CRISPR-Cas-Integrated LAMP
Abstract
:1. Introduction
2. LAMP Technology and Its Modifiability
3. CRISPR/Cas Technology in Nucleic Acid Amplification Detection
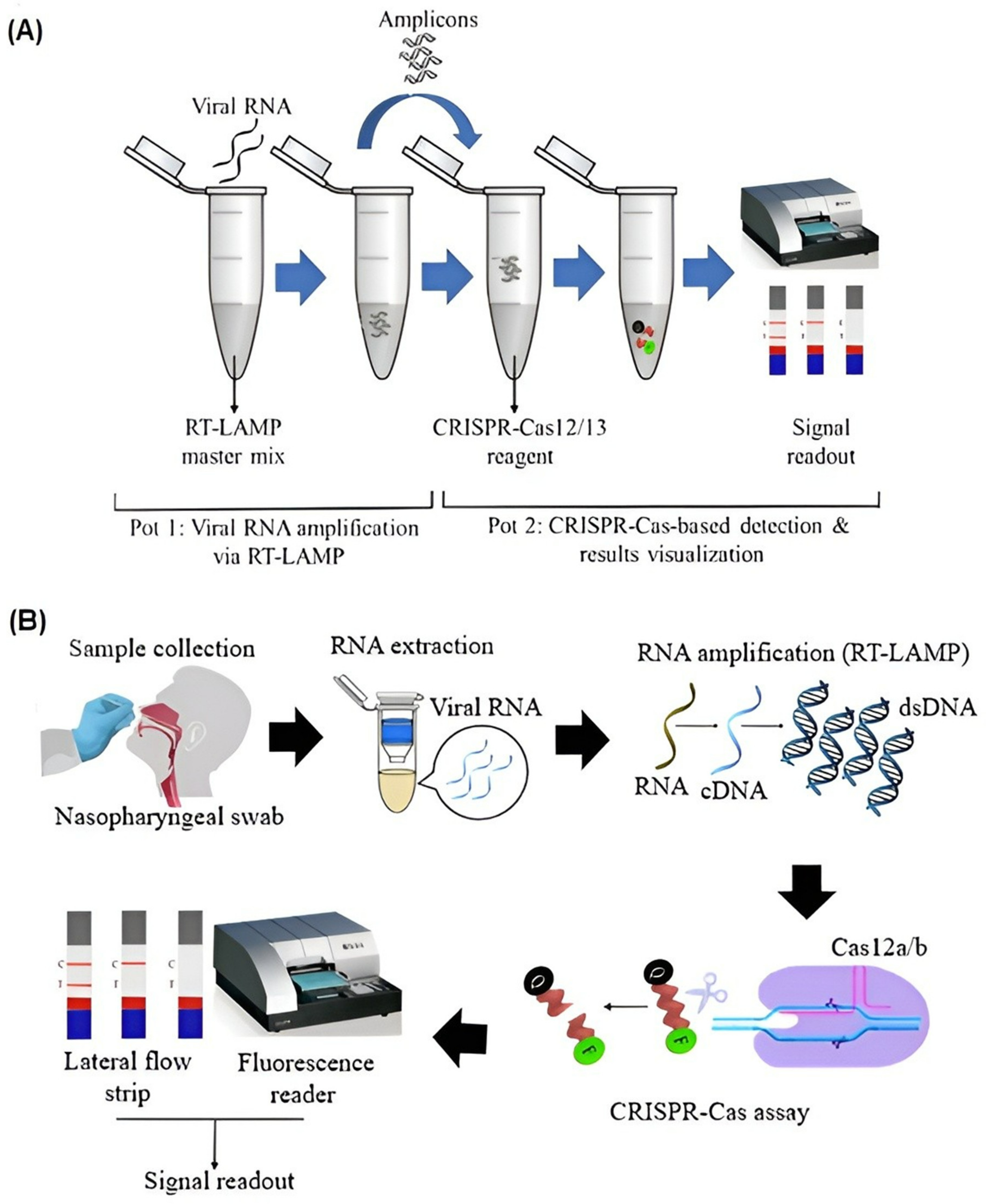
4. CRISPR/Cas Combined LAMP
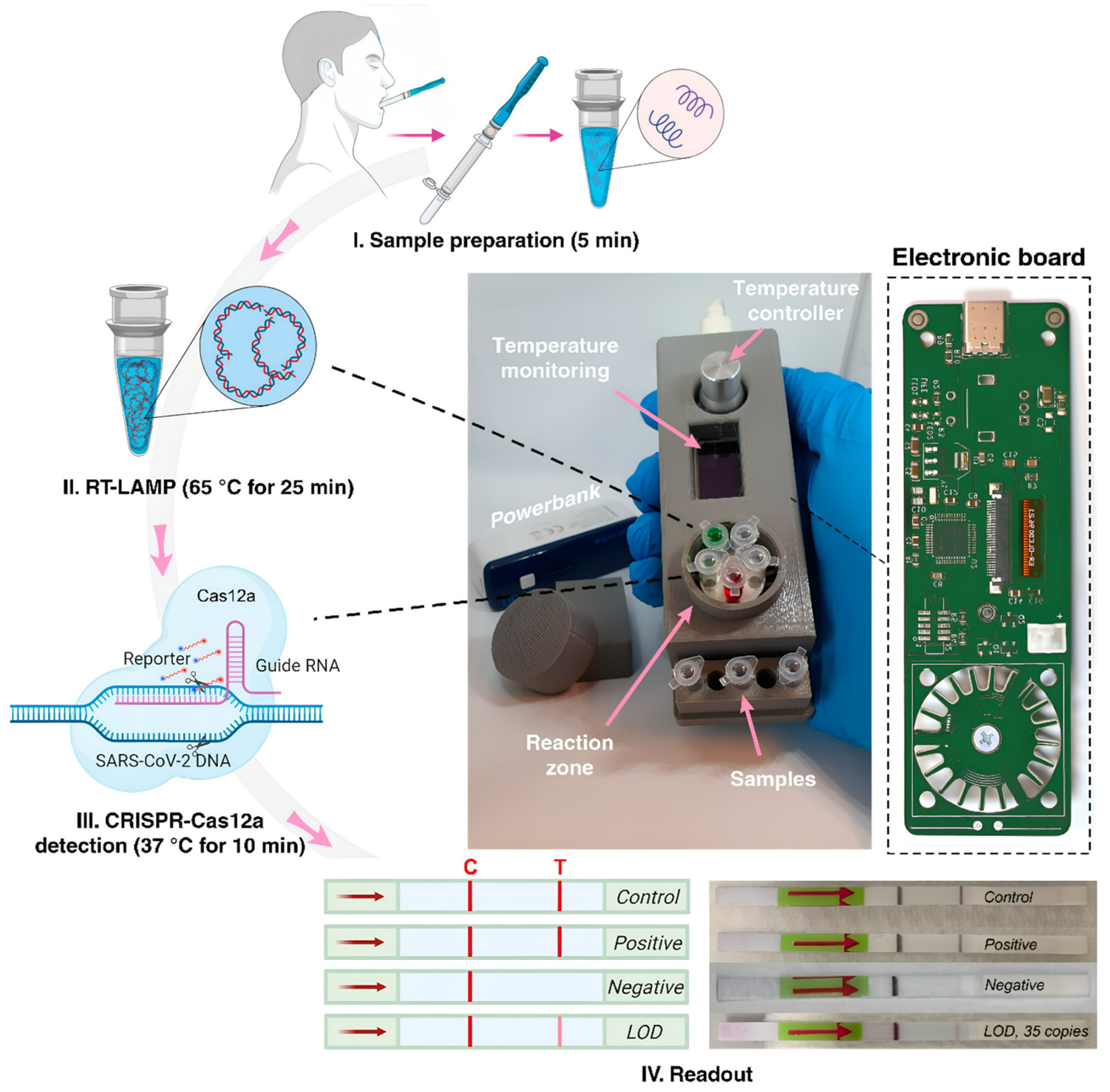
5. Point-of-Care Platforms
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhardwaj, P.; Kant, R.; Behera, S.P.; Dwivedi, G.R.; Singh, R. Next-Generation Diagnostic with CRISPR/Cas: Beyond Nucleic Acid Detection. Int. J. Mol. Sci. 2022, 23, 6052. [Google Scholar] [CrossRef] [PubMed]
- Aman, R.; Mahas, A.; Mahfouz, M. Nucleic Acid Detection Using CRISPR/Cas Biosensing Technologies. ACS Synth. Biol. 2020, 9, 1226–1233. [Google Scholar] [CrossRef] [PubMed]
- Mann, K.; Ogilvie, C.M. QF-PCR: Application, overview and review of the literature. Prenat. Diagn. 2012, 32, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Freije, C.A.; Sabeti, P.C. Detect and destroy: CRISPR-based technologies for the response against viruses. Cell Host Microbe 2021, 29, 689–703. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, P.; Zhou, S.; Zhang, L. Loop-mediated isothermal amplification (LAMP): A novel rapid detection platform for pathogens. Microb. Pathog. 2017, 107, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [Green Version]
- Hong, T.C.; Mai, Q.L.; Cuong, D.V.; Parida, M.; Minekawa, H.; Notomi, T.; Hasebe, F.; Morita, K. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004, 42, 1956–1961. [Google Scholar] [CrossRef] [Green Version]
- Parida, M.; Posadas, G.; Inoue, S.; Hasebe, F.; Morita, K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J. Clin. Microbiol. 2004, 42, 257–263. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Tang, G.; Ismail, N.; Wang, X. Developing RT-LAMP Assays for Detection of SARS-CoV-2 in Saliva. medRxiv 2021. [Google Scholar] [CrossRef]
- García-Bernalt Diego, J.; Fernández-Soto, P.; Muro, A. LAMP in Neglected Tropical Diseases: A Focus on Parasites. Diagnostics 2021, 11, 521. [Google Scholar] [CrossRef]
- Parida, M.; Sannarangaiah, S.; Dash, P.K.; Rao, P.V.; Morita, K. Loop mediated isothermal amplification (LAMP): A new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 2008, 18, 407–421. [Google Scholar] [CrossRef]
- Gill, P.; Ghaemi, A. Nucleic acid isothermal amplification technologies: A review. Nucleosides Nucleotides Nucleic Acids 2008, 27, 224–243. [Google Scholar] [CrossRef]
