(Pentamethylcyclopentadienyl)chloridoiridium(III) Complex Bearing Bidentate Ph2PCH2CH2SPh-κP,κS Ligand
Abstract
1. Introduction
2. Materials and Methods
2.1. General Comments
2.2. Preparation of [IrCl2(η5-C5Me5){Ph2P(CH2)2SPh-κP,κS}]PF6 (1)
2.3. X-ray Crystallography
2.4. Hirshfeld Surface Analysis
2.5. Theoretical Analysis of Structure
2.6. In Vitro Investigations
2.6.1. General Remarks
2.6.2. SRB Assay
2.6.3. Flow Cytometry Analyses
3. Results and Discussion
3.1. Preparation and Characterization of [Ir(η5-C5Me5)Cl(Ph2P(CH2)2SPh-κP,κS)][PF6] (1)
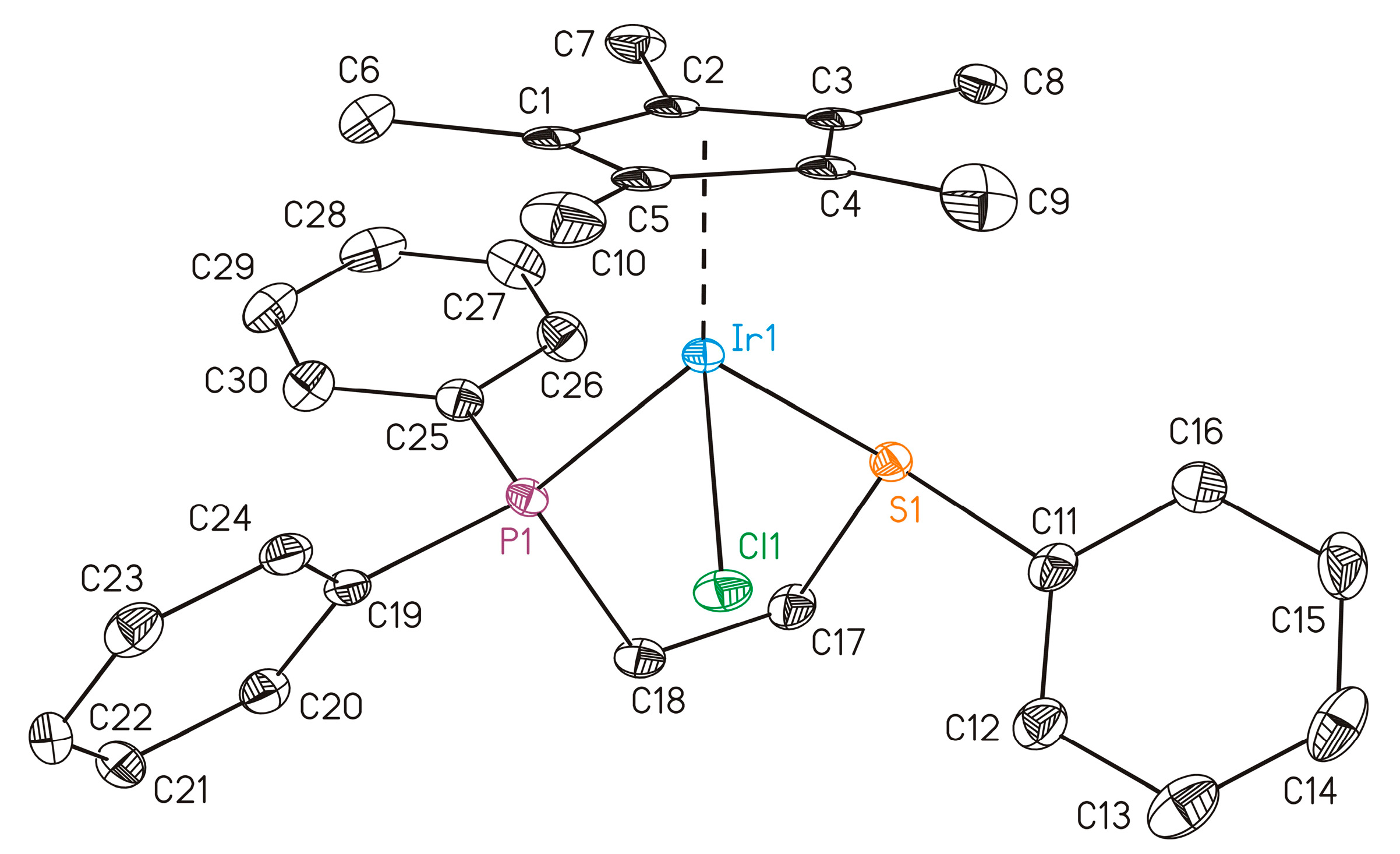
3.2. Molecular Structure of 1
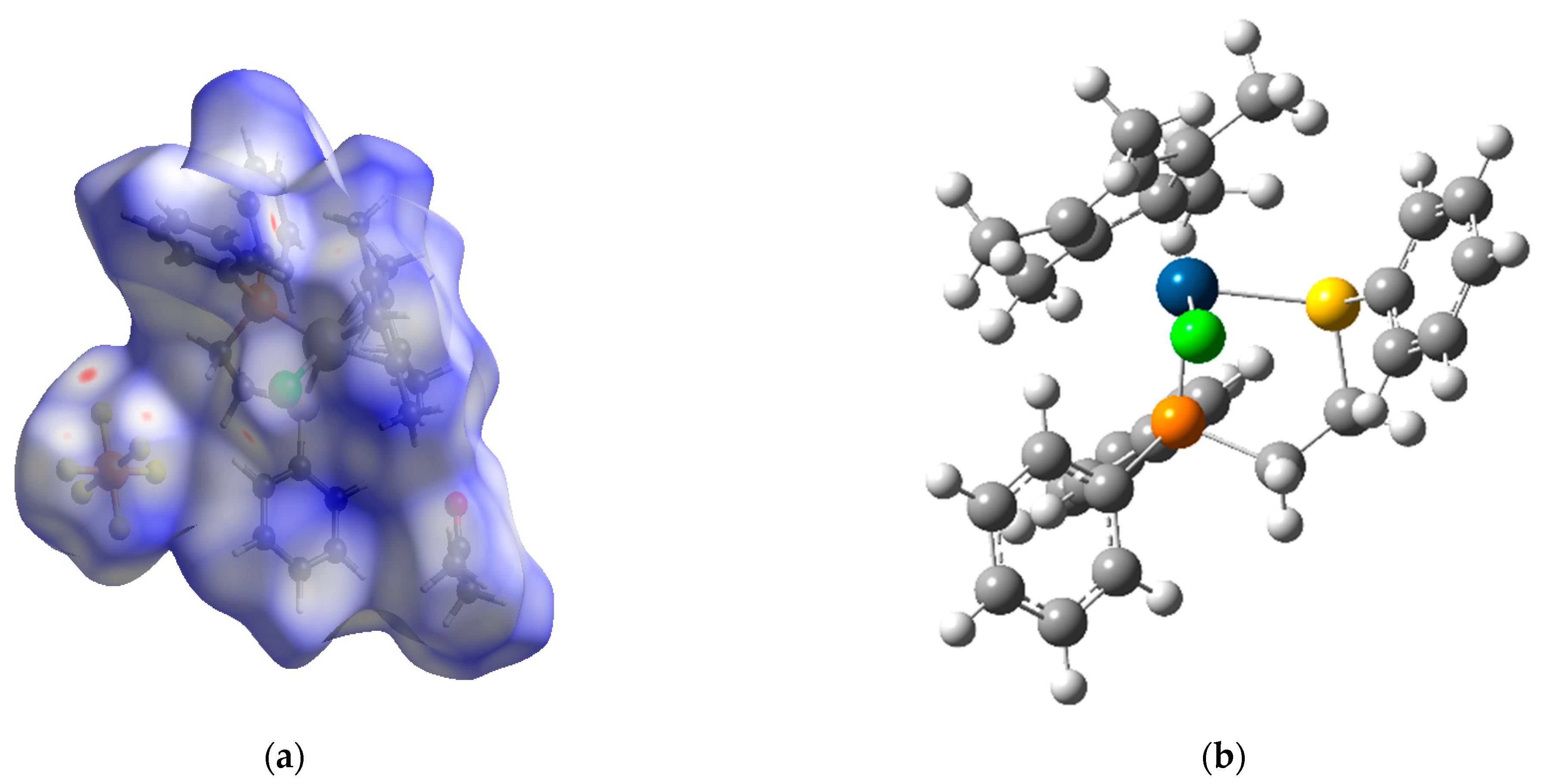
3.3. Hishfeld Surface Analysis
3.4. Theoretical Analysis
3.5. Experimental and Theoretical NMR Characterization
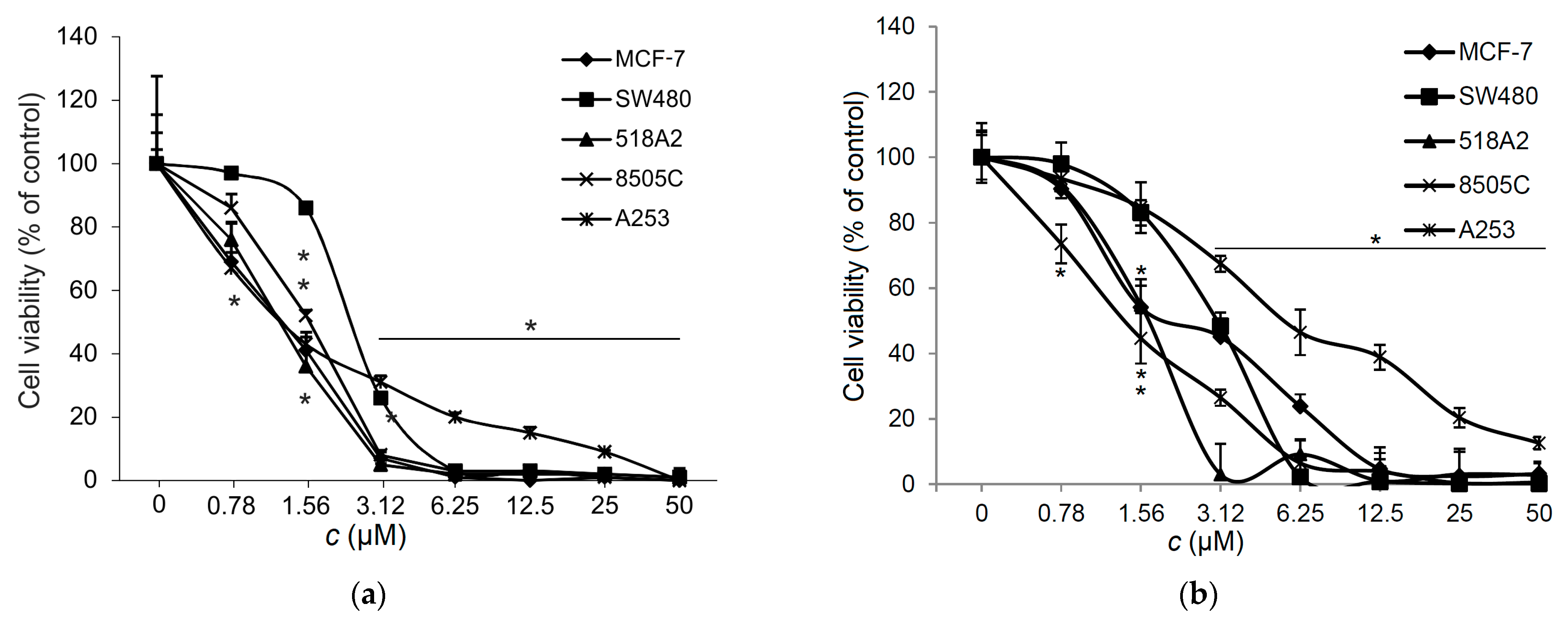
3.6. Effect of 1 on the Viability of Tumor Cells
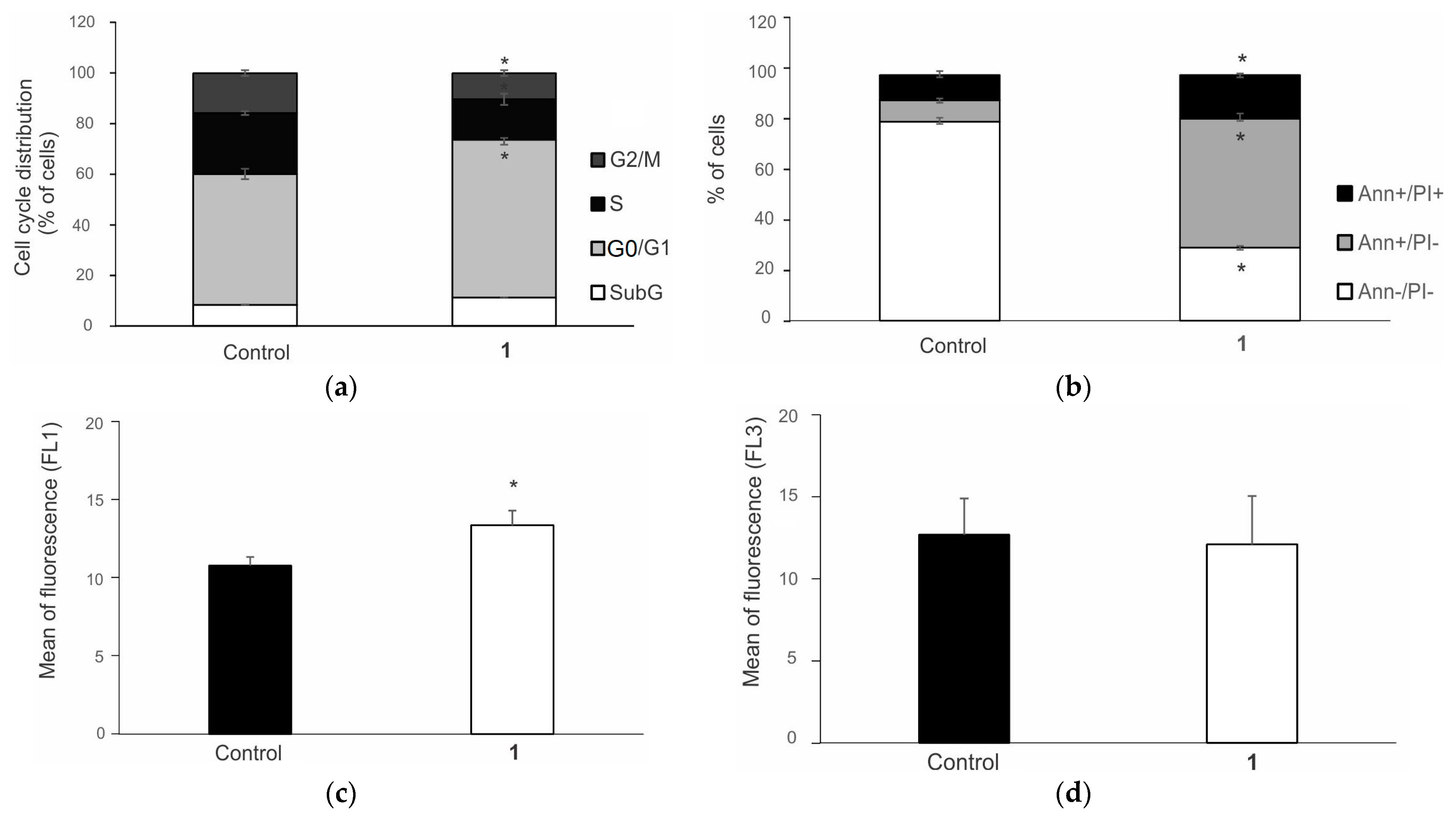
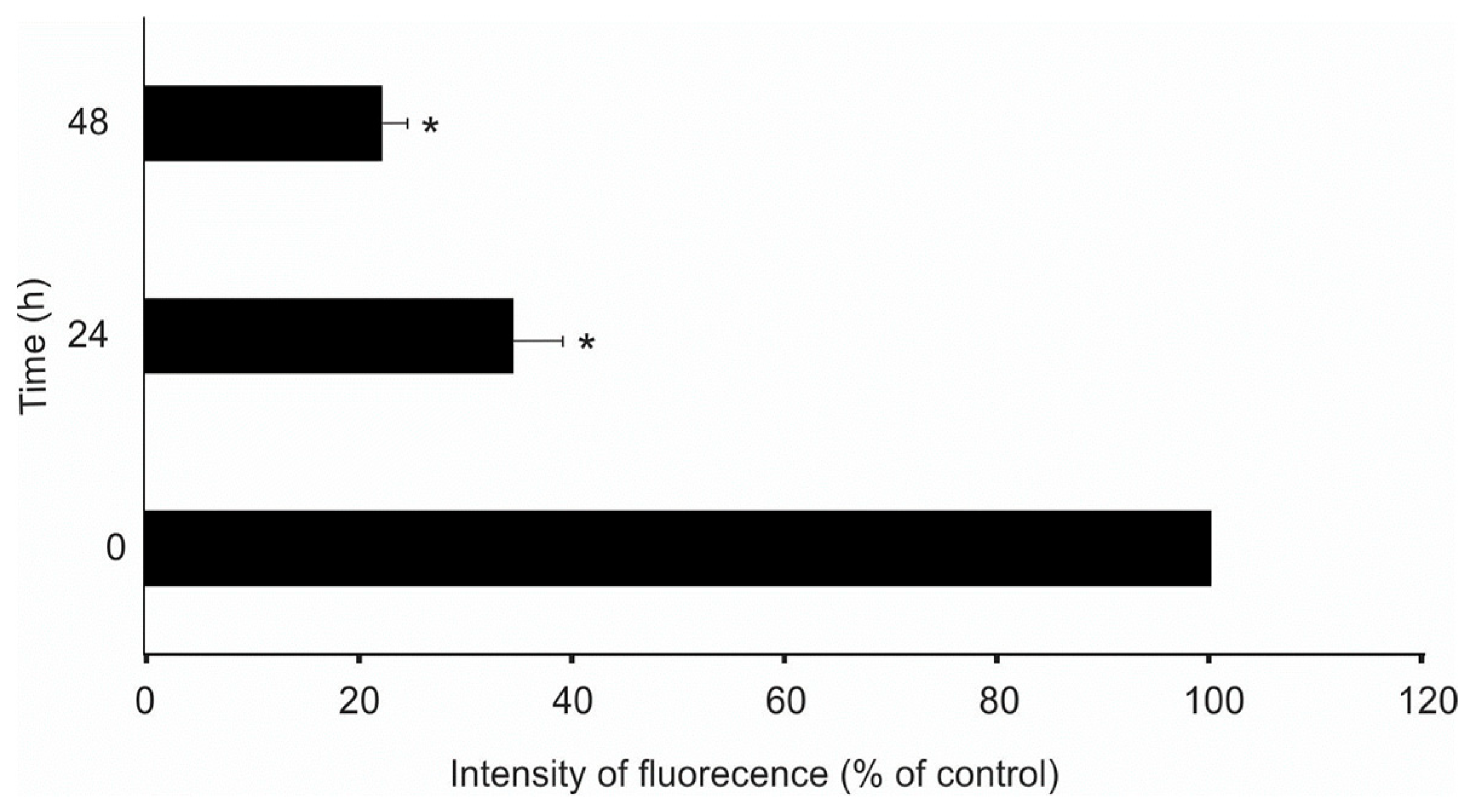
3.7. Complex 1 Triggers Apoptosis in 8505C Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zoń, A.; Bednarek, I. Cisplatin in Ovarian Cancer Treatment—Known Limitations in Therapy Force New Solutions. Int. J. Mol. Sci. 2023, 24, 7585. [Google Scholar] [CrossRef] [PubMed]
- Coffetti, G.; Moraschi, M.; Facchetti, G.; Rimoldi, I. The Challenging Treatment of Cisplatin-Resistant Tumors: State of the Art and Future Perspectives. Molecules 2023, 28, 3407. [Google Scholar] [CrossRef] [PubMed]
- Kaluderovic, G.N.; Paschke, R. Anticancer Metallotherapeutics in Preclinical Development. Curr. Med. Chem. 2011, 18, 4738–4752. [Google Scholar] [CrossRef] [PubMed]
- Abdolmaleki, S.; Khaksar, S.; Aliabadi, A.; Panjehpour, A.; Motieiyan, E.; Marabello, D.; Faraji, M.H.; Beihaghi, M. Cytotoxicity and Mechanism of Action of Metal Complexes: An Overview. Toxicology 2023, 492, 153516. [Google Scholar] [CrossRef] [PubMed]
- Biswal, S.; Panda, M.; Sahoo, R.K.; Tripathi, S.K.; Biswal, B.K. Tumour Microenvironment and Aberrant Signaling Pathways in Cisplatin Resistance and Strategies to Overcome in Oral Cancer. Arch. Oral Biol. 2023, 151, 105697. [Google Scholar] [CrossRef] [PubMed]
