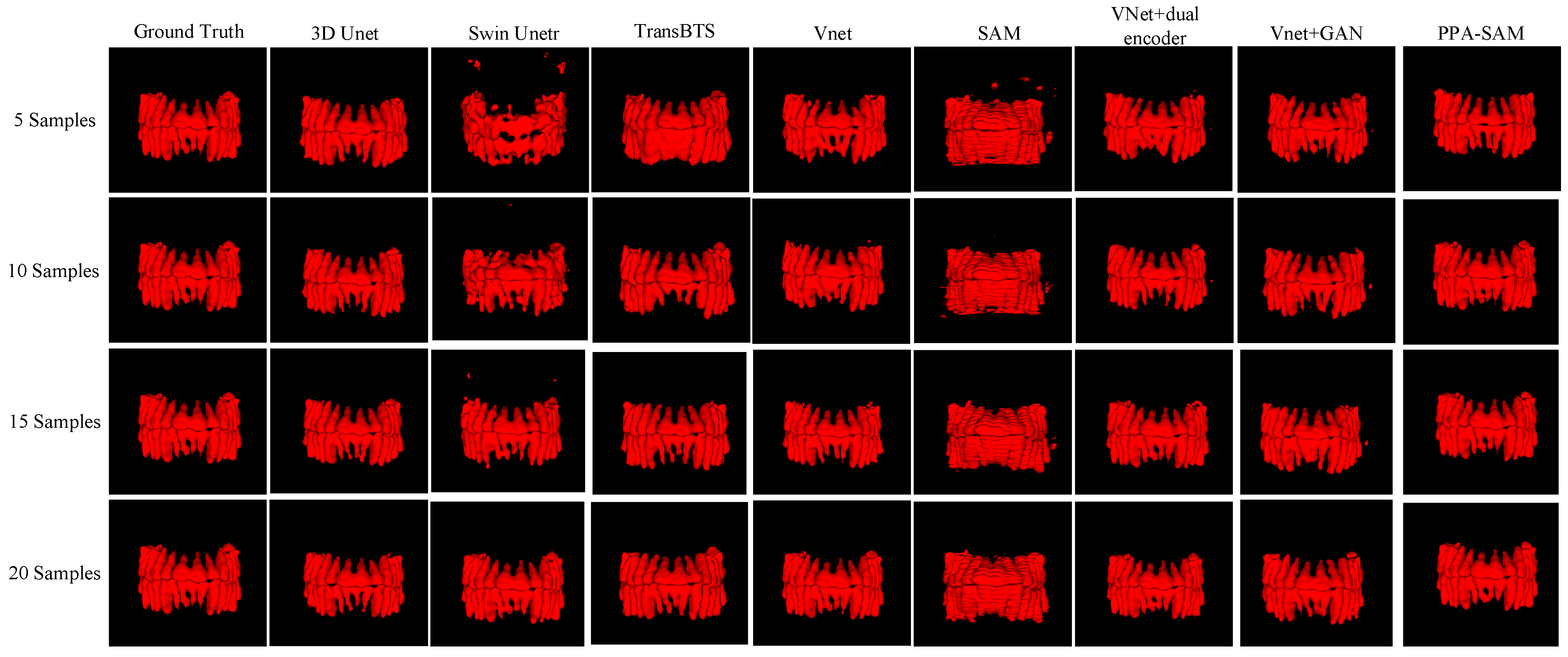
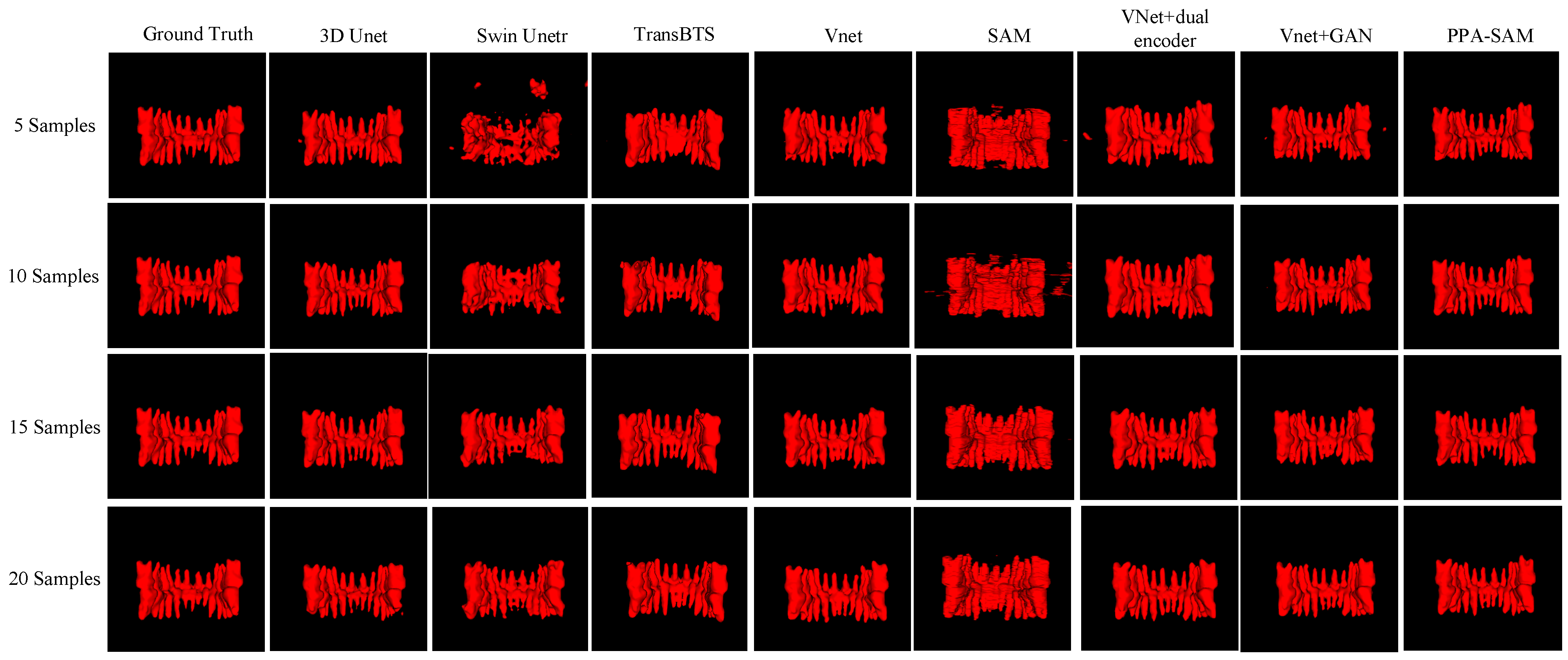
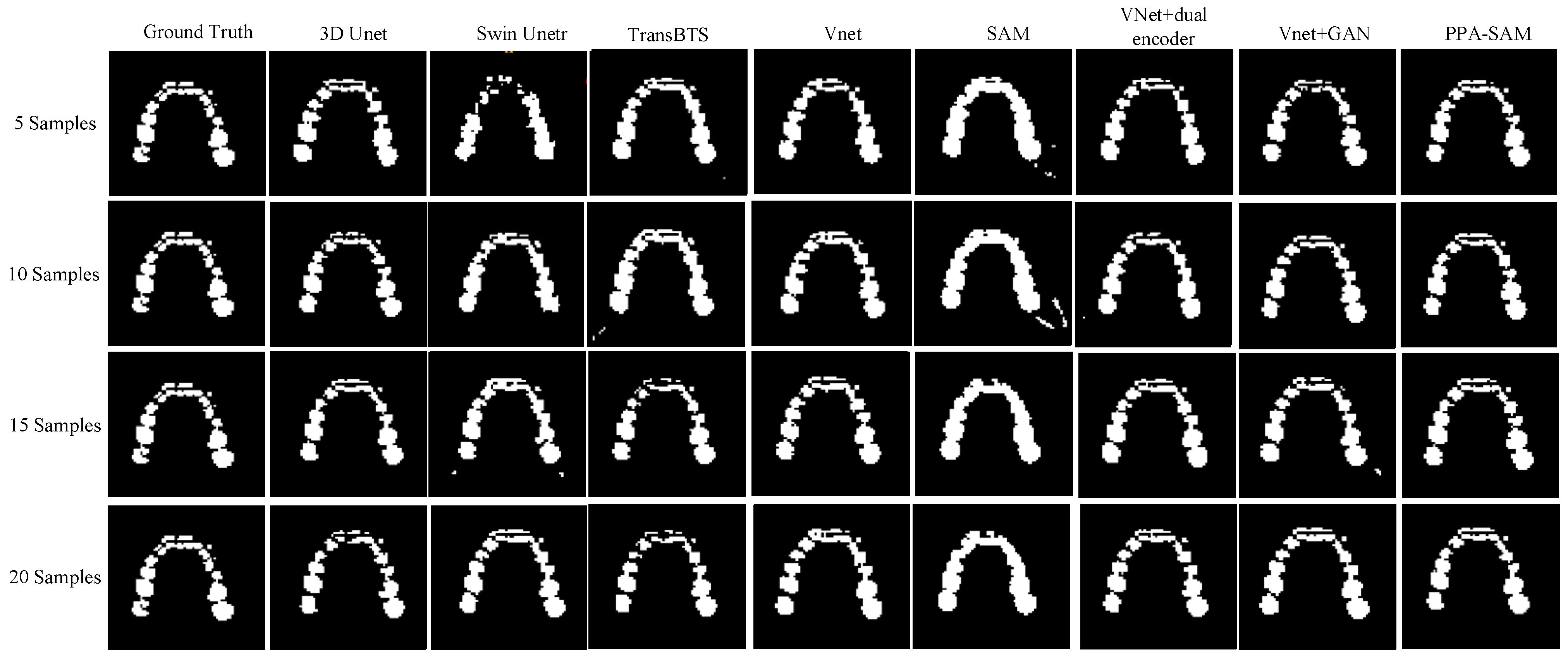
We performed three-dimensional visualization on samples numbered 11, 23, and 32, and selected a slice for two-dimensional visualization on sample number 40, as shown in Figure 5, Figure 6, Figure 7 and Figure 8, revealing that PPA-SAM’s segmentation results closely approximated the ground truth labels. In contrast, Swin Unetr exhibited lower accuracy in segmentation with only 5 