- Augustine, R.; Hasan, A.; Das, S.; Ahmed, R.; Mori, Y.; Notomi, T.; Kevadiya, B.D.; Thakor, A.S. Loop-Mediated Isothermal Amplification (LAMP): A Rapid, Sensitive, Specific, and Cost-Effective Point-of-Care Test for Coronaviruses in the Context of COVID-19 Pandemic. Biology 2020, 9, 182. [Google Scholar] [CrossRef]
- Sakurai, A.; Shibasaki, F. Updated values for molecular diagnosis for highly pathogenic avian influenza virus. Viruses 2012, 4, 1235–1257. [Google Scholar] [CrossRef] [Green Version]
- Wong, Y.P.; Othman, S.; Lau, Y.L.; Radu, S.; Chee, H.Y. Loop-mediated isothermal amplification (LAMP): A versatile technique for detection of micro-organisms. J. Appl. Microbiol. 2018, 124, 626–643. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.; Wang, R.; Li, B.; Liu, P.; Weng, Q.; Chen, Q. Comparative Evaluation of the LAMP Assay and PCR-Based Assays for the Rapid Detection of Alternaria solani. Front. Microbiol. 2018, 9, 2089. [Google Scholar] [CrossRef] [Green Version]
- Varona, M.; Anderson, J.L. Advances in Mutation Detection Using Loop-Mediated Isothermal Amplification. ACS Omega 2021, 6, 3463–3469. [Google Scholar] [CrossRef]
- Higgins, O.; Smith, T.J. Loop-Primer Endonuclease Cleavage-Loop-Mediated Isothermal Amplification Technology for Multiplex Pathogen Detection and Single-Nucleotide Polymorphism Identification. J. Mol. Diagn. 2020, 22, 640–651. [Google Scholar] [CrossRef]
- Liu, W.; Huang, S.; Liu, N.; Dong, D.; Yang, Z.; Tang, Y.; Ma, W.; He, X.; Ao, D.; Xu, Y.; et al. Establishment of an accurate and fast detection method using molecular beacons in loop-mediated isothermal amplification assay. Sci. Rep. 2017, 7, 40125. [Google Scholar] [CrossRef] [Green Version]
- Carlos, F.F.; Veigas, B.; Matias, A.S.; Doria, G.; Flores, O.; Baptista, P.V. Allele specific LAMP-gold nanoparticle for characterization of single nucleotide polymorphisms. Biotechnol. Rep. 2017, 16, 21–25. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Karvelis, T.; Gasiunas, G.; Miksys, A.; Barrangou, R.; Horvath, P.; Siksnys, V. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol. 2013, 10, 841–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, P.; Chou, S.J.; Li, J.; Hui, W.; Liu, W.; Sun, N.; Zhang, R.Y.; Zhu, Y.; Tsai, M.L.; Lai, H.I.; et al. Supramolecular nanosubstrate-mediated delivery system enables CRISPR-Cas9 knockin of hemoglobin beta gene for hemoglobinopathies. Sci. Adv. 2020, 6, eabb7107. [Google Scholar] [CrossRef] [PubMed]
