Chemotaxis and Related Signaling Systems in Vibrio cholerae
Abstract
:1. Introduction
2. A Brief Overview of Motility and Chemotaxis of Vibrio cholerae
2.1. Single, Sheathed Polar Flagellum
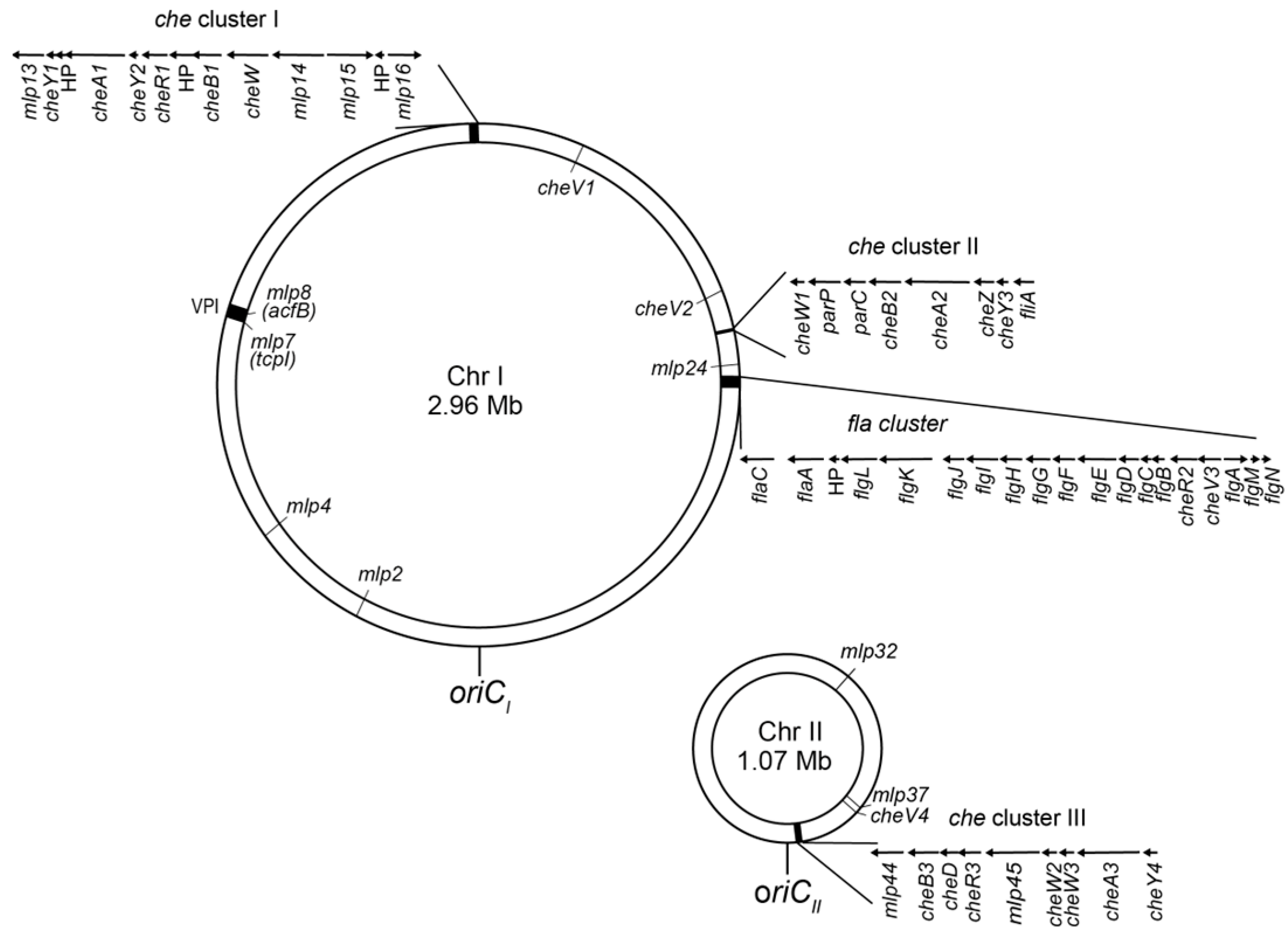
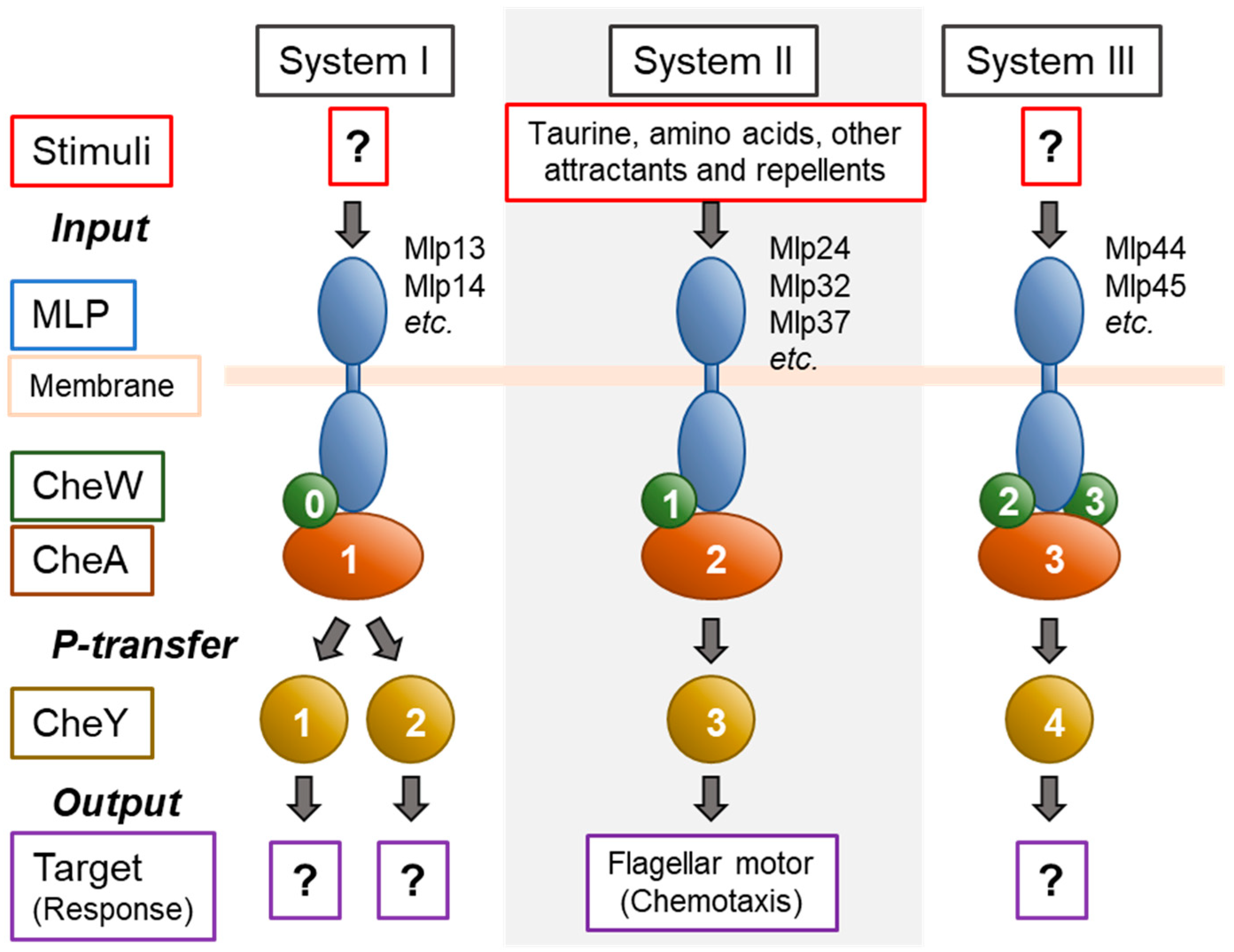
2.2. Chemotaxis and Related Signaling Systems
- Cluster I: cheY1, cheA1, cheY2, cheR1, cheB1, and putative cheW
- Cluster II: cheW1, cheB2, cheA2, cheZ and cheY3
- Cluster III: cheB3, cheD, cheR3, cheW2, cheW3, cheA3 and cheY4.
2.3. MLPs (Methyl-Accepting Chemotaxis Protein-like Proteins)
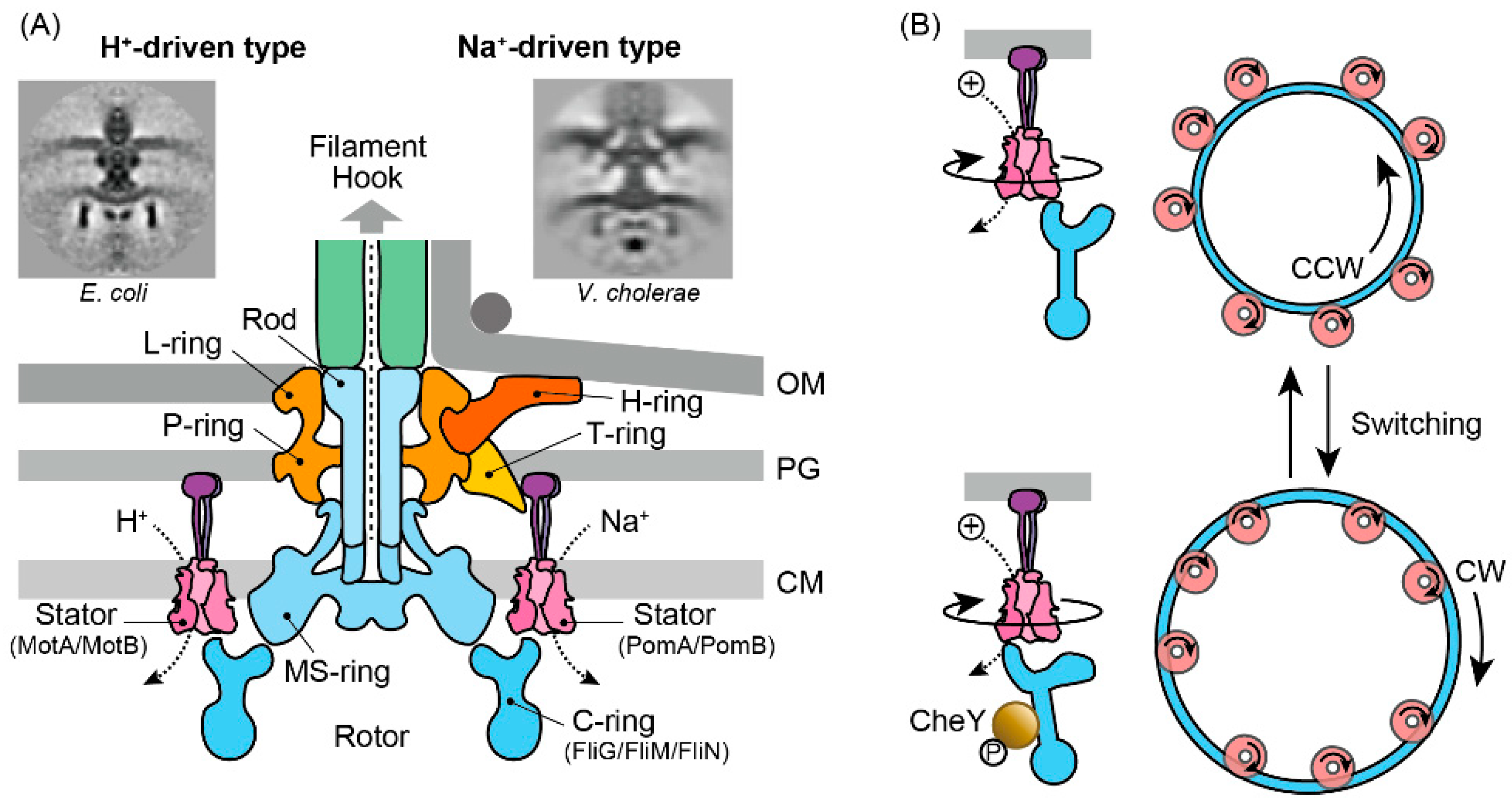
3. Flagellar Motor as an Actuator for the Chemotaxis Sensory System
3.1. Molecular Architecture
3.2. Rotor–Stator Interaction
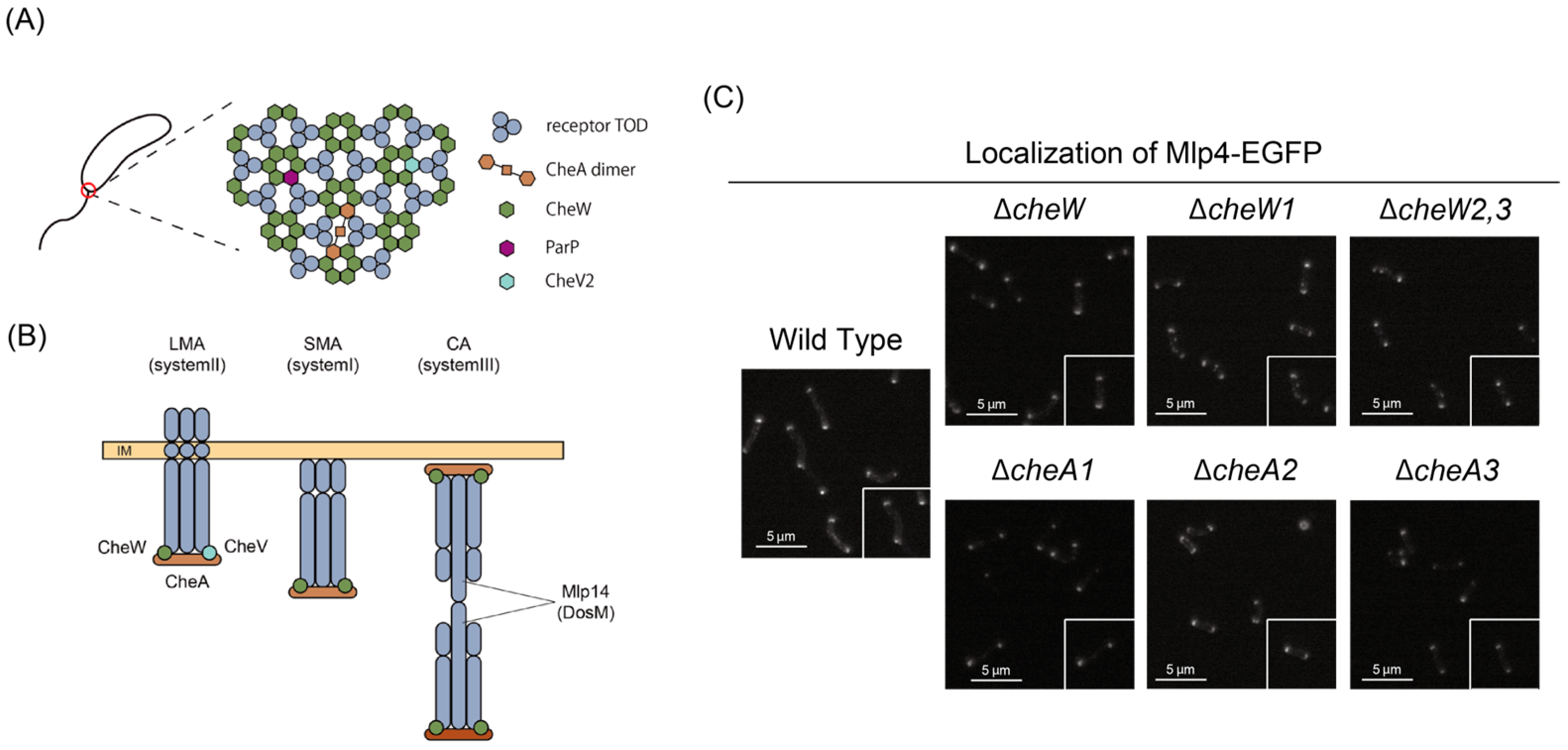
4. MLPs as Sensors/Transducers for the Chemotaxis Sensory System
4.1. Molecular Architectures
4.2. Polar Clustering
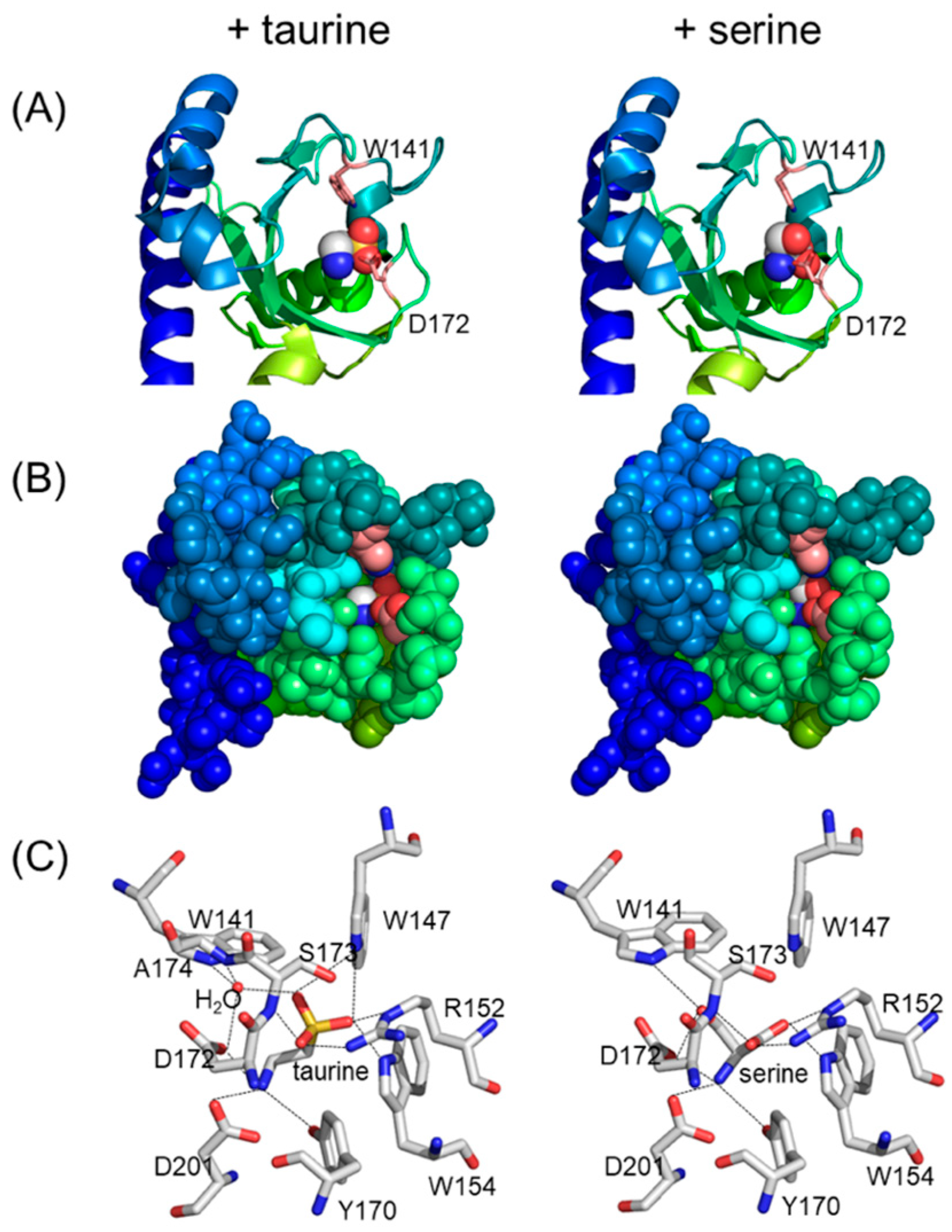
4.3. The Periplasmic Sensor Domain and Stimuli Sensed
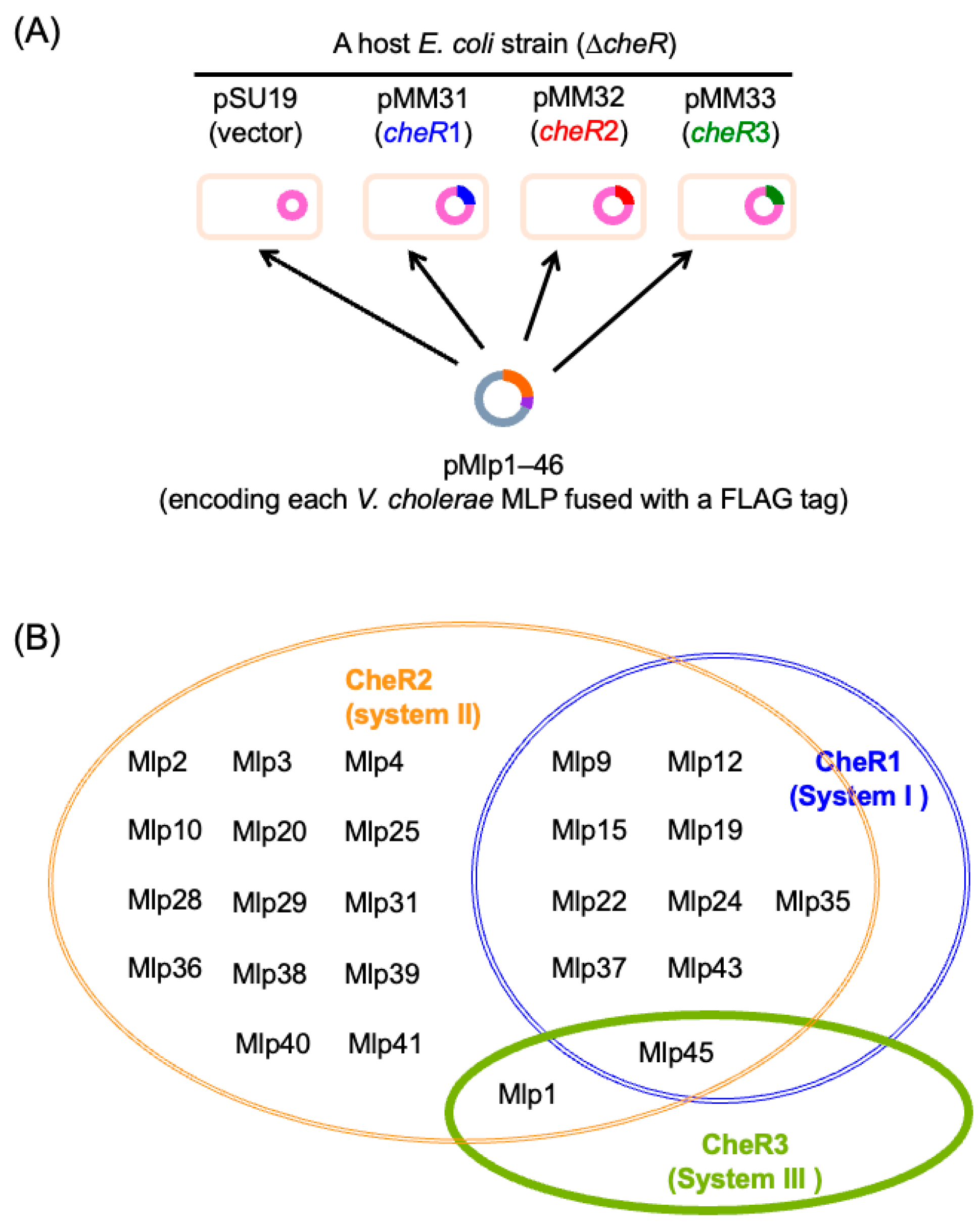
4.4. Assignment of MLPs to Three Che Systems
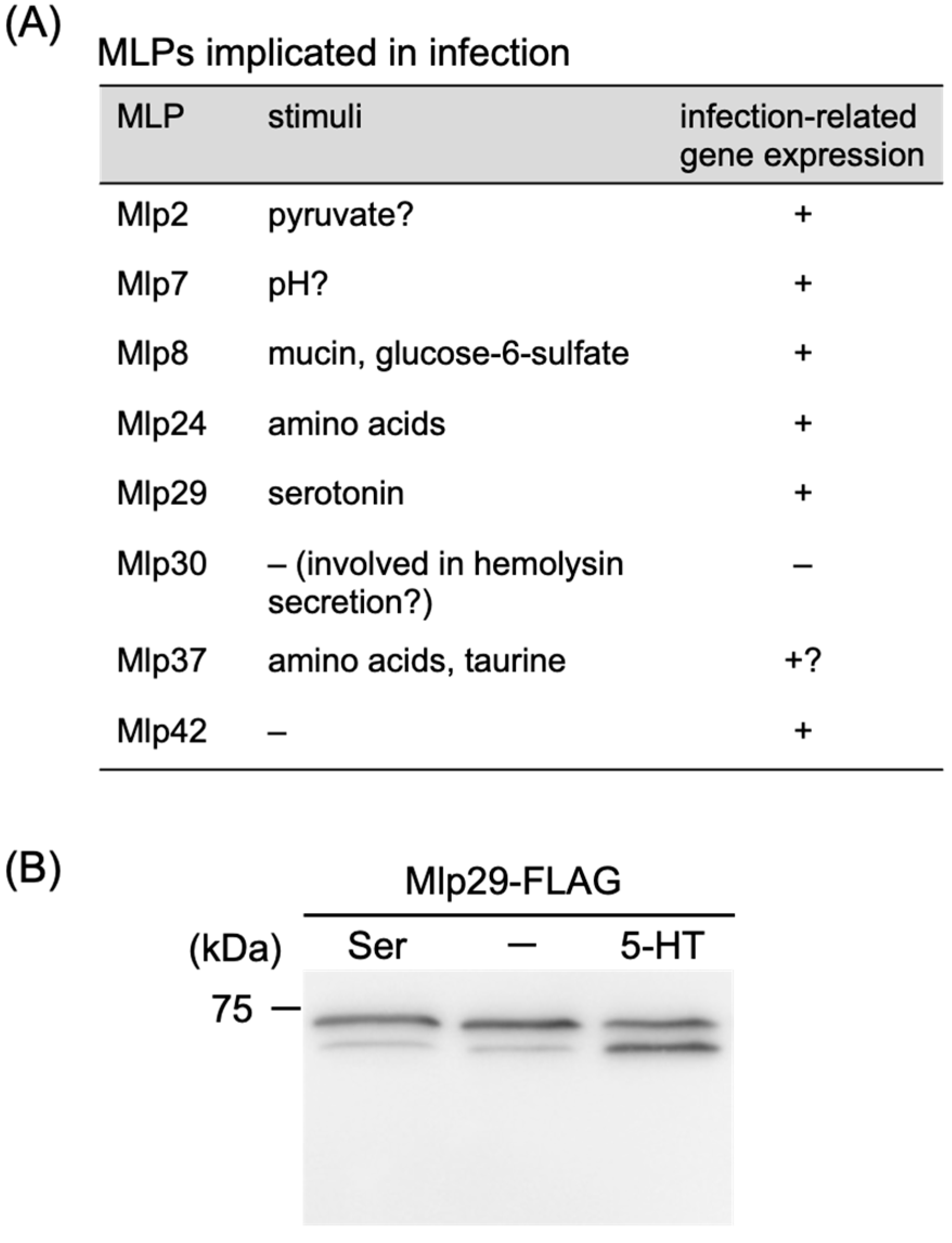
4.5. Roles in Infection and Pathogenicity
5. Concluding Remarks and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Conner, J.G.; Teschler, J.K.; Jones, C.J.; Yildiz, F.H. Staying Alive: Vibrio cholerae’s Cycle of Environmental Survival, Transmission, and Dissemination. Microbiol. Spectr. 2016, 4, 593–633. [Google Scholar] [CrossRef]
