Identification and Characterization of a Novel Thermostable GDSL Lipase LipGt6 from Geobacillus thermoleovorans H9
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, and Chemicals
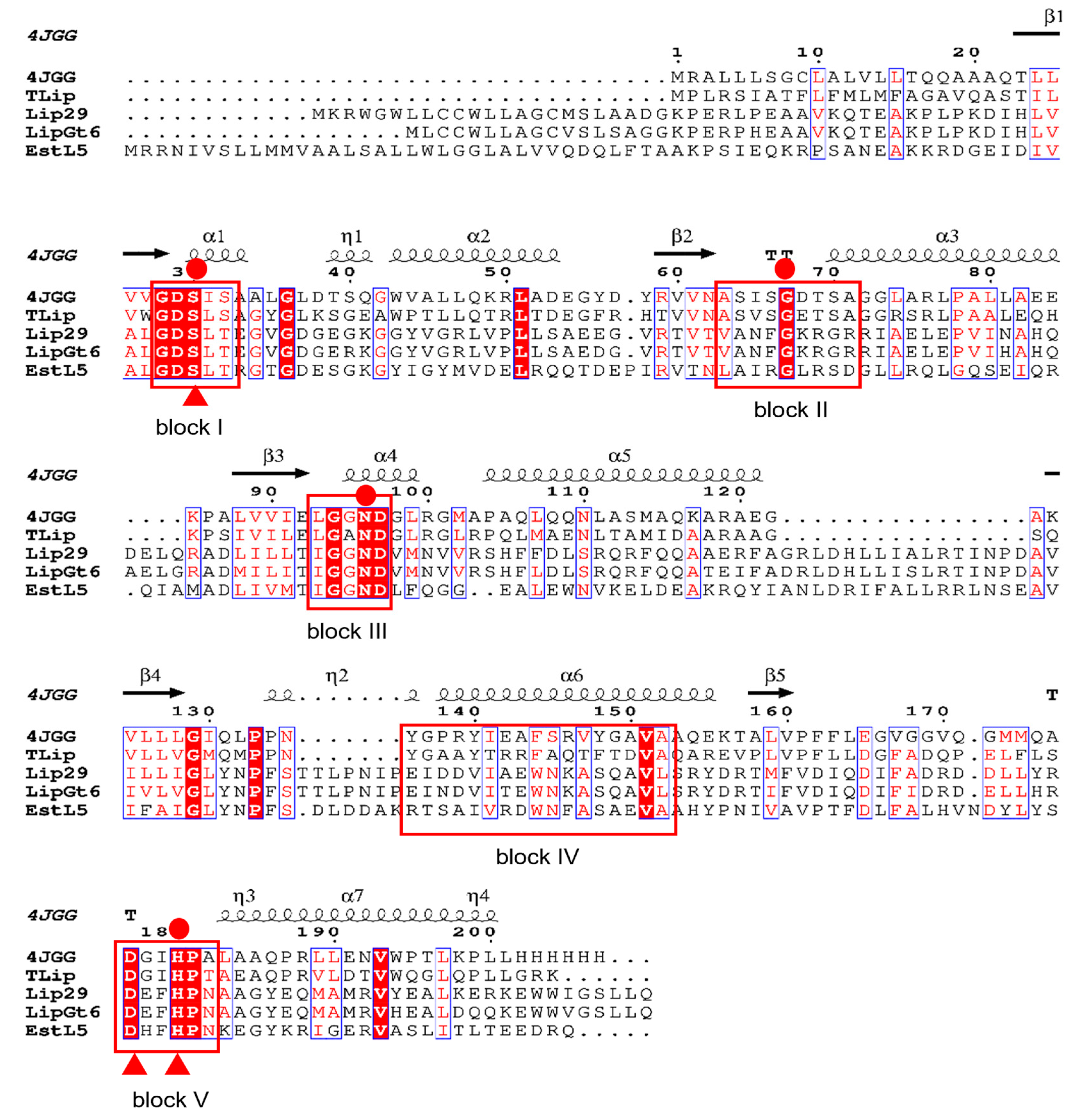
2.2. Homology Modelling and Sequence Alignment
2.3. Construction of the Expression Vector
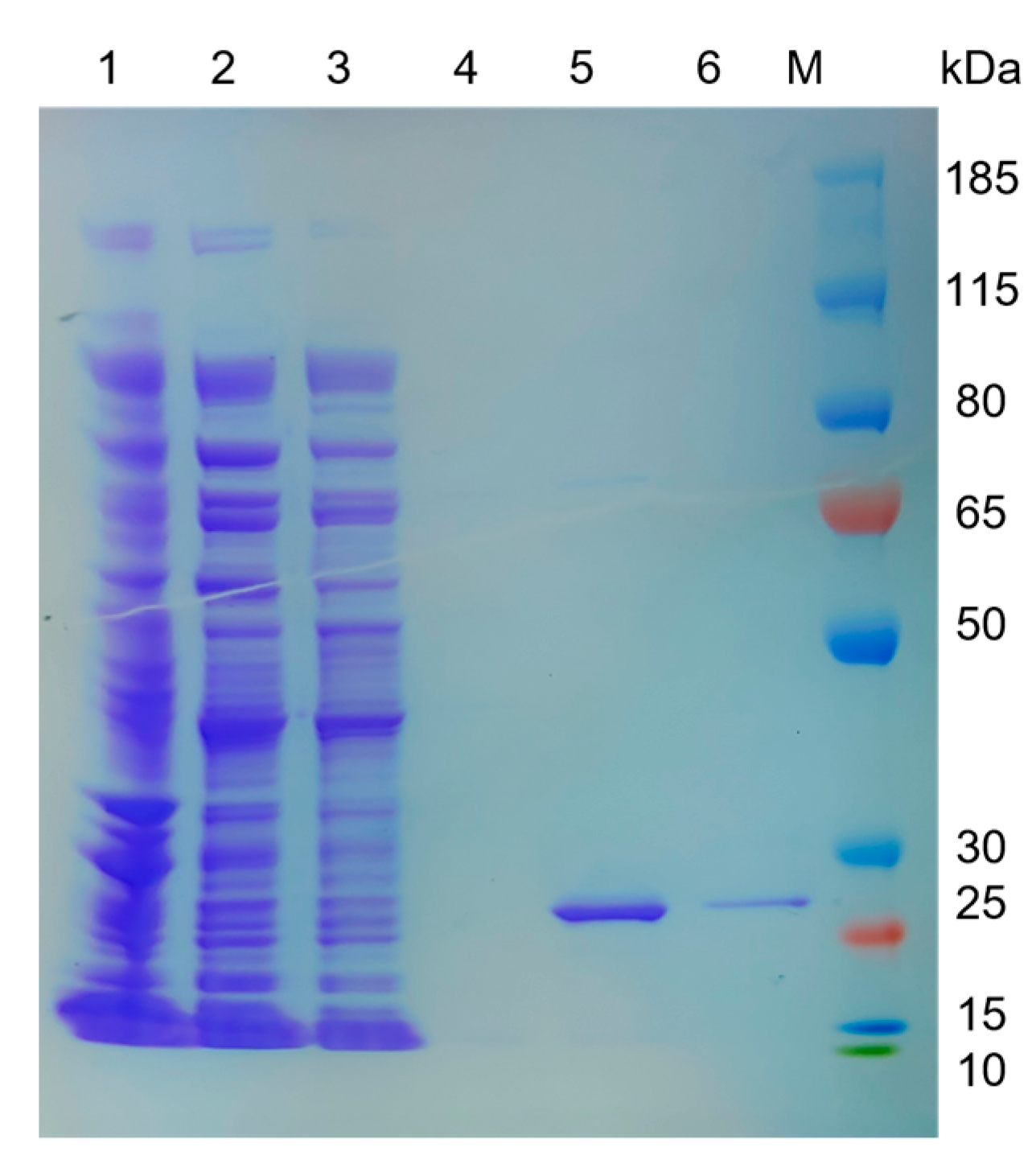
2.4. Expression and Purification of LipGt6
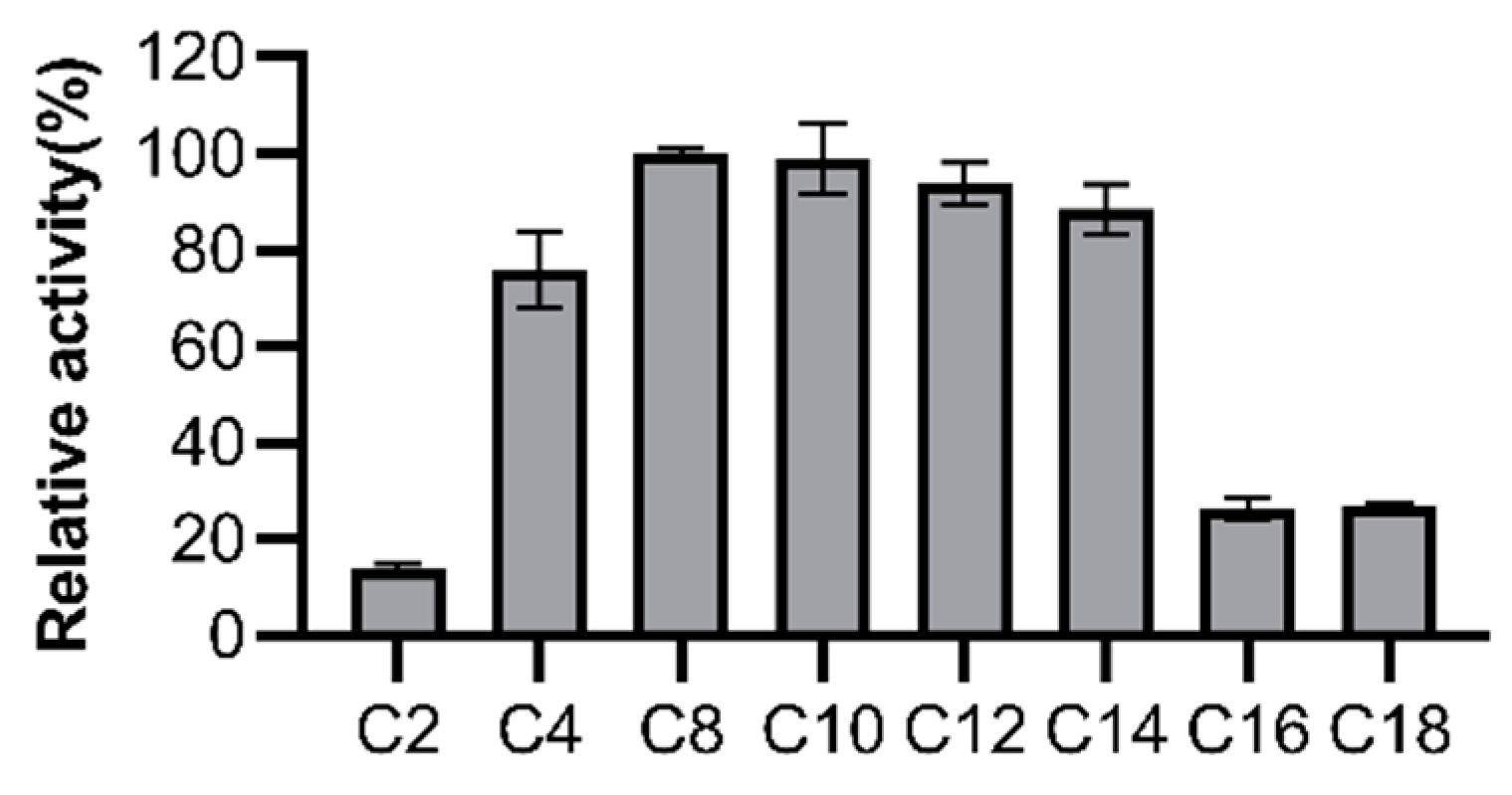
2.5. Substrate Specificity
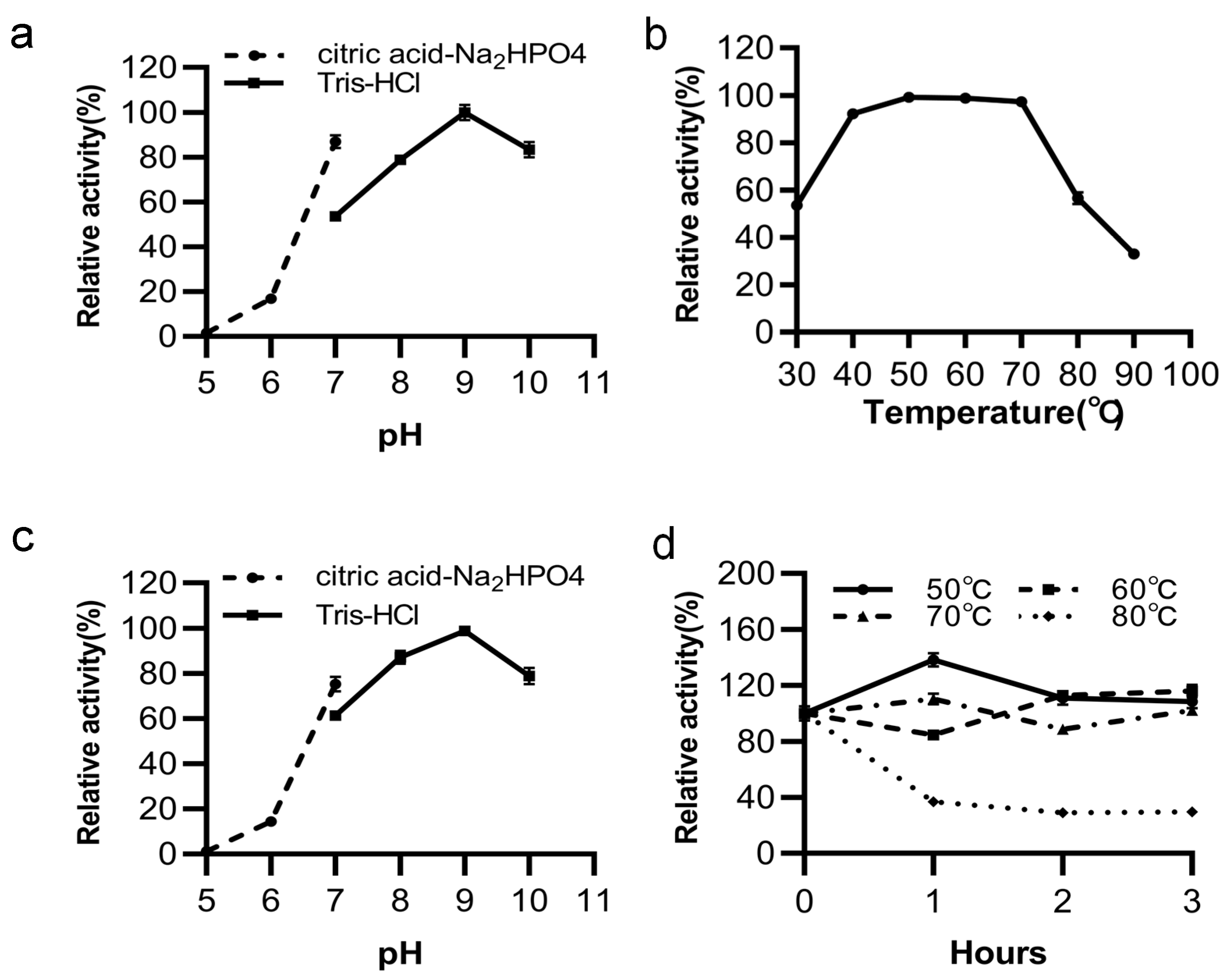
2.6. Effects of Temperature and pH on Enzyme Activity and Stability
2.7. Effects of Metal Ions, Surfactants, and Oxidizing Agents on Enzyme Activity
2.8. Site-Directed Mutagenesis
2.9. Statistical Analysis
3. Results
3.1. Sequence Analysis of the Enzyme LipGt6
3.2. Cloning, Expression, and Purification of LipGt6
3.3. Substrate Specificity of LipGt6
3.4. Effects of Temperature and pH on LipGt6
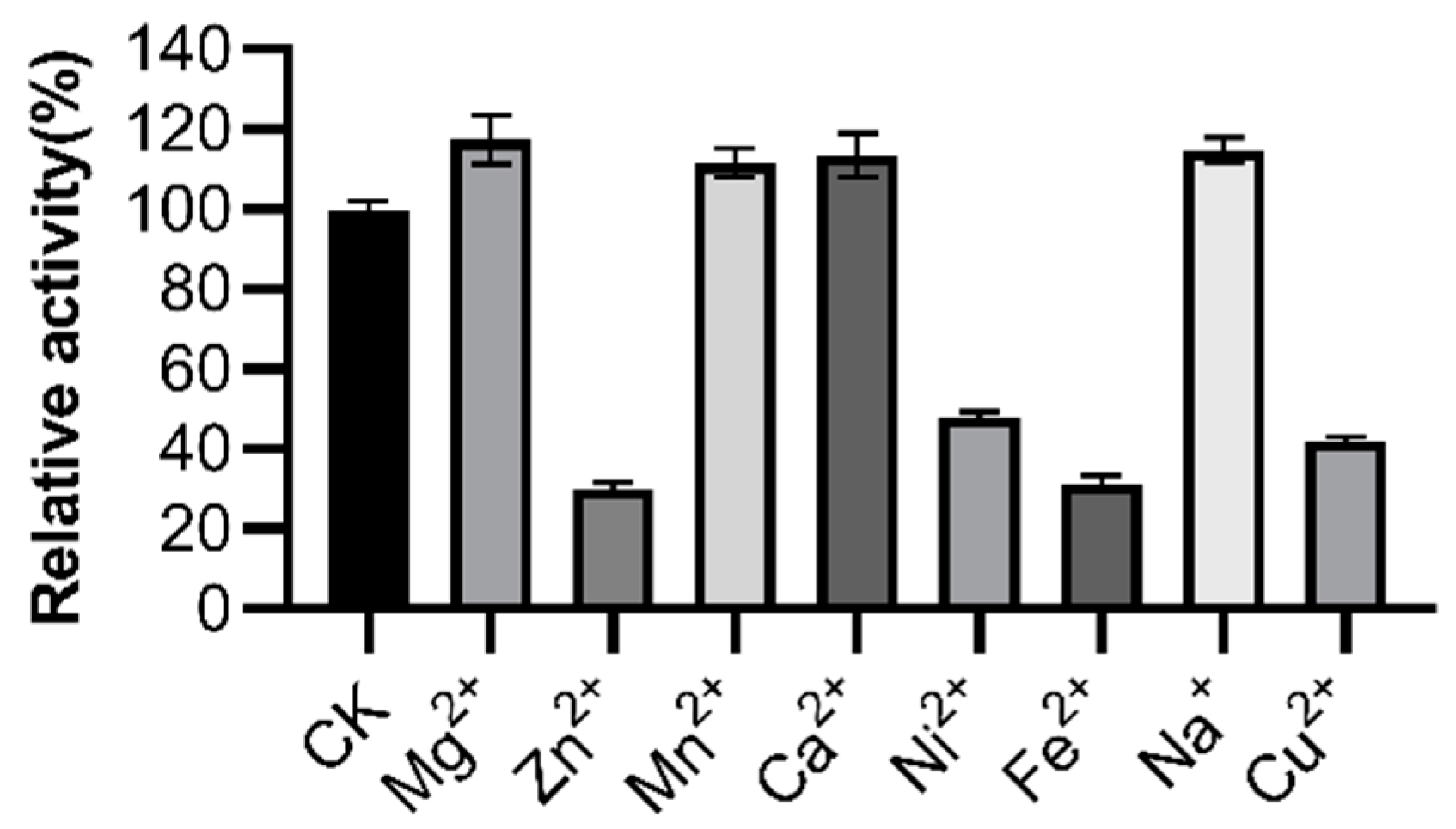
3.5. Effects of Metal Ions, Surfactants, and Oxidizing Agents on LipGt6
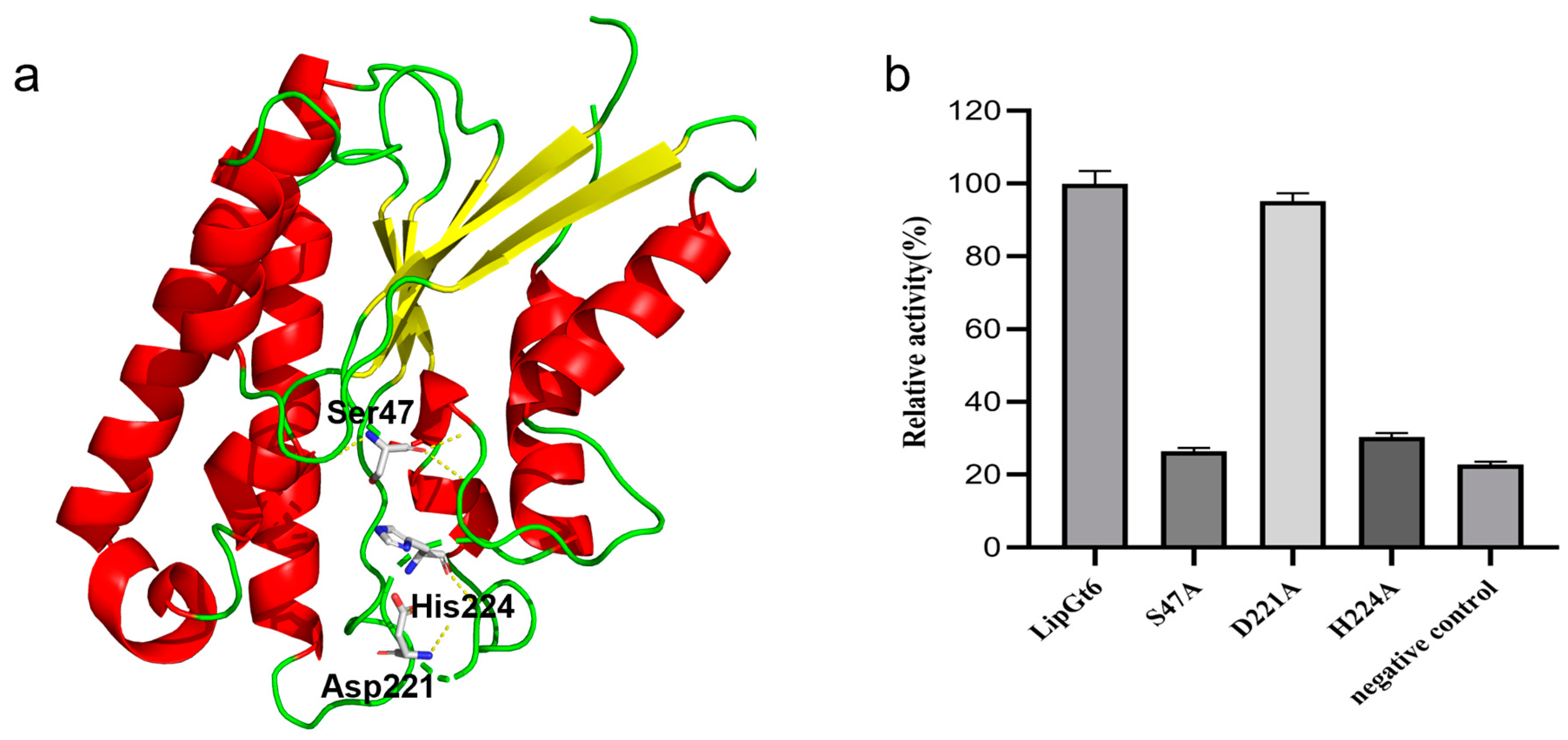
3.6. Analysis of the LipGt6 Catalytic Triad
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patel, N.; Rai, D.; Shivam; Shahane, S.; Mishra, U. Lipases: Sources, Production, Purification, and Applications. Recent Pat. Biotechnol. 2019, 13, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Melani, N.B.; Tambourgi, E.B.; Silveira, E. Lipases: From production to applications. Sep. Purif. Rev. 2020, 49, 143–158. [Google Scholar] [CrossRef]