- Kostyusheva, A.; Brezgin, S.; Babin, Y.; Vasilyeva, I.; Glebe, D.; Kostyushev, D.; Chulanov, V. CRISPR-Cas systems for diagnosing infectious diseases. Methods 2022, 203, 431–446. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shang, X.; Huang, X. Next-generation pathogen diagnosis with CRISPR/Cas-based detection methods. Emerg. Microbes Infect. 2020, 9, 1682–1691. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-based diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Wang, Q.; Xu, X.; Xia, Q.; Long, F.; Li, W.; Shui, Y.; Xia, X.; Wang, J. Detection of target DNA with a novel Cas9/sgRNAs-associated reverse PCR (CARP) technique. Anal. Bioanal. Chem. 2018, 410, 2889–2900. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, B.; Xu, X.; Long, F.; Wang, J. CRISPR-typing PCR (ctPCR), a new Cas9-based DNA detection method. Sci. Rep. 2018, 8, 14126. [Google Scholar] [CrossRef] [Green Version]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [Green Version]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef] [Green Version]
- Li, S.Y.; Cheng, Q.X.; Wang, J.M.; Li, X.Y.; Zhang, Z.L.; Gao, S.; Cao, R.B.; Zhao, G.P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 20. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, A.; Fatima, Z.; Ruwali, M.; Misra, C.S.; Rangu, S.S.; Rath, D.; Rattan, A.; Hameed, S. CLEVER assay: A visual and rapid RNA extraction-free detection of SARS-CoV-2 based on CRISPR-Cas integrated RT-LAMP technology. J. Appl. Microbiol. 2022, 133, 410–421. [Google Scholar] [CrossRef]
- Chou, P.H.; Lin, Y.C.; Teng, P.H.; Chen, C.L.; Lee, P.Y. Real-time target-specific detection of loop-mediated isothermal amplification for white spot syndrome virus using fluorescence energy transfer-based probes. J. Virol. Methods. 2011, 173, 67–74. [Google Scholar] [CrossRef]
- Nagai, K.; Horita, N.; Yamamoto, M.; Tsukahara, T.; Nagakura, H.; Tashiro, K.; Shibata, Y.; Watanabe, H.; Nakashima, K.; Ushio, R.; et al. Diagnostic test accuracy of loop-mediated isothermal amplification assay for Mycobacterium tuberculosis: Systematic review and meta-analysis. Sci. Rep. 2016, 6, 39090. [Google Scholar] [CrossRef] [Green Version]
- Hardinge, P.; Murray, J.A.H. Reduced False Positives and Improved Reporting of Loop-Mediated Isothermal Amplification using Quenched Fluorescent Primers. Sci. Rep. 2019, 9, 7400. [Google Scholar] [CrossRef] [Green Version]