- Reidl, J.; Klose, K.E. Vibrio cholerae and cholera: Out of the water and into the host. FEMS Microbiol. Rev. 2002, 26, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Tabassum, N.; Anand, R.; Kim, Y.M. Motility of Vibrio spp.: Regulation and controlling strategies. Appl. Microbiol. Biotechnol. 2020, 104, 8187–8208. [Google Scholar] [CrossRef]
- Alm, R.A.; Manning, P.A. Characterization of the hlyB gene and its role in the production of the El Tor haemolysin of Vibrio cholerae O1. Mol. Microbiol. 1990, 4, 413–425. [Google Scholar] [CrossRef]
- Banerjee, R.; Das, S.; Mukhopadhyay, K.; Nag, S.; Chakrabortty, A.; Chaudhuri, K. Involvement of in vivo induced cheY-4 gene of Vibrio cholerae in motility, early adherence to intestinal epithelial cells and regulation of virulence factors. FEBS Lett. 2002, 532, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Echazarreta, M.A.; Klose, K.E. Vibrio Flagellar Synthesis. Front. Cell Infect. Microbiol. 2019, 9, 131. [Google Scholar] [CrossRef] [PubMed]
- Hang, L.; John, M.; Asaduzzaman, M.; Bridges, E.A.; Vanderspurt, C.; Kirn, T.J.; Taylor, R.K.; Hillman, J.D.; Progulske-Fox, A.; Handfield, M.; et al. Use of in vivo-induced antigen technology (IVIAT) to identify genes uniquely expressed during human infection with Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2003, 100, 8508–8513. [Google Scholar] [CrossRef]