- Hitch, T.C.A.; Clavel, T. A proposed update for the classification and description of bacterial lipolytic enzymes. PeerJ 2019, 7, e7249. [Google Scholar] [CrossRef] [PubMed]
- Mlgaard, A.; Kauppinen, S.; Larsen, S. Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Structure 2000, 8, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Bharathi, D.; Rajalakshmi, G. Microbial lipases: An overview of screening, production and purification. Biocatal. Agric. Biotechnol. 2019, 22, 101368. [Google Scholar] [CrossRef]
- Abdella, B.; Youssif, A.M.; Sabry, S.A.; Ghozlan, H.A. Production, purification, and characterization of cold-active lipase from the psychrotroph Pseudomonas sp. A6. Braz. J. Microbiol. 2023, 54, 1623–1633. [Google Scholar] [CrossRef]
- Li, J.; Shen, W.; Fan, G.; Li, X. Screening, purification and characterization of lipase from Burkholderia pyrrocinia B1213. 3 Biotech 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Cho, A.-R.; Yoo, S.-K.; Kim, E.-J. Cloning, sequencing and expression in Escherichia coli of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiol. Lett. 2000, 186, 235–238. [Google Scholar] [CrossRef]
- Bacha, A.B.; Moubayed, N.M.S.; Abid, I. Thermostable, alkaline and detergent-tolerant lipase from a newly isolated thermophilic Bacillus stearothermophilus. Indian J. Biochem. Biophys. 2015, 52, 179–188. [Google Scholar]
- Cao, Y.; Huang, H.; Qian, Y.; Wang, L.; Xu, Y.; Jin, H.; Sun, J.; Chang, Z. Hyperthermophilic pretreatment device and its application on improving decomposition effect for chicken manure and rice straw aerobic composting. Trans. Chin. Soc. Agric. Eng. 2017, 33, 243–250. [Google Scholar]
- Yu, Z.; Zhou, S. Hyperthermophilic composting of organic solid wastes: Accelerated humification and pollution control mechanisms. J. Nanjing Agric. Univ. 2020, 43, 781–789. [Google Scholar]
- Burgess, S.; Flint, S.; Lindsay, D.; Cox, M.; Biggs, P. Insights into the Geobacillus stearothermophilus species based on phylogenomic principles. BMC Microbiol. 2017, 17, 140. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhu, H.; Kong, Z.; Li, T.; Ma, L.; Liu, D.; Shen, Q. Pan-Genome Analyses of Geobacillus spp. Reveal Genetic Characteristics and Composting Potential. Int. J. Mol. Sci. 2020, 21, 3393. [Google Scholar] [CrossRef]
- Jo, E.; Kim, J.; Lee, A.; Moon, K.; Cha, J. Identification and Characterization of a Novel Thermostable GDSL-Type Lipase from Geobacillus thermocatenulatus. J. Microbiol. Biotechnol. 2021, 31, 483–491. [Google Scholar] [CrossRef]