training samples, making it less suitable for medical-assistive applications. SAM’s approach of segmenting individual slices and then combining them resulted in noticeable deviations in each slice, leading to a less smooth surface in the visualized results. In summary, PPA-SAM excelled in the small-sample three-dimensional tooth segmentation task, highlighting its robust generalization and adaptability, making it suitable for scenarios with limited data.
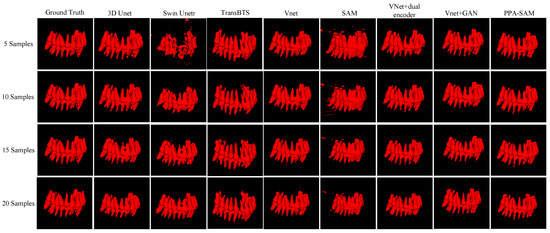
Figure 5.
Qualitative Visualization Comparison of PPA-SAM and other networks on the 11th sample in the small-sample tooth segmentation task.
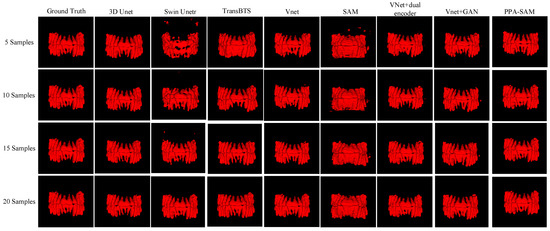
Figure 6.
Qualitative Visualization Comparison of PPA-SAM and other networks on the 23rd sample in the small-sample tooth segmentation task.

Figure 7.
Qualitative Visualization Comparison of PPA-SAM and other networks on the 32nd sample in the small-sample tooth segmentation task.
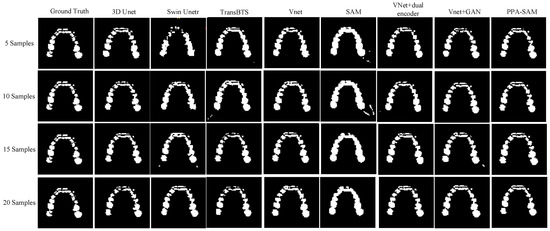
Figure 8.
