Dietary Stimuli, Intestinal Bacteria and Peptide Hormones Regulate Female Drosophila Defecation Rate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of LB and BHI Agar Plates with or without Rifampicin
2.2. Bacteria Strains, Infection and Determination of Bacterial Load
2.2.1. Bacteria Strains
2.2.2. Bacterial Infection
2.2.3. Bacterial Load
2.2.4. Isolation of Bacterial Species from Drosophila
2.2.5. Single Colony PCR and Analysis of 16S rRNA Genes
2.2.6. DNA Extraction from Bacterial Species for Sequencing
2.3. Drosophila Melanogaster Diet, Maintenance, Strains and Experiments
2.3.1. Drosophila Diet and Maintenance
2.3.2. Germ-Free Flies
2.3.3. Drosophila Strains
2.3.4. Defecation Assay
2.3.5. Survival Assay
2.3.6. Determination of Narrow-Sense Heritability (h2)
2.3.7. Fly Midgut and Head Dissections
2.4. RNA Extraction, cDNA Synthesis and RT-qPCR
2.5. Immunohistochemistry
2.6. Image Acquisition and Analysis
2.7. Statistical Analysis
3. Results
3.1. Specific Gut Microbiota Composition and Virulent Bacteria Ingestion Increase Defecation Rate in Drosophila Females
3.2. Water-Only and Ethanol-Containing Food Reduce Lifespan and Defecation Rate in Female Adults
3.3. Genome Wide Analysis Reveals the Genetic Basis of Female Drosophila Defecation
3.4. Defecation Rate Is Controlled in Female Midgut EEs and Neurons by GWAS-Identified and Hormone-Encoding Genes
3.5. Higher Expression of NPF and CG11307 in DGRP Lines Displaying High Defecation Rates
3.6. GWAS-Identified and Hormonal Genes Participate in a Defecation Rate Signaling Network
3.7. GWAS-Identified Inducers of Defecation Induce Dh31 Expression in the Midgut and Reduce it in the Head
3.8. Water-Only Consumption Reduces Dh31, AstC, AstA and Pmp70 Expression in the Midgut and Reduced Expression of meso18E in the Head
3.9. Downregulation of Dh31 in the Midgut EEs or in Neurons Reduces Dh31 Expression in Both the Midgut and Head
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kitazawa, T.; Kaiya, H. Regulation of Gastrointestinal Motility by Motilin and Ghrelin in Vertebrates. Front. Endocrinol. 2019, 10, 278. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, M.P.; Batterham, R.L. The Importance of the Gastrointestinal Tract in Controlling Food Intake and Regulating Energy Balance. Gastroenterology 2017, 152, 1707–1717.e2. [Google Scholar] [CrossRef] [PubMed]