- Abol-Fotouh, D.; AlHagar, O.E.; Hassan, M.A. Optimization, purification, and biochemical characterization of thermoalkaliphilic lipase from a novel Geobacillus stearothermophilus FMR12 for detergent formulations. Int. J. Biol. Macromol. 2021, 181, 125–135. [Google Scholar] [CrossRef]
- Bayoumi, R.A.; El-Louboudey, S.S.; Sidkey, N.M.; Abd-El-Rahman, M.A. Production, Purification and Characterization of Thermoalkalophilic Lipase for Application in Bio-detergent Industry. J. Appl. Sci. Res. 2007, 3, 1752–1765. [Google Scholar]
- Xiao, Y.; Liu, Y.-D.; Yuan, G.; Mao, R.-Q.; Li, G. An uncharacterized protein from the metagenome with no obvious homology to known lipases shows excellent alkaline lipase properties and potential applications in the detergent industry. Biotechnol. Lett. 2021, 43, 2311–2325. [Google Scholar] [CrossRef]
- Salihu, A.; Alam, M.Z. Solvent tolerant lipases: A review. Process Biochem. 2015, 50, 86–96. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Kordel, M.; Hofmann, B.; Schomburg, D.; Schmid, R.D. Extracellular lipase of Pseudomonas sp. strain ATCC 21808: Purification, characterization, crystallization, and preliminary X-ray diffraction data. J. Bacteriol. 1991, 173, 4836–4841. [Google Scholar] [CrossRef] [PubMed]