- Krukonis, E.S.; DiRita, V.J. From motility to virulence: Sensing and responding to environmental signals in Vibrio cholerae. Curr. Opin. Microbiol. 2003, 6, 186–190. [Google Scholar] [CrossRef]
- Lee, S.H.; Butler, S.M.; Camilli, A. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. USA 2001, 98, 6889–6894. [Google Scholar] [CrossRef]
- Peterson, K.M.; Gellings, P.S. Multiple intraintestinal signals coordinate the regulation of Vibrio cholerae virulence determinants. Pathog. Dis. 2018, 76, ftx126. [Google Scholar] [CrossRef]
- Armitage, J.P. Microbial Primer: The bacterial flagellum—How bacteria swim. Microbiology 2024, 170, 001406. [Google Scholar] [CrossRef] [PubMed]
- Berg, H.C. E. coli in Motion; Springer: New York, NY, USA, 2004; pp. 131–133. [Google Scholar]
- Karmakar, R. State of the art of bacterial chemotaxis. J. Basic. Microbiol. 2021, 61, 366–379. [Google Scholar] [CrossRef]
- McCarter, L.L. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 2001, 65, 445–462. [Google Scholar] [CrossRef]
- Correa, N.E.; Peng, F.; Klose, K.E. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J. Bacteriol. 2005, 187, 6324–6332. [Google Scholar] [CrossRef]