- Daniel, N.; Lécuyer, E.; Chassaing, B. Host/Microbiota Interactions in Health and Diseases-Time for Mucosal Microbiology! Mucosal Immunol. 2021, 14, 1006–1016. [Google Scholar] [CrossRef] [PubMed]
- Iacob, S.; Iacob, D.G.; Luminos, L.M. Intestinal Microbiota as a Host Defense Mechanism to Infectious Threats. Front. Microbiol. 2018, 9, 3328. [Google Scholar] [CrossRef]
- Rozé, C. Neurohumoral Control of Gastrointestinal Motility. Reprod. Nutr. Dev. 1980, 20, 1125–1141. [Google Scholar] [CrossRef]
- Daniel, E.; Tougas, G.; Allescher, H.D.; Vergara, P.; Fox-Threlkeld, J.A. Mediators and Enteric Nerve Pathways Controlling Gastric Emptying. Dig. Dis. Sci. 1994, 39, 63S–68S. [Google Scholar] [CrossRef]
- Travagli, R.A.; Anselmi, L. Vagal Neurocircuitry and Its Influence on Gastric Motility. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 389–401. [Google Scholar] [CrossRef]
- Boeckxstaens, G.; Camilleri, M.; Sifrim, D.; Houghton, L.A.; Elsenbruch, S.; Lindberg, G.; Azpiroz, F.; Parkman, H.P. Fundamentals of Neurogastroenterology: Physiology/Motility—Sensation. Gastroenterology 2016, 150, 1292–1304.e2. [Google Scholar] [CrossRef]
- Apidianakis, Y.; Rahme, L.G. Drosophila Melanogaster as a Model for Human Intestinal Infection and Pathology. Dis. Model. Mech. 2011, 4, 21–30. [Google Scholar] [CrossRef]
- Hadjieconomou, D.; King, G.; Gaspar, P.; Mineo, A.; Blackie, L.; Ameku, T.; Studd, C.; de Mendoza, A.; Diao, F.; White, B.H.; et al. Enteric Neurons Increase Maternal Food Intake during Reproduction. Nature 2020, 587, 455–459. [Google Scholar] [CrossRef]
- Charalambous, A.; Grivogiannis, E.; Dieronitou, I.; Michael, C.; Rahme, L.; Apidianakis, Y. Proteobacteria and Firmicutes Secreted Factors Exert Distinct Effects on Pseudomonas aeruginosa Infection under Normoxia or Mild Hypoxia. Metabolites 2022, 12, 449. [Google Scholar] [CrossRef] [PubMed]
- Pitsouli, C.; Apidianakis, Y.; Perrimon, N. Homeostasis in Infected Epithelia: Stem Cells Take the Lead. Cell Host Microbe 2009, 6, 301–307. [Google Scholar] [CrossRef] [PubMed]
- LaJeunesse, D.R.; Johnson, B.; Presnell, J.S.; Catignas, K.K.; Zapotoczny, G. Peristalsis in the Junction Region of the Drosophila Larval Midgut is Modulated by DH31 Expressing Enteroendocrine Cells. BMC Physiol. 2010, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Benguettat, O.; Jneid, R.; Soltys, J.; Loudhaief, R.; Brun-Barale, A.; Osman, D.; Gallet, A. The DH31/CGRP Enteroendocrine Peptide Triggers Intestinal Contractions Favoring the Elimination of Opportunistic Bacteria. PLoS Pathog. 