- Ali, Z.; Aman, R.; Mahas, A.; Rao, G.S.; Tehseen, M.; Marsic, T.; Salunke, R.; Subudhi, A.K.; Hala, S.M.; Hamdan, S.M.; et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020, 288, 198129. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shi, Y.; Chen, Y.; Yang, Z.; Wu, H.; Zhou, Z.; Li, J.; Ping, J.; He, L.; Shen, H.; et al. Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: A promising method in the point-of-care detection. Biosens. Bioelectron. 2020, 169, 112642. [Google Scholar] [CrossRef]
- Shi, Y.; Kang, L.; Mu, R.; Xu, M.; Duan, X.; Li, Y.; Yang, C.; Ding, J.W.; Wang, Q.; Li, S. CRISPR/Cas12a-Enhanced Loop-Mediated Isothermal Amplification for the Visual Detection of Shigella flexneri. Front. Bioeng. Biotechnol. 2022, 10, 845688. [Google Scholar] [CrossRef]
- Selvam, K.; Najib, M.A.; Khalid, M.F.; Mohamad, S.; Palaz, F.; Ozsoz, M.; Aziah, I. RT-LAMP CRISPR-Cas12/13-Based SARS-CoV-2 Detection Methods. Diagnostics 2021, 11, 1646. [Google Scholar] [CrossRef] [PubMed]
- Jolany vangah, S.; Katalani, C.; Boone, H.A.; Hajizade, A.; Sijercic, A.; Ahmadian, G. CRISPR-Based Diagnosis of Infectious and Noninfectious Diseases. Biol. Proced. Online 2020, 22, 22. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.X.; Hunnewell, P.; Alfonse, L.E.; Carte, J.M.; Keston-Smith, E.; Sothiselvam, S.; Garrity, A.J.; Chong, S.; Makarova, K.S.; Koonin, E.V.; et al. Functionally diverse type V CRISPR-Cas systems. Science 2019, 363, 88–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [Green Version]
- Pang, B.; Xu, J.; Liu, Y.; Peng, H.; Feng, W.; Cao, Y.; Wu, J.; Xiao, H.; Pabbaraju, K.; Tipples, G.; et al. Isothermal Amplification and Ambient Visualization in a Single Tube for the Detection of SARS-CoV-2 Using Loop-Mediated Amplification and CRISPR Technology. Anal. Chem. 2020, 92, 16204–16212. [Google Scholar] [CrossRef]
- Joung, J.; Ladha, A.; Saito, M.; Kim, N.G.; Woolley, A.E.; Segel, M.; Barretto, R.P.J.; Ranu, A.; Macrae, R.K.; Faure, G.; et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N. Engl. J. Med. 2020, 383, 1492–1494. [Google Scholar] [CrossRef]
- Li, S.; Huang, J.; Ren, L.; Jiang, W.; Wang, M.; Zhuang, L.; Zheng, Q.; Yang, R.; Zeng, Y.; Luu, L.D.W.; et al. A one-step, one-pot CRISPR nucleic acid detection platform (CRISPR-top): Application for the diagnosis of COVID-19. Talanta 2021, 233, 122591. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [Green Version]