- Arjmand, F.; Yasir Khan, H.; Tabassum, S. Progress of Metal-Based Anticancer Chemotherapeutic Agents in Last Two Decades and Their Comprehensive Biological (DNA/RNA Binding, Cleavage and Cytotoxicity Activity) Studies. Chem. Rec. 2023, 23, e202200247. [Google Scholar] [CrossRef] [PubMed]
- Gandin, V.; Hoeschele, J.D.; Margiotta, N. Special Issue “Cisplatin in Cancer Therapy: Molecular Mechanisms of Action 3.0.”. Int. J. Mol. Sci. 2023, 24, 7917. [Google Scholar] [CrossRef]
- Todorov, L.; Kostova, I. Recent Trends in the Development of Novel Metal-Based Antineoplastic Drugs. Molecules 2023, 28, 1959. [Google Scholar] [CrossRef] [PubMed]
- Kumar Singh, A.; Kumar, A.; Singh, H.; Sonawane, P.; Pathak, P.; Grishina, M.; Pal Yadav, J.; Verma, A.; Kumar, P. Metal Complexes in Cancer Treatment: Journey So Far. Chem. Biodivers. 2023, 20, e202300061. [Google Scholar] [CrossRef] [PubMed]
- Predarska, I.; Saoud, M.; Morgan, I.; Lönnecke, P.; Kaluđerović, G.N.; Hey-Hawkins, E. Triphenyltin(IV) Carboxylates with Exceptionally High Cytotoxicity against Different Breast Cancer Cell Lines. Biomolecules 2023, 13, 595. [Google Scholar] [CrossRef]
- Ansari, M.F.; Khan, H.Y.; Tabassum, S.; Arjmand, F. Advances in Anticancer Alkaloid-Derived Metallo-Chemotherapeutic Agents in the Last Decade: Mechanism of Action and Future Prospects. Pharmacol. Ther. 2023, 241, 108335. [Google Scholar] [CrossRef] [PubMed]
- Schatzschneider, U. Metallointercalators and Metalloinsertors: Structural Requirements for DNA Recognition and Anticancer Activity. In Metallo-Drugs: Development and Action of Anticancer Agents; De Gruyter: Berlin/Heidelberg, Germany, 2018; pp. 387–436. [Google Scholar]
- Ferraro, M.G.; Piccolo, M.; Misso, G.; Santamaria, R.; Irace, C. Bioactivity and Development of Small Non-Platinum Metal-Based Chemotherapeutics. Pharmaceutics 2022, 14, 954. [Google Scholar] [CrossRef] [PubMed]
- Borutzki, Y.; Skos, L.; Gerner, C.; Meier-Menches, S.M. Exploring the Potential of Metal-Based Candidate Drugs as Modulators of the Cytoskeleton. ChemBioChem 2023, 24, e202300178. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Aodeng, G.; Ga, L.; Hai, W.; Ai, J. Research Progress of Metal Anticancer Drugs. Pharmaceutics 2023, 15, 2750. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Zheng, Y.; Xia, W.; Mao, Z.