2018, 14, e1007279. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Kim, S.-M.; Kwon, J.Y. A Systematic Analysis of Drosophila Regulatory Peptide Expression in Enteroendocrine Cells. Mol. Cells 2016, 39, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Coast, G.M.; Webster, S.G.; Schegg, K.M.; Tobe, S.S.; Schooley, D.A. The Drosophila Melanogaster Homologue of an Insect Calcitonin-like Diuretic Peptide Stimulates V-ATPase Activity in Fruit Fly Malpighian Tubules. J. Exp. Biol. 2001, 204, 1795–1804. [Google Scholar] [CrossRef]
- Wu, Q.; Zhao, Z.; Shen, P. Regulation of Aversion to Noxious Food by Drosophila Neuropeptide Y- and Insulin-like Systems. Nat. Neurosci. 2005, 8, 1350–1355. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, Y.; Xu, J.; Shen, P. Regulation of Hunger-Driven Behaviors by Neural Ribosomal S6 Kinase in Drosophila. Proc. Natl. Acad. Sci. USA 2005, 102, 13289–13294. [Google Scholar] [CrossRef]
- Wu, Q.; Wen, T.; Lee, G.; Park, J.H.; Cai, H.N.; Shen, P. Developmental Control of Foraging and Social Behavior by the Drosophila Neuropeptide Y-like System. Neuron 2003, 39, 147–161. [Google Scholar] [CrossRef]
- Yoshinari, Y.; Kosakamoto, H.; Kamiyama, T.; Hoshino, R.; Matsuoka, R.; Kondo, S.; Tanimoto, H.; Nakamura, A.; Obata, F.; Niwa, R. The Sugar-Responsive Enteroendocrine Neuropeptide F Regulates Lipid Metabolism through Glucagon-like and Insulin-like Hormones in Drosophila Melanogaster. Nat. Commun. 2021, 12, 4818. [Google Scholar] [CrossRef]
- Chen, J.; Reiher, W.; Hermann-Luibl, C.; Sellami, A.; Cognigni, P.; Kondo, S.; Helfrich-Förster, C.; Veenstra, J.A.; Wegener, C. Allatostatin A Signalling in Drosophila Regulates Feeding and Sleep and is Modulated by PDF. PLoS Genet. 2016, 12, e1006346. [Google Scholar]
- Hergarden, A.C.; Tayler, T.D.; Anderson, D.J. Allatostatin-A Neurons Inhibit Feeding Behavior in Adult Drosophila. Proc. Natl. Acad. Sci. USA 2012, 109, 3967–3972. [Google Scholar] [CrossRef]
- Hentze, J.L.; Carlsson, M.A.; Kondo, S.; Nässel, D.R.; Rewitz, K.F. The Neuropeptide Allatostatin A Regulates Metabolism and Feeding Decisions in Drosophila. Sci. Rep. 2015, 5, 11680. [Google Scholar] [CrossRef]
- Kubrak, O.; Koyama, T.; Ahrentløv, N.; Jensen, L.; Malita, A.; Naseem, M.T.; Lassen, M.; Nagy, S.; Texada, M.J.; Halberg, K.V.; et al. The Gut Hormone Allatostatin C/Somatostatin Regulates Food Intake and Metabolic Homeostasis under Nutrient Stress. Nat. Commun. 2022, 13, 692. [Google Scholar] [CrossRef]