- Ramachandran, A.; Huyke, D.A.; Sharma, E.; Sahoo, M.K.; Huang, C.; Banaei, N.; Pinsky, B.A.; Santiago, J.G. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 29518–29525. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Macaluso, N.C.; Pizzano, B.L.M.; Cash, M.N.; Spacek, J.; Karasek, J.; Miller, M.R.; Lednicky, J.A.; Dinglasan, R.R.; Salemi, M.; et al. A thermostable Cas12b from Brevibacillus leverages one-pot discrimination of SARS-CoV-2 variants of concern. eBioMedicine 2022, 77, 103926. [Google Scholar] [CrossRef]
- Rezaei, M.; Razavi Bazaz, S.; Morshedi Rad, D.; Shimoni, O.; Jin, D.; Rawlinson, W.; Ebrahimi Warkiani, M. A Portable RT-LAMP/CRISPR Machine for Rapid COVID-19 Screening. Biosensors 2021, 11, 369. [Google Scholar] [CrossRef]
- Garcia-Venzor, A.; Rueda-Zarazua, B.; Marquez-Garcia, E.; Maldonado, V.; Moncada-Morales, A.; Olivera, H.; Lopez, I.; Zuñiga, J.; Melendez-Zajgla, J. SARS-CoV-2 Direct Detection Without RNA Isolation With Loop-Mediated Isothermal Amplification (LAMP) and CRISPR-Cas12. Front. Med. 2021, 8, 627679. [Google Scholar] [CrossRef]
- Park, B.J.; Park, M.S.; Lee, J.M.; Song, Y.J. Specific Detection of Influenza A and B Viruses by CRISPR-Cas12a-Based Assay. Biosensors 2021, 11, 88. [Google Scholar] [CrossRef]
- Mukama, O.; Yuan, T.; He, Z.; Li, Z.; Habimana, J.d.D.; Hussain, M.; Li, W.; Yi, Z.; Liang, Q.; Zeng, L. A high fidelity CRISPR/Cas12a based lateral flow biosensor for the detection of HPV16 and HPV18. Sens. Actuators B Chem. 2020, 316, 128119. [Google Scholar] [CrossRef]
- Kham-Kjing, N.; Ngo-Giang-Huong, N.; Tragoolpua, K.; Khamduang, W.; Hongjaisee, S. Highly Specific and Rapid Detection of Hepatitis C Virus Using RT-LAMP-Coupled CRISPR-Cas12 Assay. Diagnostics 2022, 12, 1524. [Google Scholar] [CrossRef]
- Trung, N.T.; Son, L.H.P.; Hien, T.X.; Quyen, D.T.; Bang, M.H.; Song, L.H. CRISPR-Cas12a combination to alleviate the false-positive in loop-mediated isothermal amplification-based diagnosis of Neisseria meningitidis. BMC Infect. Dis. 2022, 22, 429. [Google Scholar] [CrossRef]
- Xu, H.; Tang, H.; Li, R.; Xia, Z.; Yang, W.; Zhu, Y.; Liu, Z.; Lu, G.; Ni, S.; Shen, J. A New Method Based on LAMP-CRISPR-Cas12a-Lateral Flow Immunochromatographic Strip for Detection. Infect. Drug Resist. 2022, 15, 685–696. [Google Scholar] [CrossRef]
- Zanoli, L.M.; Spoto, G. Isothermal amplification methods for the detection of nucleic acids in microfluidic devices. Biosensors 2013, 3, 18–43. [Google Scholar] [CrossRef] [Green Version]