-W. Organometallic Anti-Tumor Agents: Targeting from Biomolecules to Dynamic Bioprocesses. Chem. Soc. Rev. 2023, 52, 2790–2832. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Zhu, M.; Jiang, M.; Yang, F.; Zhang, Z. Current Status of Iridium-Based Complexes against Lung Cancer. Front. Pharmacol. 2022, 13, 1025544. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Shen, Q.; Mao, Z.; Tan, C. Rising Interest in the Development of Metal Complexes in Cancer Immunotherapy. Chem. Asian J. 2022, 17, e202200270. [Google Scholar] [CrossRef]
- Raza, M.K.; Noor, A.; Samantaray, P.K. Ir(III) and Ru(II) Complexes in Photoredox Catalysis and Photodynamic Therapy: A New Paradigm towards Anticancer Applications. ChemBioChem 2021, 22, 3270–3272. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-H.; Chang, J.-Y. New Insights into Mechanisms of Cisplatin Resistance: From Tumor Cell to Microenvironment. Int. J. Mol. Sci. 2019, 20, 4136. [Google Scholar] [CrossRef]
- Graf, M.; Ochs, J.; Metzler-Nolte, N.; Mayer, P.; Böttcher, H. Synthesis, Characterization and Cytotoxic Activities of Half-sandwich Pentamethylcyclopentadienyl Iridium(III) Complexes Containing 4,4’-substituted 2,2’-Bipyridine Ligands. Z. Für Anorg. Und Allg. Chem. 2023, 649, e202200382. [Google Scholar] [CrossRef]
- Li, J.; Guo, L.; Tian, Z.; Tian, M.; Zhang, S.; Xu, K.; Qian, Y.; Liu, Z. Novel Half-Sandwich Iridium(III) Imino-Pyridyl Complexes Showing Remarkable in Vitro Anticancer Activity. Dalton Trans. 2017, 46, 15520–15534. [Google Scholar] [CrossRef] [PubMed]
- Prathima, T.S.; Choudhury, B.; Ahmad, M.G.; Chanda, K.; Balamurali, M.M. Recent Developments on Other Platinum Metal Complexes as Target-Specific Anticancer Therapeutics. Coord. Chem. Rev. 2023, 490, 215231. [Google Scholar] [CrossRef]
- Ma, D.-L.; Wu, C.; Wu, K.-J.; Leung, C.-H. Iridium(III) Complexes Targeting Apoptotic Cell Death in Cancer Cells. Molecules 2019, 24, 2739. [Google Scholar] [CrossRef]
- Almodares, Z.; Lucas, S.J.; Crossley, B.D.; Basri, A.M.; Pask, C.M.; Hebden, A.J.; Phillips, R.M.; McGowan, P.C. Rhodium, Iridium, and Ruthenium Half-Sandwich Picolinamide Complexes as Anticancer Agents. Inorg. Chem. 2014, 53, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Rayne, S.; Forest, K. A Comparative Examination of Density Functional Performance against the ISOL24/11 Isomerization Energy Benchmark. Comput. Theor. Chem. 2016, 1090, 147–152. [Google Scholar] [CrossRef]