- Akoh, C.C.; Lee, G.-C.; Liaw, Y.-C.; Huang, T.-H.; Shaw, J.-F. GDSL family of serine esterases/lipases. Prog. Lipid Res. 2004, 43, 534–552. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhu, H.; Ye, Y.; Li, J.; Han, N.; Wu, Q.; Huang, Z.; Meng, Z. A thermostable and alkaline GDSL-motif esterase from Bacillus sp. K91: Crystallization and X-ray crystallographic analysis. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2018, 74, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, K.E.; Dijkstra, B.W.; Reetz, M.T. Bacterial Biocatalysts: Molecular Biology, Three-Dimensional Structures, and Biotechnological Applications of Lipases. Annu. Rev. Microbiol. 1999, 53, 315–351. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, K.-E.; Reetz, M. Microbial lipases form versatile tools for biotechnology. Trends Biotechnol. 1998, 16, 369–403. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Ding, J.; Zheng, Q.; Han, N.; Yu, J.; Yang, Y.; Li, J.; Mu, Y.; Wu, Q.; Huang, Z. Identification and characterization of a new alkaline SGNH hydrolase from a thermophilic bacterium Bacillus sp. K91. J. Microbiol. Biotechnol. 2016, 26, 730–738. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, Y.; Shen, T.; Xie, Y.; Mao, Y.; Ji, C. Cloning, expression and biochemical characterization of a novel, moderately thermostable GDSL family esterase from Geobacillus thermodenitrificans T2. J. Biosci. Bioeng. 