- Van Dongen, J.E.; Berendsen, J.T.W.; Steenbergen, R.D.M.; Wolthuis, R.M.F.; Eijkel, J.C.T.; Segerink, L.I. Point-of-care CRISPR/Cas nucleic acid detection: Recent advances, challenges and opportunities. Biosens. Bioelectron. 2020, 166, 112445. [Google Scholar] [CrossRef]
- Sarabi, M.R.; Yigci, D.; Alseed, M.M.; Mathyk, B.A.; Ata, B.; Halicigil, C.; Tasoglu, S. Disposable paper-based microfluidics for fertility testing. iScience 2022, 25, 104986. [Google Scholar] [CrossRef]
- Yigci, D.; Sarabi, M.R.; Ustun, M.; Atceken, N.; Sokullu, E.; Bagci-Onder, T.; Tasoglu, S. 3D bioprinted glioma models. Prog. Biomed. Eng. 2022, 4, 042001. [Google Scholar] [CrossRef]
- Nasseri, B.; Soleimani, N.; Rabiee, N.; Kalbasi, A.; Karimi, M.; Hamblin, M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018, 117, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Asiello, P.J.; Baeumner, A.J. Miniaturized isothermal nucleic acid amplification, a review. Lab Chip 2011, 11, 1420–1430. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Johnson, M.; Hill, P.; Gale, B.K. Microfluidic sample preparation: Cell lysis and nucleic acid purification. Integr. Biol. 2009, 1, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.-J.; Wang, L.; Chen, J.; Wang, R.-N.; Shi, Y.-H.; Li, C.-H.; Zhang, D.-M.; Yan, X.-J.; Zhang, Y.-J. Development and evaluation of a real-time fluorogenic loop-mediated isothermal amplification assay integrated on a microfluidic disc chip (on-chip LAMP) for rapid and simultaneous detection of ten pathogenic bacteria in aquatic animals. J. Microbiol. Methods 2014, 104, 26–35. [Google Scholar] [CrossRef]
- Liu, C.; Geva, E.; Mauk, M.; Qiu, X.; Abrams, W.R.; Malamud, D.; Curtis, K.; Owen, S.M.; Bau, H.H. An isothermal amplification reactor with an integrated isolation membrane for point-of-care detection of infectious diseases. Analyst 2011, 136, 2069–2076. [Google Scholar] [CrossRef] [Green Version]
- Hu, F.; Li, J.; Zhang, Z.; Li, M.; Zhao, S.; Li, Z.; Peng, N. Smartphone-Based Droplet Digital LAMP Device with Rapid Nucleic Acid Isolation for Highly Sensitive Point-of-Care Detection. Anal. Chem. 2020, 92, 2258–2265. [Google Scholar] [CrossRef]

| Method | Pathogen Targeted | Cas Protein | Advantages | Limitations | References |
|---|---|---|---|---|---|
| iSCAN | SARS-CoV-2 | Cas 12a Cas 12b | High specificity (100%), single tube | Low sensitivity (50% for fluorescence, 86% for lateral flow) | [38] |