- Ma, W.; Zhang, S.; Tian, Z.; Xu, Z.; Zhang, Y.; Xia, X.; Chen, X.; Liu, Z. Potential Anticancer Agent for Selective Damage to Mitochondria or Lysosomes: Naphthalimide-Modified Fluorescent Biomarker Half-Sandwich Iridium (III) and Ruthenium (II) Complexes. Eur. J. Med. Chem. 2019, 181, 111599. [Google Scholar] [CrossRef]
- Li, J.; Tian, M.; Tian, Z.; Zhang, S.; Yan, C.; Shao, C.; Liu, Z. Half-Sandwich Iridium(III) and Ruthenium(II) Complexes Containing P^P-Chelating Ligands: A New Class of Potent Anticancer Agents with Unusual Redox Features. Inorg. Chem. 2018, 57, 1705–1716. [Google Scholar] [CrossRef]
- Liu, Z.; Habtemariam, A.; Pizarro, A.M.; Fletcher, S.A.; Kisova, A.; Vrana, O.; Salassa, L.; Bruijnincx, P.C.A.; Clarkson, G.J.; Brabec, V.; et al. Organometallic Half-Sandwich Iridium Anticancer Complexes. J. Med. Chem. 2011, 54, 3011–3026. [Google Scholar] [CrossRef]
- Casini, A.; Edafe, F.; Erlandsson, M.; Gonsalvi, L.; Ciancetta, A.; Re, N.; Ienco, A.; Messori, L.; Peruzzini, M.; Dyson, P.J. Rationalization of the Inhibition Activity of Structurally Related Organometallic Compounds against the Drug Target Cathepsin B by DFT. Dalton Trans. 2010, 39, 5556. [Google Scholar] [CrossRef]
- Ludwig, G.; Kaluđerović, G.N.; Bette, M.; Block, M.; Paschke, R.; Steinborn, D. Highly Active Neutral Ruthenium(II) Arene Complexes: Synthesis, Characterization, and Investigation of Their Anticancer Properties. J. Inorg. Biochem. 2012, 113, 77–82. [Google Scholar] [CrossRef]
- Ludwig, G.; Kaluđerović, G.N.; Rüffer, T.; Bette, M.; Korb, M.; Block, M.; Paschke, R.; Lang, H.; Steinborn, D. Cationic Arene Ruthenium(Ii) Complexes with Chelating P-Functionalized Alkyl Phenyl Sulfide and Sulfoxide Ligands as Potent Anticancer Agents. Dalton Trans. 2013, 42, 3771. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, G.; Mijatović, S.; Ranđelović, I.; Bulatović, M.; Miljković, D.; Maksimović-Ivanić, D.; Korb, M.; Lang, H.; Steinborn, D.; Kaluđerović, G.N. Biological Activity of Neutral and Cationic Iridium(III) Complexes with κP and κP,κS Coordinated Ph2PCH2CH2CH2S(O)xPh (x = 0–2) Ligands. Eur. J. Med. Chem. 2013, 69, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, G.; Ranđelović, I.; Maksimović-Ivanić, D.; Mijatović, S.; Bulatović, M.Z.; Miljković, D.; Korb, M.; Lang, H.; Steinborn, D.; Kaluđerović, G.N. Anticancer Potential of (Pentamethylcyclopentadienyl)Chloridoiridium(III) Complexes Bearing κ P and κP,κS -Coordinated Ph2PCH2CH2CH2S(O)xPh (x = 0–2) Ligands. ChemMedChem 2014, 9, 1586–1593. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, G.; Mojić, M.; Bulatović, M.Z.; Mijatović, S.; Maksimović-Ivanić, D.; Steinborn, D.; Kaluđerović, G.N. Biological Potential of Halfsandwich Ruthenium(II) and Iridium(III) Complexes. Anticancer Agents Med. Chem. 2016, 16, 1455–1460. [Google Scholar] [CrossRef] [PubMed]
- Arlt, S.; Petković, V.; Ludwig, G.; Eichhorn, T.; Lang, H.; Rüffer, T.; Mijatović, S.; Maksimović-Ivanić, D.; Kaluđerović, G.N. Arene Ruthenium(II) Complexes Bearing the κ-P or κ-P, κ-S Ph2P(CH2)3SPh Ligand. Molecules 2021, 26, 1860. [Google Scholar] [CrossRef] [PubMed]
- Block, M.; Bette, M.; Wagner, C.; Schmidt, J.; Steinborn, D. Rhodium(I) Complexes with κP Coordinated ω-Phosphinofunctionalized Alkyl Phenyl Sulfide, Sulfoxide and Sulfone Ligands and Their Reactions with Sodium Bis(Trimethylsilyl)Amide and Ag[BF4]. J. Organomet. Chem. 2011, 696, 1768–1781. [Google Scholar] [CrossRef]
- Ball, R.G.; Graham, W.A.G.; Heinekey, D.M.; Hoyano, J.K.; McMaster, A.D.; Mattson, B.M.; Michel, S.T. Synthesis and Structure of Dicarbonylbis(.Eta.-Pentamethylcyclopentadienyl)Diiridium. Inorg. Chem. 1990, 29, 2023–2025. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT—Integrated Space-Group and Crystal-Structure Determination. Acta Cryst. A 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer17; University of Western Australia, Perth, Australia, 2017. In CrystalExplorer17; University of Western Australia: Perth, Australia, 2017. [Google Scholar]
- Karrouchi, K.; Brandán, S.A.; Sert, Y.; El Karbane, M.; Radi, S.; Ferbinteanu, M.; Garcia, Y.; Ansar, M. Synthesis, Structural, Molecular Docking and Spectroscopic Studies of (E)-N’-(4-Methoxybenzylidene)-5-Methyl-1H-Pyrazole-3-Carbohydrazide. J. Mol. Struct. 2021, 1225, 129072. [Google Scholar] [CrossRef]