- Green, J.C.; Kahramanoglou, C.; Rahman, A.; Pender, A.M.; Charbonnel, N.; Fraser, G.M. Recruitment of the earliest component of the bacterial flagellum to the old cell division pole by a membrane-associated signal recognition particle family GTP-binding protein. J. Mol. Biol. 2009, 391, 679–690. [Google Scholar] [CrossRef]
- Hranitzky, K.W.; Mulholland, A.; Larson, A.D.; Eubanks, E.R.; Hart, L.T. Characterization of a flagellar sheath protein of Vibrio cholerae. Infect. Immun. 1980, 27, 597–603. [Google Scholar] [CrossRef]
- Kojima, S.; Yamamoto, K.; Kawagishi, I.; Homma, M. The polar flagellar motor of Vibrio cholerae is driven by an Na+ motive force. J. Bacteriol. 1999, 181, 1927–1930. [Google Scholar] [CrossRef] [PubMed]
- Yorimitsu, T.; Homma, M. Na+-driven flagellar motor of Vibrio. Biochim. Biophys. Acta 2001, 1505, 82–93. [Google Scholar] [CrossRef]
- Grognot, M.; Mittal, A.; Mah’moud, M.; Taute, K.M. Vibrio cholerae Motility in Aquatic and Mucus-Mimicking Environments. Appl. Env. Microbiol. 2021, 87, e0129321. [Google Scholar] [CrossRef]
- Xie, L.; Altindal, T.; Chattopadhyay, S.; Wu, X.L. Bacterial flagellum as a propeller and as a rudder for efficient chemotaxis. Proc. Natl. Acad. Sci. USA 2011, 108, 2246–2251. [Google Scholar] [CrossRef]
- Syed, K.A.; Beyhan, S.; Correa, N.; Queen, J.; Liu, J.; Peng, F.; Satchell, K.J.; Yildiz, F.; Klose, K.E. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J. Bacteriol. 2009, 191, 6555–6570. [Google Scholar] [CrossRef] [PubMed]