- Chatterjee, N.; Perrimon, N. What Fuels the Fly: Energy Metabolism in Drosophila and Its Application to the Study of Obesity and Diabetes. Sci. Adv. 2021, 7, eabg4336. [Google Scholar] [CrossRef]
- Shimizu, Y.; Matsuyama, H.; Shiina, T.; Takewaki, T.; Furness, J.B. Tachykinins and Their Functions in the Gastrointestinal Tract. Cell. Mol. Life Sci. 2008, 65, 295–311. [Google Scholar] [CrossRef]
- Apidianakis, Y.; Pitsouli, C.; Perrimon, N.; Rahme, L. Synergy between Bacterial Infection and Genetic Predisposition in Intestinal Dysplasia. Proc. Natl. Acad. Sci. USA 2009, 106, 20883–20888. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Mackay, T.F.C.; Richards, S.; Stone, E.A.; Barbadilla, A.; Ayroles, J.F.; Zhu, D.; Casillas, S.; Han, Y.; Magwire, M.M.; Cridland, J.M.; et al. The Drosophila Melanogaster Genetic Reference Panel. Nature 2012, 482, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Massouras, A.; Inoue, Y.; Peiffer, J.; Ràmia, M.; Tarone, A.M.; Turlapati, L.; Zichner, T.; Zhu, D.; Lyman, R.F.; et al. Natural Variation in Genome Architecture among 205 Drosophila Melanogaster Genetic Reference Panel Lines. Genome Res. 2014, 24, 1193–1208. [Google Scholar] [CrossRef] [PubMed]
- Heys, C.; Lizé, A.; Blow, F.; White, L.; Darby, A.; Lewis, Z.J. The Effect of Gut Microbiota Elimination in Drosophila Melanogaster: A How-to Guide for Host-Microbiota Studies. Ecol. Evol. 2018, 8, 4150–4161. [Google Scholar] [CrossRef] [PubMed]
- Ridley, E.V.; Wong, A.C.-N.; Westmiller, S.; Douglas, A.E. Impact of the Resident Microbiota on the Nutritional Phenotype of Drosophila Melanogaster. PLoS ONE 2012, 7, e36765. [Google Scholar] [CrossRef]
- Clark, A.G.; Fucito, C.D. Stress Tolerance and Metabolic Response to Stress in Drosophila Melanogaster. Heredity 1998, 81, 514–527. [Google Scholar] [CrossRef] [PubMed]
- Hill, W.G.; Goddard, M.E.; Visscher, P.M. Data and Theory Point to Mainly Additive Genetic Variance for Complex Traits. PLoS Genet. 2008, 4, e1000008. [Google Scholar] [CrossRef]
- Weng, Y.-L.; Liu, N.; DiAntonio, A.; Broihier, H.T. The Cytoplasmic Adaptor Protein Caskin Mediates Lar Signal Transduction during Drosophila Motor Axon Guidance. J. Neurosci. 2011, 31, 4421–4433. [Google Scholar] [CrossRef] [PubMed]