| SARS-CoV-2/DETECTR | SARS-CoV-2 | Cas 12a | High sensitivity (100%) and specificity (100%), short assay time (40 min), single tube | [45] | |
| STOPCovid.v2 | SARS-CoV-2 | Cas 12a | High specificity (98.5%) | Complex assay design | [46] |
| SARS-CoV-2 DETECTR | SARS-CoV-2 | Cas12a | High specificity (100%) and sensitivity (100%), short assay time (40 min) | [48] | |
| ITP-CRISPR | SARS-CoV-2 | Cas 12a | Short assay time (30–40 min), high specificity (100%), reduced cost of production | Complex assay design, relatively low sensitivity (90.6%) | [49] |
| CRISPR-SPADE | SARS-CoV-2 | Cas 12b | Short assay time (10–30 min), high specificity (99.4%), single tube assay design, variant-specific detection | [50] | |
| Portable RT-LAMP/CRISPR | SARS-CoV-2 | Cas 12a | Short assay time (35 min), portable device | Sensitivity and specificity no quantified | [51] |
| LAMP-LbCas12a Method | SARS-CoV-2 | Cas 12a | Short assay time (40 min), high specificity (100%) | [52] | |
| CRISPR/Cas12a Based Detection | Influenza A and B | Cas 12a | Relatively short assay time (75–85%) | Sensitivity and specificity no quantified | [53] |
| CIALFB | Human Papillomavirus 16 and 18 | Cas 12a | High specificity (100%) and sensitivity (100%) | [54] | |
| RT-LAMP Coupled CRISPR/Cas12 | Hepatitis C Virus | Cas 12a | High sensitivity (96%) and specificity (100%) | Longer assay time (60–90 min) | [55] |
| CRISPR/Cas12a-E-LAMP | Shigella flexneri | Cas 12a | Short assay time (40 min), single tube | Sensitivity and specificity no quantified | [40] |
| CRISPR/Cas12a-LAMP | Neisseria meningitidis | Cas 12a | No false positives produced | Sensitivity and specificity no quantified | [56] |
| CRISPR/Cas12a-LAMP | Klebsiella pneumonia carbapenemase | Cas 12a | Short assay time (30–40 min) | Sensitivity and specificity no quantified | [57] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atçeken, N.; Yigci, D.; Ozdalgic, B.; Tasoglu, S. CRISPR-Cas-Integrated LAMP. Biosensors 2022, 12, 1035. https://doi.org/10.3390/bios12111035
Atçeken N, Yigci D, Ozdalgic B, Tasoglu S. CRISPR-Cas-Integrated LAMP. Biosensors. 2022; 12(11):1035. https://doi.org/10.3390/bios12111035
Chicago/Turabian StyleAtçeken, Nazente, Defne Yigci, Berin Ozdalgic, and Savas Tasoglu. 2022. "CRISPR-Cas-Integrated LAMP" Biosensors 12, no. 11: 1035. https://doi.org/10.3390/bios12111035
APA StyleAtçeken, N., Yigci, D., Ozdalgic, B., & Tasoglu, S. (2022). CRISPR-Cas-Integrated LAMP. Biosensors, 12(11), 1035. https://doi.org/10.3390/bios12111035