2013, 115, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, Y.; Qiu, R.; Huang, J.; Ji, C. Crystallization and preliminary X-ray diffraction analysis of a thermostable GDSL-family esterase, EstL5, from Geobacillus thermodenitrificans T2. Acta Crystallogr. 2013, 69, 776–778. [Google Scholar]
- Abu, S.; Leow, B.; Chor, T.; Raja, R.; Zaliha, N. Cloning, expression and characterization of a novel cold-adapted GDSL family esterase from Photobacterium sp. strain J15. Extrem. Life Under Extrem. Cond. 2016, 20, 44–55. [Google Scholar]
- Lescic Asler, I.; Stefanic, Z.; Mar?Avelski, A.; Vianello, R.; Kojic-Prodic, B. Catalytic Dyad in the SGNH Hydrolase Superfamily: In-depth Insight into Structural Parameters Tuning the Catalytic Process of Extracellular Lipase from Streptomyces rimosus. ACS Chem. Biol. 2017, 12, 1928–1936. [Google Scholar] [CrossRef]
- Brumlik, M.J.; Buckley, J.T. Identification of the catalytic triad of the lipase/acyltransferase from Aeromonas hydrophila. J. Bacteriol. 1996, 178, 2060–2064. [Google Scholar] [CrossRef] [PubMed]
- Arpigny, J.L.; Jaeger, K.E. Bacterial lipolytic enzymes: Classification and properties. Biochem. J. 1999, 343, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Sinchaikul, S.; Sookkheo, B.; Phutrakul, S.; Pan, F.-M.; Chen, S.