- Chung, B.Y.; Ro, J.; Hutter, S.A.; Miller, K.M.; Guduguntla, L.S.; Kondo, S.; Pletcher, S.D. Drosophila Neuropeptide F Signaling Independently Regulates Feeding and Sleep-Wake Behavior. Cell Rep. 2017, 19, 2441–2450. [Google Scholar] [CrossRef]
- Veenstra, J.A.; Agricola, H.-J.; Sellami, A. Regulatory Peptides in Fruit Fly Midgut. Cell Tissue Res. 2008, 334, 499–516. [Google Scholar] [CrossRef]
- Dutta, D.; Dobson, A.J.; Houtz, P.L.; Gläßer, C.; Revah, J.; Korzelius, J.; Patel, P.H.; Edgar, B.A.; Buchon, N. Regional Cell-Specific Transcriptome Mapping Reveals Regulatory Complexity in the Adult Drosophila Midgut. Cell Rep. 2015, 12, 346–358. [Google Scholar] [CrossRef]
- Broderick, N.A.; Lemaitre, B. Gut-Associated Microbes of Drosophila Melanogaster. Gut Microbes 2012, 3, 307–321. [Google Scholar] [CrossRef]
- Chandler, J.A.; Morgan Lang, J.; Bhatnagar, S.; Eisen, J.A.; Kopp, A. Bacterial Communities of Diverse Drosophila Species: Ecological Context of a Host–Microbe Model System. PLoS Genet. 2011, 7, e1002272. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment Dominates over Host Genetics in Shaping Human Gut Microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Cognigni, P.; Bailey, A.P.; Miguel-Aliaga, I. Enteric Neurons and Systemic Signals Couple Nutritional and Reproductive Status with Intestinal Homeostasis. Cell Metab. 2011, 13, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Dus, M.; Lai, J.S.-Y.; Gunapala, K.M.; Min, S.; Tayler, T.D.; Hergarden, A.C.; Geraud, E.; Joseph, C.M.; Suh, G.S.B. Nutrient Sensor in the Brain Directs the Action of the Brain-Gut Axis in Drosophila. Neuron 2015, 87, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Senapati, B.; Tsao, C.-H. Neural Basis of Hunger-Driven Behaviour in Drosophila. Open Biol. 2019, 9, 180259. [Google Scholar] [CrossRef] [PubMed]
- Keene, A.C.; Duboué, E.R.; McDonald, D.M.; Dus, M.; Suh, G.S.B.; Waddell, S.; Blau, J. Clock and Cycle Limit Starvation-Induced Sleep Loss in Drosophila. Curr. Biol. 2010, 20, 1209–1215. [Google Scholar] [CrossRef] [PubMed]
- Chopra, G.; Kaushik, S.; Kain, P. Nutrient Sensing via Gut in Drosophila Melanogaster. Int. J. Mol. Sci. 2022, 23, 2694. [Google Scholar] [CrossRef]
- Veenstra, J.A.; Ida, T. More Drosophila Enteroendocrine Peptides: Orcokinin B and the CCHamides 1 and 2. Cell Tissue Res. 2014, 357, 607–621. [Google Scholar] [CrossRef]
- Visscher, P.M.; Hill, W.G.; Wray, N.R. Heritability in the Genomics Era—Concepts and Misconceptions. Nat. Rev. Genet. 2008, 9, 255–266. [Google Scholar] [CrossRef]