- Syed, K.A.; Klose, K.E. Vibrio Cholerae Flagellar Synthesis and Virulence. In Epidemiological and Molecular Aspects on Cholera; Ramamurthy, T., Bhattacharya, S.K., Eds.; Springer: New York, NY, USA, 2011. [Google Scholar]
- Yoon, S.S.; Mekalanos, J.J. Decreased potency of the Vibrio cholerae sheathed flagellum to trigger host innate immunity. Infect. Immun. 2008, 76, 1282–1288. [Google Scholar] [CrossRef]
- Dongre, M.; Singh, B.; Aung, K.M.; Larsson, P.; Miftakhova, R.; Persson, K.; Askarian, F.; Johannessen, M.; von Hofsten, J.; Persson, J.L.; et al. Flagella-mediated secretion of a novel Vibrio cholerae cytotoxin affecting both vertebrate and invertebrate hosts. Commun. Biol. 2018, 1, 59. [Google Scholar] [CrossRef]
- Buschiazzo, A.; Trajtenberg, F. Two-Component Sensing and Regulation: How Do Histidine Kinases Talk with Response Regulators at the Molecular Level? Annu. Rev. Microbiol. 2019, 73, 507–528. [Google Scholar] [CrossRef] [PubMed]
- Ishii, E.; Eguchi, Y. Diversity in Sensing and Signaling of Bacterial Sensor Histidine Kinases. Biomolecules 2021, 11, 1524. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, J.S.; Hazelbauer, G.L.; Falke, J.J. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015, 23, 257–266. [Google Scholar] [CrossRef]
- Briegel, A.; Ortega, D.R.; Tocheva, E.I.; Wuichet, K.; Li, Z.; Chen, S.; Muller, A.; Iancu, C.V.; Murphy, G.E.; Dobro, M.J.; et al. Universal architecture of bacterial chemoreceptor arrays. Proc. Natl. Acad. Sci. USA 2009, 106, 17181–17186. [Google Scholar] [CrossRef]