-T. Optimization of a thermostable lipase from Bacillus stearothermophilus P1: Overexpression, purification, and characterization. Protein Expr. Purif. 2001, 22, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-W.; Kim, H.-W.; Lee, K.-W.; Kim, B.-C.; Choe, E.-A.; Lee, H.-S.; Kim, D.-S.; Pyun, Y.-R. Purification and characterization of two distinct thermostable lipases from the gram-positive thermophilic bacterium Bacillus thermoleovorans ID-1. Enzym. Microb. Technol. 2001, 29, 363–371. [Google Scholar] [CrossRef]
- Lailaja, V.; Chandrasekaran, M. Detergent compatible alkaline lipase produced by marine Bacillus smithii BTMS 11. World J. Microbiol. Biotechnol. 2013, 29, 1349–1360. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Ma, M.; Zeng, Z.; Yu, P.; Deng, S. Production, purification and biochemical characterisation of a novel lipase from a newly identified lipolytic bacterium Staphylococcus caprae NCU S6. J. Enzym. Inhib. Med. Chem. 2021, 36, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.K.; Song, H.K.; Shin, D.H.; Hwang, K.Y.; Suh, S.W. The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor. Structure 1997, 5, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Jithesh, K. Expression and purification of organic solvent stable lipase from soil metagenomic library. World J. Microbiol. Biotechnol. 2012, 28, 2417–2424. [Google Scholar] [CrossRef]