- O’Connell, K.S.; Hindley, G.; Smeland, O.B.; Shadrin, A.; Wang, Y.; Frei, O.; Andreassen, O.A. Chapter 16—Shared Heritability among Psychiatric Disorders and Traits. In Translational and Applied Genomics; Tsermpini, E.E., Alda, M., Patrinos, G.P.B.T.-P.G., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 341–360. ISBN 978-0-12-819602-1. [Google Scholar]
- Martin, A.R.; Daly, M.J.; Robinson, E.B.; Hyman, S.E.; Neale, B.M. Predicting Polygenic Risk of Psychiatric Disorders. Biol. Psychiatry 2019, 86, 97–109. [Google Scholar] [CrossRef]
- Havdahl, A.; Niarchou, M.; Starnawska, A.; Uddin, M.; van der Merwe, C.; Warrier, V. Genetic Contributions to Autism Spectrum Disorder. Psychol. Med. 2021, 51, 2260–2273. [Google Scholar] [CrossRef]
- Blizard, D.A.; Adams, N. The Maudsley Reactive and Nonreactive Strains: A New Perspective. Behav. Genet. 2002, 32, 277–299. [Google Scholar] [CrossRef]
- Branicky, R.; Hekimi, S. What Keeps C. Elegans Regular: The Genetics of Defecation. Trends Genet. 2006, 22, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Bonfiglio, F.; Liu, X.; Smillie, C.; Pandit, A.; Kurilshikov, A.; Bacigalupe, R.; Zheng, T.; Nim, H.; Garcia-Etxebarria, K.; Bujanda, L.; et al. GWAS of Stool Frequency Provides Insights into Gastrointestinal Motility and Irritable Bowel Syndrome. Cell Genom. 2021, 1, 100069. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.-H.; Kuang, M.C.; Hossain, I.; Xuan, Y.; Beebe, L.; Shepherd, A.K.; Rolandi, M.; Wang, J.W. A Nutrient-Specific Gut Hormone Arbitrates between Courtship and Feeding. Nature 2022, 602, 632–638. [Google Scholar] [CrossRef]
- Jimenez-Sanchez, G.; Hebron, K.J.; Thomas, G.; Valle, D. Targeted Disruption of the 70 kDa Peroxisomal Membrane Protein (PMP70) in Mouse is Associated with an Increase in the Related P70R Protein, Deficiency of Hepatic Glycogen and a Dicarboxylic Aciduria. Pediatr. Res. 1999, 45, 139. [Google Scholar] [CrossRef]
- Imanaka, T.; Aihara, K.; Suzuki, Y.; Yokota, S.; Osumi, T. The 70-KDa Peroxisomal Membrane Protein (PMP70) an ATP-Binding Cassette Transporter. Cell Biochem. Biophys. 2000, 32, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Schnorrer, F.; Schönbauer, C.; Langer, C.C.H.; Dietzl, G.; Novatchkova, M.; Schernhuber, K.; Fellner, M.; Azaryan, A.; Radolf, M.; Stark, A.; et al. Systematic Genetic Analysis of Muscle Morphogenesis and Function in Drosophila. Nature 2010, 464, 287–291. [Google Scholar] [CrossRef]