- Spackman, M.A.; Byrom, P.G. A Novel Definition of a Molecule in a Crystal. Chem. Phys. Lett. 1997, 267, 215–220. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld Surface Analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Becke, A.D. Density-Functional Exchange-Energy Approximation with Correct Asymptotic Behavior. Phys. Rev. A 1988, 38, 3098–3100. [Google Scholar] [CrossRef] [PubMed]
- Becke, A.D.; Johnson, E.R. A Density-Functional Model of the Dispersion Interaction. J. Chem. Phys. 2005, 123, 154101. [Google Scholar] [CrossRef] [PubMed]
- Dunning, T.H. Gaussian Basis Sets for Use in Correlated Molecular Calculations. I. The Atoms Boron through Neon and Hydrogen. J. Chem. Phys. 1989, 90, 1007. [Google Scholar] [CrossRef]
- Hay, P.J.; Wadt, W.R. Ab Initio Effective Core Potentials for Molecular Calculations. Potentials for K to Au Including the Outermost Core Orbitale. J. Chem. Phys. 1985, 82, 299–310. [Google Scholar] [CrossRef]
- Hay, P.J.; Wadt, W.R. Ab Initio Effective Core Potentials for Molecular Calculations. Potentials for the Transition Metal Atoms Sc to Hg. J. Chem. Phys. 1985, 82, 270–283. [Google Scholar] [CrossRef]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [Google Scholar] [CrossRef] [PubMed]
- Bohmann, J.A.; Weinhold, F.; Farrar, T.C. Natural Chemical Shielding Analysis of Nuclear Magnetic Resonance Shielding Tensors from Gauge-Including Atomic Orbital Calculations. J. Chem. Phys. 1997, 107, 1173. [Google Scholar] [CrossRef]
- Foster, J.P.; Weinhold, F. Natural Hybrid Orbitals. J. Am. Chem. Soc. 1980, 102, 7211–7218. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]
- Quah, B.J.C.; Parish, C.R. The Use of Carboxyfluorescein Diacetate Succinimidyl Ester (CFSE) to Monitor Lymphocyte Proliferation. J. Vis. Exp. 2010, 44, 2259. [Google Scholar] [CrossRef]
- Fan, Z.; Xie, J.; Sadhukhan, T.; Liang, C.; Huang, C.; Li, W.; Li, T.; Zhang, P.; Banerjee, S.; Raghavachari, K.; et al. Highly Efficient Ir(III)-Coumarin Photo-Redox Catalyst for Synergetic Multi-Mode Cancer Photo-Therapy. Chem. A Eur. J. 2022, 28, e202103346. [Google Scholar] [CrossRef] [PubMed]
- Demissie, T.B.; Hansen, J.H. Mechanism and Site Selectivity in Visible-Light Photocatalyzed C–H Functionalization: Insights from DFT Calculations. J. Org. Chem. 2016, 81, 7110–7120. [Google Scholar] [CrossRef] [PubMed]