Qualitative Visualization Comparison of PPA-SAM and other networks on the 40th sample in the small-sample tooth segmentation task.
6. Discussion
The experimental results demonstrate that the PPA-SAM model exhibits outstanding segmentation performance and generalization ability on a small-sample CBCT dental dataset, showcasing significant clinical application value. The generator of PPA-SAM combines an optimizer-equipped SAM encoder with a VNet encoder, forming a dual-encoder structure that facilitates the acquisition of both global and local information. In dental images, global information focuses on overall structure and positioning, while local information involves the details and edges of each tooth. This combination enables the model to more accurately segment each tooth. The features from the dual-encoder entering the decoder provide it with more information, covering features at different scales and levels. This allows the decoder to better accomplish the three-dimensional tooth segmentation task using multi-level, rich features.
We utilize a VNet network with a dual-encoder as the generator, combined with a discriminator containing a three-layer convolutional network, forming the conditional generative adversarial network known as PPA-SAM. Simultaneously, CBCT images are integrated as conditions with labels and input into the discriminator. This conditional information helps enhance the model’s generalization ability, enabling it to comprehensively and representatively learn features and better understand the relationship between teeth and their surrounding environment, resulting in segmented three-dimensional tooth images that closely resemble real data.
In the tooth segmentation task of this study, PPA-SAM demonstrates higher Dice coefficients compared to other comparative networks across different scales of training datasets. Specifically, for training set sizes of 5, 10, 15, and 20, the PPA-SAM shows an improvement in Dice coefficients over the second-ranking model by 1.85%, 2.41%, 3.08%, and 0.87%, respectively. These results indicate that even with smaller amounts of data, the PPA-SAM can achieve superior segmentation performance, highlighting its significant potential for clinical applications. Traditional convolutional networks [6,7,8,9,10,31,32,33] typically require large-scale datasets and perform poorly in situations with small samples, especially in medical segmentation tasks where datasets are often lim-ited. For example, 3D Unet [33] has long training times and faces significant computa-tional pressure. Transformer-based segmentation networks, such as Swin Unetr [32], perform poorly and have limited generalization capability when trained with limited data.
Visualizing the three-dimensional segmentation results from Figure 5, Figure 6 and Figure 7, irregular fragmented regions are observed in the segmented 3D models generated by Swin Unetr, while TransBTS exhibits contiguous phenomena in the root regions of the teeth, with a lower accuracy in detail. This suggests potential overfitting issues of Swin Unetr and TransBTS in small-sample segmentation tasks, leading to decreased seg-mentation accuracy. However, due to the two-dimensional shape of input and output data for the SAM, the inter-slice correlation in the SAM’s three-dimensional segmenta-tion results is relatively low. When segmenting individual slices, it is prone to misiden-tifying surrounding tissues as teeth, resulting in fine fragments around the tooth mod-el, and the weak correlation between slices leads to a staircase-like appearance on the surface of the 3D segmentation result. In comparison, 3D Unet and VNet, as traditional segmentation models, demonstrate good segmentation performance. However, through three-dimensional and two-dimensional visualizations, it can be observed that the segmentation accuracy in detail areas such as between the roots and teeth remains low. Through the quantitative analysis presented in Table 1, it can be concluded that PPA-SAM exhibits more robust performance by incorporating dual encoders and gen-erative adversarial networks into VNet. PPA-SAM combines the advantages of the SAM’s large data model, possessing a strong generalization capability and better ad-dressing the issue of small sample sizes in medical image segmentation.Our research still encounters certain limitations:(1) The training process of the generative network exhibits some instability, potentially leading to an imbalance between the generator and discriminator, ultimately resulting in training breakdown. (2)The current network’s training still relies on a considerable number of samples for support, making it challenging to achieve high-quality zero-shot or one-shot experiments. In future investigations, we aim to delve into innovative approaches that involve training the CBCT three-dimensional tooth segmentation network with only one or zero samples. For instance, in situations with only a single training sample, we utilize a Generative Adversarial Network to generate synthetic medical images and labels that closely resemble the existing sample and exhibit high quality. This method allows for the synthesis of additional training data, integrating the generated samples into the original training set for model training. This enhances the model’s generalization ability when dealing with a limited number of samples. This exploration in the direction of using minimal or no samples seeks to challenge traditional training frameworks and promote a more flexible and adaptive learning approach for networks in medical image segmentation tasks.