- Khursigara, C.M.; Wu, X.; Zhang, P.; Lefman, J.; Subramaniam, S. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc. Natl. Acad. Sci. USA 2008, 105, 16555–16560. [Google Scholar] [CrossRef]
- Zhang, P.; Khursigara, C.M.; Hartnell, L.M.; Subramaniam, S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc. Natl. Acad. Sci. USA 2007, 104, 3777–3781. [Google Scholar] [CrossRef]
- Zhang, P.; Weis, R.M.; Peters, P.J.; Subramaniam, S. Electron tomography of bacterial chemotaxis receptor assemblies. Methods Cell Biol. 2007, 79, 373–384. [Google Scholar] [CrossRef]
- Gosink, K.K.; Kobayashi, R.; Kawagishi, I.; Hase, C.C. Analyses of the roles of the three cheA homologs in chemotaxis of Vibrio cholerae. J. Bacteriol. 2002, 184, 1767–1771. [Google Scholar] [CrossRef]
- Heidelberg, J.F.; Eisen, J.A.; Nelson, W.C.; Clayton, R.A.; Gwinn, M.L.; Dodson, R.J.; Haft, D.H.; Hickey, E.K.; Peterson, J.D.; Umayam, L.; et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 2000, 406, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Hess, J.F.; Oosawa, K.; Kaplan, N.; Simon, M.I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell 1988, 53, 79–87. [Google Scholar] [CrossRef]