- Eichhorn, T.; Kolbe, F.; Mišić, S.; Dimić, D.; Morgan, I.; Saoud, M.; Milenković, D.; Marković, Z.; Rüffer, T.; Dimitrić Marković, J.; et al. Synthesis, Crystallographic Structure, Theoretical Analysis, Molecular Docking Studies, and Biological Activity Evaluation of Binuclear Ru(II)-1-Naphthylhydrazine Complex. Int. J. Mol. Sci. 2023, 24, 689. [Google Scholar] [CrossRef] [PubMed]
- Avdović, E.H.; Milenković, D.; Dimitrić Marković, J.M.; Đorović, J.; Vuković, N.; Vukić, M.D.; Jevtić, V.V.; Trifunović, S.R.; Potočňák, I.; Marković, Z. Synthesis, Spectroscopic Characterization (FT-IR, FT-Raman, and NMR), Quantum Chemical Studies and Molecular Docking of 3-(1-(Phenylamino)Ethylidene)-Chroman-2,4-Dione. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 195, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Vichai, V.; Kirtikara, K. Sulforhodamine B Colorimetric Assay for Cytotoxicity Screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef] [PubMed]
- Mijatović, S.; Bramanti, A.; Nicoletti, F.; Fagone, P.; Kaluđerović, G.N.; Maksimović-Ivanić, D. Naturally Occurring Compounds in Differentiation Based Therapy of Cancer. Biotechnol. Adv. 2018, 36, 1622–1632. [Google Scholar] [CrossRef] [PubMed]
- Parrish, A.B.; Freel, C.D.; Kornbluth, S. Cellular Mechanisms Controlling Caspase Activation and Function. Cold Spring Harb. Perspect. Biol. 2013, 5, a008672. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Mao, Z.; Kang, Y.; Zhang, W.; Mei, L.; Ji, X. Redox Regulation and Its Emerging Roles in Cancer Treatment. Coord. Chem. Rev. 2023, 475, 214897. [Google Scholar] [CrossRef]
- Goel, S.; Singh, R.; Singh, V.; Singh, H.; Kumari, P.; Chopra, H.; Sharma, R.; Nepovimova, E.; Valis, M.; Kuca, K.; et al. Metformin: Activation of 5′ AMP-Activated Protein Kinase and Its Emerging Potential beyond Anti-Hyperglycemic Action. Front. Genet 2022, 13, 1022739. [Google Scholar] [CrossRef]
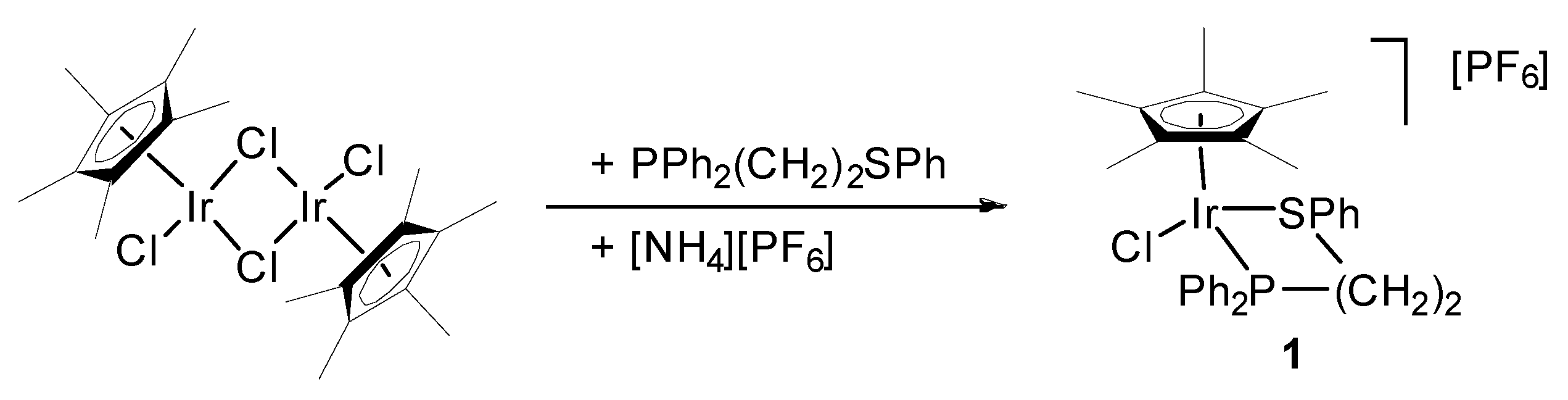
| 1H NMR | Experimental | Theoretical | 13C NMR | Experimental | Theoretical |
|---|---|---|---|---|---|
| CH3 | 1.50 | 1.69 | CCH3 | 8.1 | 10.0 |
| CH2PPh2 | 2.87 | 2.58 | CH2PPh2 | 25.2 | 36.2 |
| CH2PPh2 | 3.13 | 3.14 | SCH2 | 34.7 | 40.8 |