- Zheng, J.; Liu, C.; Liu, L.; Jin, Q. Characterisation of a thermo-alkali-stable lipase from oil-contaminated soil using a metagenomic approach. Syst. Appl. Microbiol. 2013, 36, 197–204. [Google Scholar] [CrossRef]
| Chemical Reagents | Relative Activity (%) |
|---|---|
| CK | 100.00 ± 3.93 |
| 1 mM SDS | 40.05 ± 1.94 |
| 1% Triton X-100 | 41.70 ± 2.18 |
| 1% Tween-80 | 28.03 ± 1.55 |
| 1% Tween-20 | 29.26 ± 1.68 |
| 1% sodium deoxycholate | 75.72 ± 5.76 |
| 1% H2O2 | 69.50 ± 16.61 |
| 5% NaClO | 4.09 ± 0.17 |
| 1% NaClO | 10.70 ± 0.18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, L.; Lin, M.; Zhan, Y.; Jiang, S.; Zhou, Z.; Wang, J. Identification and Characterization of a Novel Thermostable GDSL Lipase LipGt6 from Geobacillus thermoleovorans H9. Appl. Sci. 2024, 14, 3279. https://doi.org/10.3390/app14083279
Qin L, Lin M, Zhan Y, Jiang S, Zhou Z, Wang J. Identification and Characterization of a Novel Thermostable GDSL Lipase LipGt6 from Geobacillus thermoleovorans H9. Applied Sciences. 2024; 14(8):3279. https://doi.org/10.3390/app14083279
Chicago/Turabian StyleQin, Lirong, Min Lin, Yuhua Zhan, Shijie Jiang, Zhengfu Zhou, and Jin Wang. 2024. "Identification and Characterization of a Novel Thermostable GDSL Lipase LipGt6 from Geobacillus thermoleovorans H9" Applied Sciences 14, no. 8: 3279. https://doi.org/10.3390/app14083279