- Hudson, A.M.; Petrella, L.N.; Tanaka, A.J.; Cooley, L. Mononuclear Muscle Cells in Drosophila Ovaries Revealed by GFP Protein Traps. Dev. Biol. 2008, 314, 329–340. [Google Scholar] [CrossRef]
- Vasmer, D.; Pooryasin, A.; Riemensperger, T.; Fiala, A. Induction of Aversive Learning through Thermogenetic Activation of Kenyon Cell Ensembles in Drosophila. Front. Behav. Neurosci. 2014, 8, 174. [Google Scholar] [CrossRef]
- Pang, T.-L.; Ding, Z.; Liang, S.-B.; Li, L.; Zhang, B.; Zhang, Y.; Fan, Y.-J.; Xu, Y.-Z. Comprehensive Identification and Alternative Splicing of Microexons in Drosophila. Front. Genet. 2021, 12, 642602. [Google Scholar] [CrossRef]
- Tsao, C.-H.; Chen, C.-C.; Lin, C.-H.; Yang, H.-Y.; Lin, S. Drosophila Mushroom Bodies Integrate Hunger and Satiety Signals to Control Innate Food-Seeking Behavior. Elife 2018, 7, e35264. [Google Scholar] [CrossRef] [PubMed]
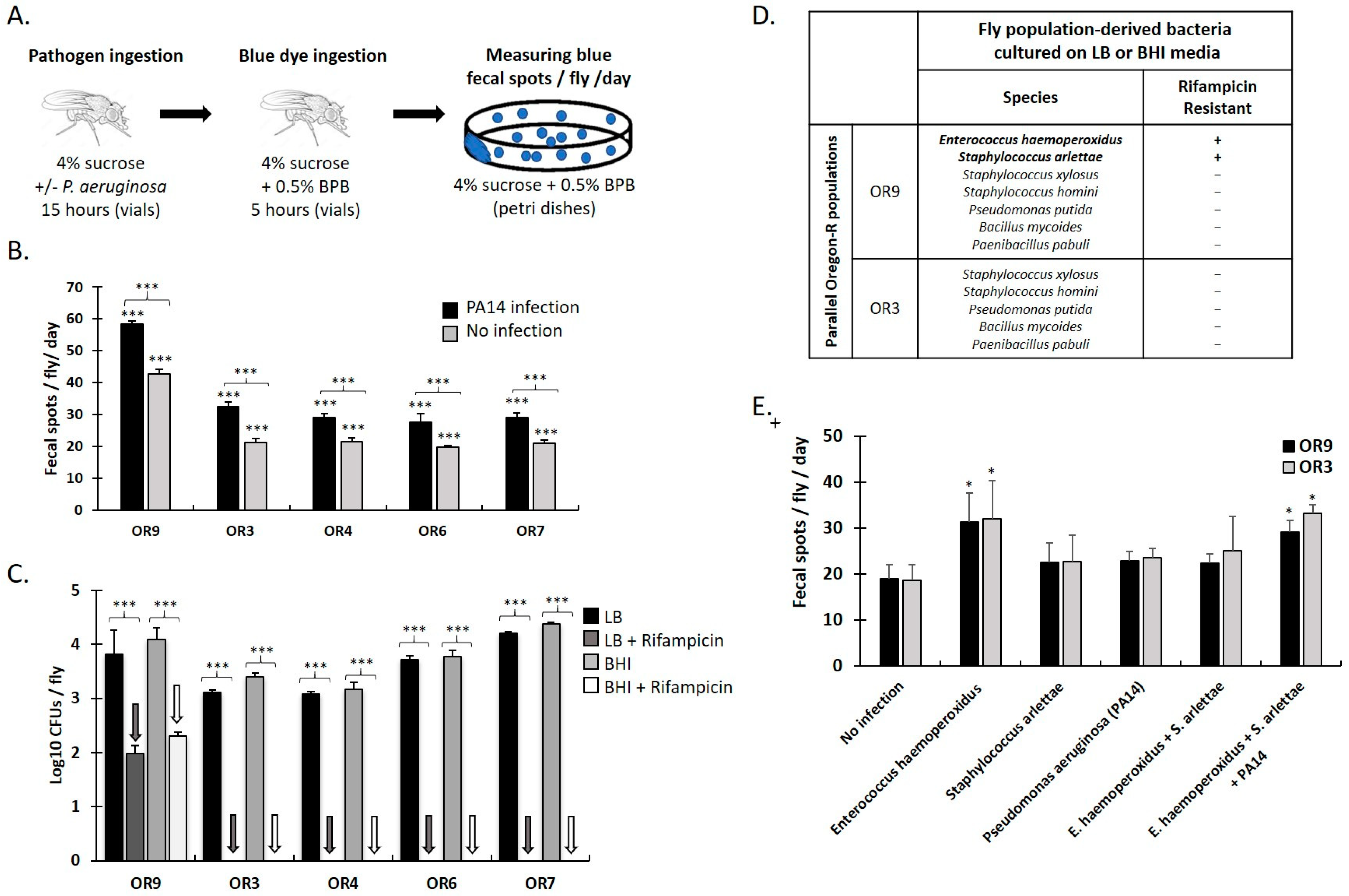
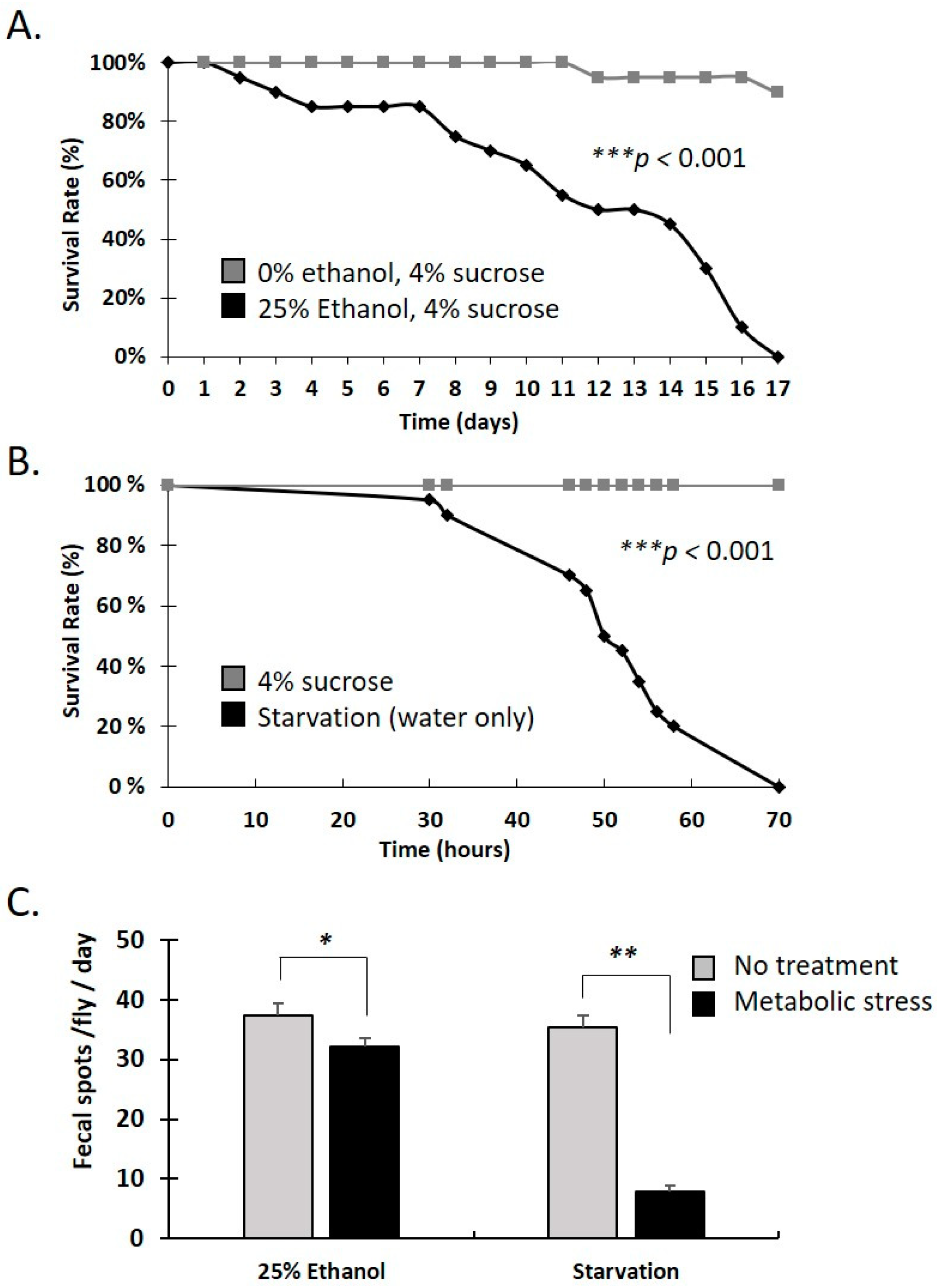

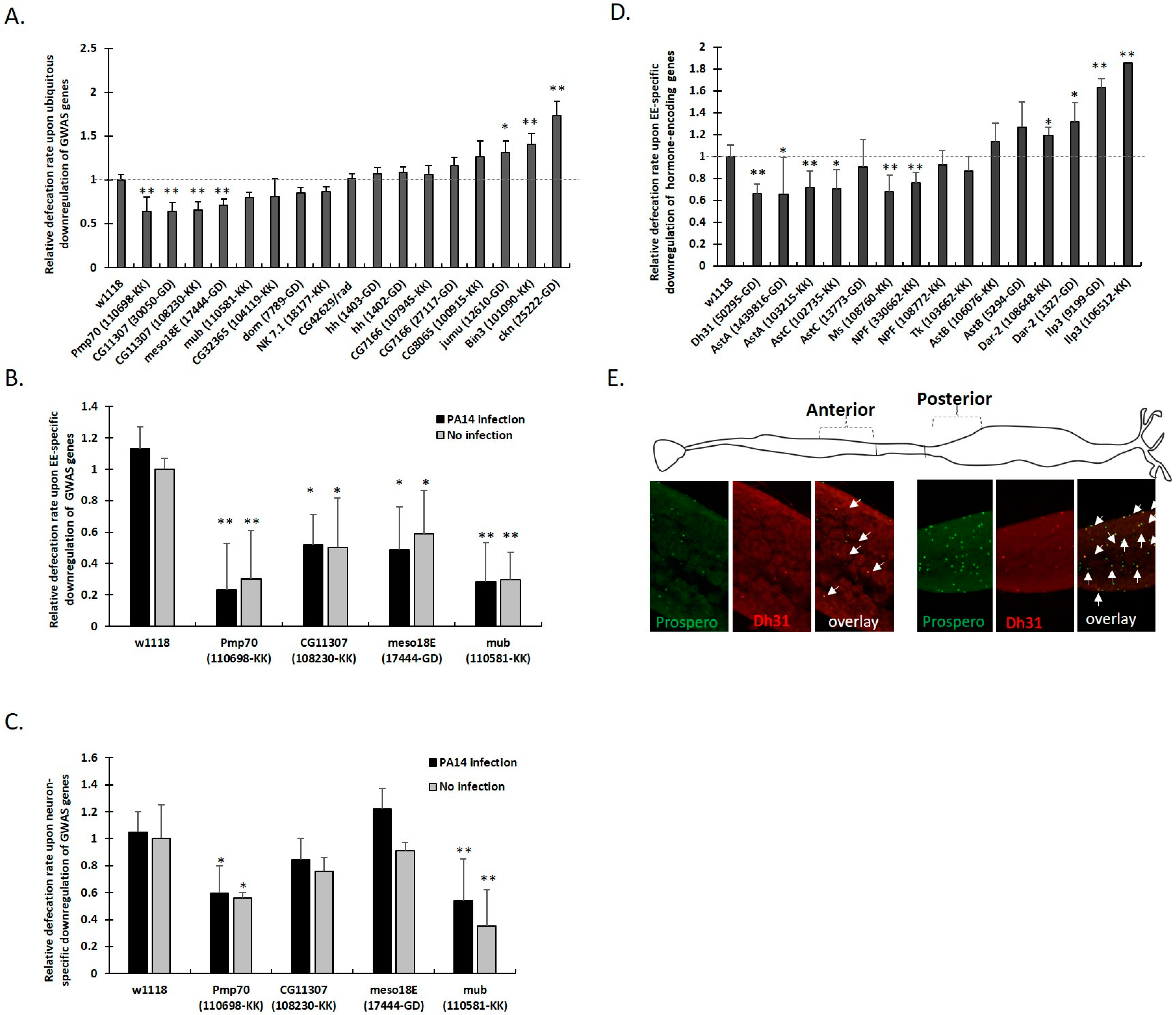






| Gene Affected (FlyBase ID, Name) | Associated Variant ID (SNPs) | Variant Hits per Gene | Gene Function and Midgut Expression (Flybase and FlyGutSeq) |
|---|---|---|---|
| FBgn0016075, vkg | 2L_5025290_SNP 2L_5025252_SNP 2L_5025248_SNP | 3 | Subunit of Collagen IV, a major component of basement membranes. Anterior midgut visceral muscle expression and enteroendocrine cell induction by infection. |
| FBgn0261388, CG42629 | X_12955420_SNP X_12955430_SNP | 2 | radish (rad) encodes a protein involved in anesthesia-resistant memory, heart contraction regulation, and small GTPase mediated signal transduction. |
| FBgn0262222, mir-971 | X_12955420_SNP X_12955430_SNP | 2 | Micro-RNA gene (within the rad locus) of unknown function. Enterocyte and visceral muscle expression. |
| FBgn0020306, dom | 2R_17221075_SNP 2R_17220640_SNP 2R_17220646_SNP | 3 | Tip60 acetyltransferase complex and functions in the exchange of histone His2Av. Uniform expression and stem cell induction upon infection. |
| FBgn0031069, Pmp70 | X_19647084_SNP | 1 | ATPase-coupled peroxisomal transmembrane transporter of long-chain fatty acids. High enteroendocrine cell and visceral muscle expression. |
| FBgn0037106, CG11307 | 3L_21612306_SNP 3L_21612307_SNP | 2 | Unknown function. Enteroendocrine cell expression. |