- Kuo, S.C.; Koshland, D.E., Jr. Roles of cheY and cheZ gene products in controlling flagellar rotation in bacterial chemotaxis of Escherichia coli. J. Bacteriol. 1987, 169, 1307–1314. [Google Scholar] [CrossRef] [PubMed]
- Kristich, C.J.; Ordal, G.W. Bacillus subtilis CheD is a chemoreceptor modification enzyme required for chemotaxis. J. Biol. Chem. 2002, 277, 25356–25362. [Google Scholar] [CrossRef]
- Alexander, R.P.; Lowenthal, A.C.; Harshey, R.M.; Ottemann, K.M. CheV: CheW-like coupling proteins at the core of the chemotaxis signaling network. Trends Microbiol. 2010, 18, 494–503. [Google Scholar] [CrossRef]
- Yang, W.; Alvarado, A.; Glatter, T.; Ringgaard, S.; Briegel, A. Baseplate variability of Vibrio cholerae chemoreceptor arrays. Proc. Natl. Acad. Sci. USA 2018, 115, 13365–13370. [Google Scholar] [CrossRef]
- Hiremath, G.; Nishiyama, S.; Kawagishi, I. CheV1 plays an important role in chemotaxis of Vibrio cholerae. Int. J. Biosci. Biochem. Bioinform. 2013, 3, 71–74. [Google Scholar] [CrossRef]
- Hyakutake, A.; Homma, M.; Austin, M.J.; Boin, M.A.; Häse, C.C.; Kawagishi, I. Only one of the five CheY homologs in Vibrio cholerae directly switches flagellar rotation. J. Bacteriol. 2005, 187, 8403–8410. [Google Scholar] [CrossRef]
- Hiremath, G.; Hyakutake, A.; Yamamoto, K.; Ebisawa, T.; Nakamura, T.; Nishiyama, S.; Homma, M.; Kawagishi, I. Hypoxia-induced localization of chemotaxis-related signaling proteins in Vibrio cholerae. Mol. Microbiol. 2015, 95, 780–790. [Google Scholar] [CrossRef]