| SCH2 | 3.66 | 3.45 | CCH3 | 96.9 | 96.3 |
| SCH2 | 4.20 | 3.50 | CPh | 96.9 | 96.3 |
| HPh | 7.12–7.70 | 7.06–8.18 | R | 0.99 | |
| R | 0.97 | MAE [ppm] | 4.1 | ||
| MAE [ppm] | 0.9 | ||||
| MCF-7 | SW480 | 518A2 | 8505C | A253 | MRC-5 | |
|---|---|---|---|---|---|---|
| IC50 [µM] | ||||||
| Ph2P(CH2)2SPh [31] | 6.3 ± 0.2 | 11.3 ± 1.2 | 20.5 ± 1.9 | 11.0 ± 1.2 | 9.5 ± 1.1 | n.d. |
| [Ir(η5C5Me5)Cl{Ph2PCH2SPh-κP,κS}][PF6] [33] | 16.0 ± 1.9 | 14.6 ± 3.4 | 9.8 ± 0.5 | 16.8 ± 3.5 | 10.2 ± 2.8 | n.d. |
| [Ir(η5C5Me5)Cl{Ph2P(CH2)2SPh-κP,κS}][PF6] | 1.3 ± 0.3 | 2.5 ± 0.5 | 1.3 ± 0.4 | 1.6 ± 0.2 | 1.7 ± 0.2 | 15.3 ± 2.3 |
| [Ir(η5C5Me5)Cl{Ph2P(CH2)3SPhκP,κS}][PF6] [34,35] | 0.4 ± 0.1 | 1.0 ± 0.3 | 1.0 ± 0.6 | 0.5 ± 0.2 | 0.4 ± 0.0 | 6.25 ± 0.31 |
| [Ru(p-cym)Cl{Ph2P(CH2)2SPh-κP,κS}][PF6] [32] | 0.26 ± 0.03 | 1.73 ± 0.14 | 1.70 ± 0.06 | 1.70 ± 0.06 | 0.93 ± 0.04 | n.d. |
| Cisplatin | 2.3 ± 0.2 | 3.0 ± 0.2 | 1.7 ± 0.2 | 5.5 ± 0.3 | 1.1 ± 0.2 | 0.65 ± 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ludwig, G.; Ranđelović, I.; Dimić, D.; Komazec, T.; Maksimović-Ivanić, D.; Mijatović, S.; Rüffer, T.; Kaluđerović, G.N. (Pentamethylcyclopentadienyl)chloridoiridium(III) Complex Bearing Bidentate Ph2PCH2CH2SPh-κP,κS Ligand. Biomolecules 2024, 14, 420. https://doi.org/10.3390/biom14040420
Ludwig G, Ranđelović I, Dimić D, Komazec T, Maksimović-Ivanić D, Mijatović S, Rüffer T, Kaluđerović GN. (Pentamethylcyclopentadienyl)chloridoiridium(III) Complex Bearing Bidentate Ph2PCH2CH2SPh-κP,κS Ligand. Biomolecules. 2024; 14(4):420. https://doi.org/10.3390/biom14040420
Chicago/Turabian StyleLudwig, Gerd, Ivan Ranđelović, Dušan Dimić, Teodora Komazec, Danijela Maksimović-Ivanić, Sanja Mijatović, Tobias Rüffer, and Goran N. Kaluđerović. 2024. "(Pentamethylcyclopentadienyl)chloridoiridium(III) Complex Bearing Bidentate Ph2PCH2CH2SPh-κP,κS Ligand" Biomolecules 14, no. 4: 420. https://doi.org/10.3390/biom14040420
APA StyleLudwig, G., Ranđelović, I., Dimić, D., Komazec, T., Maksimović-Ivanić, D., Mijatović, S., Rüffer, T., & Kaluđerović, G. N. (2024). (Pentamethylcyclopentadienyl)chloridoiridium(III) Complex Bearing Bidentate Ph2PCH2CH2SPh-κP,κS Ligand. Biomolecules, 14(4), 420. https://doi.org/10.3390/biom14040420