7. Conclusions
In our study, we introduce an innovative CBCT three-dimensional tooth segmentation network named PPA-SAM. The design philosophy of PPA-SAM leverages the sensitivity of traditional convolutional networks to detailed features and the generalization ability of large models, showcasing outstanding performance in the small-sample task of three-dimensional CBCT tooth image segmentation.
However, despite the excellent performance of PPA-SAM and its broad prospects in small-sample medical image segmentation tasks, its task transfer across different data types may appear less than ideal. For example, whe n using CBCT dental images as the training set and X-ray dental images as the testing set for segmentation, its performance may not meet expectations. Through further research and experimentation, we aspire to optimize this network, making it a robust tool for addressing challenges in small-sample medical image segmentation. This not only holds significant practical value in the domain of tooth segmentation but also has the potential to play a positive role in other medical image segmentation tasks. The successful application of PPA-SAM provides new insights and methodologies for small-sample medical image segmentation tasks.
Author Contributions
Conceptualization, J.L. and H.W.; Methodology, H.G.; Software, J.L.; Validation, J.L. and Y.C.; Formal analysis, H.W. and H.G.; Investigation, J.L.; Project administration, J.L.; Data curation, Y.C.; Visualization, J.L.; Writing—original draft preparation, J.L.; Writing—review and editing, H.W. and H.G.; funding acquisition, H.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Leading Talents of Science and Technology Innovation in Zhejiang Province (Grant No. 2020R52042), Zhejiang-Netherlands Joint Laboratory for Digital Diagnosis and Treatment of oral diseases, The Key research and development program of Zhejiang (Grant No. 2021C01189), National Natural Science Foundation of China (Grant No. 82011530399), and Zhejiang Provincial Natural Science Foundation of China (Grant No. LGG20F020015).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gan, Y.; Xia, Z.; Xiong, J.; Li, G.; Zhao, Q. Tooth and alveolar bone segmentation from dental computed tomography images. IEEE J. Biomed. Health Inform. 2017, 22, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Gan, Y.; Xiong, J.; Zhang, D.; Zhao, Q.; Xia, Z. A method for tooth model reconstruction based on integration of multimodal images. J. Healthc. Eng. 2018, 2018, 4950131. [Google Scholar] [CrossRef] [PubMed]
- Borzabadi-Farahani, A. Systematic review and meta-analysis of the index of orthognathic functional treatment need for detecting subjects with great need for orthognathic surgery. Cleft Palate Craniofacial J. 2023, 10556656231216833. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, C.; Shan, Z. Application of artificial intelligence in orthodontics: Current state and future perspectives. Healthcare 2023, 11, 2760. [Google Scholar] [CrossRef]
- Gao, H.; Chae, O. Individual tooth segmentation from CT images using level set method with shape and intensity prior. Pattern Recognit. 2010, 43, 2406–2417. [Google Scholar] [CrossRef]
- Gan, Y.; Xia, Z.; Xiong, J.; Zhao, Q.; Hu, Y.; Zhang, J. Toward accurate tooth segmentation from computed tomography images using a hybrid level set model. Med. Phys. 2015, 42, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Sahiner, B.; Pezeshk, A.; Hadjiiski, L.M.; Wang, X.; Drukker, K.; Cha, K.H.; Summers, R.M.; Giger, M.L. Deep learning in medical imaging and radiation therapy. Med. Phys. 2019, 46, e1–e36. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Woo, S.; Yu, J.; Seo, J.; Lee, J.; Lee, C. Automated CNN-based tooth segmentation in cone-beam CT for dental implant planning. IEEE Access 2020, 8, 50507–50518. [Google Scholar] [CrossRef]