| FBgn0037107, CG7166 | 3L_21612306_SNP 3L_21612307_SNP | 2 | Predicted to be involved in homophilic cell adhesion. Enteroendocrine cell expression. |
| FBgn0262737, mub | 3L_21916473_SNP 3L_21853239_SNP | 2 | Regulation of alternative mRNA splicing. |
| FBgn0033987, ckn | 2R_10853015_SNP | 1 | Cell contact and tyrosine phosphatase signaling pathway. Enteroendocrine cell expression. |
| FBgn0024321, NK7.1 | 3R_10187071_SNP | 1 | Regulation of transcription by RNA polymerase II. Uniform expression. |
| FBgn0040089, meso18E | X_19604135_SNP | 1 | Visceral muscle development. Uniform expression and progenitor cell bias. |
| FBgn0004644, hh | 3R_18968168_SNP | 1 | Morphogen. Uniform expression and enteroblast bias. |
| FBgn0016075, CG8065 | 3L_10208120_SNP | 1 | Unknown function. Enterocyte lineage expression. |
| FBgn00263144, Bin3 | 2R_2126406_SNP | 1 | mRNA translation inhibition. Uniform expression and enterocyte bias. |
| FBgn0052365, CG32365 | 3L_7873543_SNP | 1 | Unknown function. Uniform expression and progenitor cell bias. |
| FBgn0001316, klar | 3L_461631_SNP | 1 | Organelle movement via microtubules. Uniform expression and enteroblast bias. |
| FBgn0011828, pxn | 3L_2603619_SNP | 1 | Collagen IV cross-linking. Uniform expression, enteroendocrine cell induction by infection. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kotronarou, K.; Charalambous, A.; Evangelou, A.; Georgiou, O.; Demetriou, A.; Apidianakis, Y. Dietary Stimuli, Intestinal Bacteria and Peptide Hormones Regulate Female Drosophila Defecation Rate. Metabolites 2023, 13, 264. https://doi.org/10.3390/metabo13020264
Kotronarou K, Charalambous A, Evangelou A, Georgiou O, Demetriou A, Apidianakis Y. Dietary Stimuli, Intestinal Bacteria and Peptide Hormones Regulate Female Drosophila Defecation Rate. Metabolites. 2023; 13(2):264. https://doi.org/10.3390/metabo13020264
Chicago/Turabian StyleKotronarou, Katerina, Anna Charalambous, Amalia Evangelou, Olympiada Georgiou, Andri Demetriou, and Yiorgos Apidianakis. 2023. "Dietary Stimuli, Intestinal Bacteria and Peptide Hormones Regulate Female Drosophila Defecation Rate" Metabolites 13, no. 2: 264. https://doi.org/10.3390/metabo13020264
APA StyleKotronarou, K., Charalambous, A., Evangelou, A., Georgiou, O., Demetriou, A., & Apidianakis, Y. (2023). Dietary Stimuli, Intestinal Bacteria and Peptide Hormones Regulate Female Drosophila Defecation Rate. Metabolites, 13(2), 264. https://doi.org/10.3390/metabo13020264