- Rao, Y.; Wang, Y.; Meng, F.; Pu, J.; Sun, J.; Wang, Q. A symmetric fully convolutional residual network with DCRF for accurate tooth segmentation. IEEE Access 2020, 8, 92028–92038. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Cui, Z.; Li, C.; Wang, W. ToothNet: Automatic tooth instance segmentation and identification from cone beam CT images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 6368–6377. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.-Y. Segment anything. arXiv 2023, arXiv:2304.02643. [Google Scholar]
- Deng, R.; Cui, C.; Liu, Q.; Yao, T.; Remedios, L.W.; Bao, S.; Landman, B.A.; Wheless, L.E.; Coburn, L.A.; Wilson, K.T. Segment anything model (sam) for digital pathology: Assess zero-shot segmentation on whole slide imaging. arXiv 2023, arXiv:2304.04155. [Google Scholar]
- He, S.; Bao, R.; Li, J.; Grant, P.E.; Ou, Y. Accuracy of segment-anything model (sam) in medical image segmentation tasks. arXiv 2023, arXiv:2304.09324. [Google Scholar]
- Wu, J.; Fu, R.; Fang, H.; Liu, Y.; Wang, Z.; Xu, Y.; Jin, Y.; Arbel, T. Medical sam adapter: Adapting segment anything model for medical image segmentation. arXiv 2023, arXiv:2304.12620. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Springer International Publishing: Cham, Switzerland, 2018; pp. 3–11. [Google Scholar]
- Tang, Y.; Yang, D.; Li, W.; Roth, H.R.; Landman, B.; Xu, D.; Nath, V.; Hatamizadeh, A. Self-supervised pre-training of swin transformers for 3d medical image analysis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 20730–20740. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Mirza, M.; Osindero, S. Conditional generative adversarial nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Zhao, H.; Zhang, Z.; Yang, Y.; Xiao, J.; Chen, J. A Dynamic Monitoring Method of Temperature Distribution for Cable Joints Based on Thermal Knowledge and Conditional Generative Adversarial Network. IEEE Trans. Instrum. Meas. 2023, 72, 4507014. [Google Scholar] [CrossRef]
- Isola, P.; Zhu, J.-Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Radford, A.; Kim, J.W.; Hallacy, C.; Ramesh, A.; Goh, G.; Agarwal, S.; Sastry, G.; Askell, A.; Mishkin, P.; Clark, J. Learning transferable visual models from natural language supervision. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 8748–8763. [Google Scholar]
- Ramesh, A.; Dhariwal, P.; Nichol, A.; Chu, C.; Chen, M. Hierarchical text-conditional image generation with clip latents. arXiv 2022, arXiv:2204.06125. [Google Scholar]
- Radford, A.; Narasimhan, K.; Salimans, T.; Sutskever, I. Improving language understanding by generative pre-training. 2018. Available online: https://www.mikecaptain.com/resources/pdf/GPT-1.pdf (accessed on 12 March 2024).
- Chen, Z.; Duan, Y.; Wang, W.; He, J.; Lu, T.; Dai, J.; Qiao, Y. Vision transformer adapter for dense predictions. arXiv 2022, arXiv:2205.08534. [Google Scholar]
- Ma, J.; He, Y.; Li, F.; Han, L.; You, C.; Wang, B. Segment anything in medical images. Nat. Commun. 2024, 15, 654. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Song, L.; Liu, S.; Zhang, Y. A review of deep-learning-based medical image segmentation methods. Sustainability 2021, 13, 1224. [Google Scholar] [CrossRef]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. Unetr: Transformers for 3d medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 574–584. [Google Scholar]
- Hatamizadeh, A.; Nath, V.; Tang, Y.; Yang, D.; Roth, H.R.; Xu, D. Swin unetr: Swin transformers for semantic segmentation of brain tumors in mri images. In Proceedings of International MICCAI Brainlesion Workshop; Springer International Publishing: Cham, Switzerland, 2021; pp. 272–284. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference, Athens, Greece, 17–21 October 2016; Springer International Publishing: Cham, Switzerland, 2016; pp. 424–432. [Google Scholar]
- Wang, W.; Chen, C.; Ding, M.; Yu, H.; Zha, S.; Li, J. Transbts: Multimodal brain tumor segmentation using transformer. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Springer International Publishing: Cham, Switzerland, 2021; pp. 109–119. [Google Scholar]
- Liu, W.; Shen, X.; Pun, C.-M.; Cun, X. Explicit visual prompting for low-level structure segmentations. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 19434–19445. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).