- Greer-Phillips, S.E.; Sukomon, N.; Chua, T.K.; Johnson, M.S.; Crane, B.R.; Watts, K.J. The Aer2 receptor from Vibrio cholerae is a dual PAS-heme oxygen sensor. Mol. Microbiol. 2018, 109, 209–224. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, S.; Suzuki, D.; Itoh, Y.; Suzuki, K.; Tajima, H.; Hyakutake, A.; Homma, M.; Butler-Wu, S.M.; Camilli, A.; Kawagishi, I. Mlp24 (McpX) of Vibrio cholerae implicated in pathogenicity functions as a chemoreceptor for multiple amino acids. Infect. Immun. 2012, 80, 3170–3178. [Google Scholar] [CrossRef]
- Le Moual, H.; Koshland, D.E., Jr. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J. Mol. Biol. 1996, 261, 568–585. [Google Scholar] [CrossRef]
- Zhulin, I.B. The superfamily of chemotaxis transducers: From physiology to genomics and back. Adv. Microb. Physiol. 2001, 45, 157–198. [Google Scholar] [CrossRef]
- Berg, H.C. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 2003, 72, 19–54. [Google Scholar] [CrossRef]
- Nakamura, S.; Minamino, T. Flagella-Driven Motility of Bacteria. Biomolecules 2019, 9, 279. [Google Scholar] [CrossRef]
- Sowa, Y.; Berry, R.M. Bacterial flagellar motor. Q. Rev. Biophys. 2008, 41, 103–132. [Google Scholar] [CrossRef]
- Chen, S.; Beeby, M.; Murphy, G.E.; Leadbetter, J.R.; Hendrixson, D.R.; Briegel, A.; Li, Z.; Shi, J.; Tocheva, E.I.; Muller, A.; et al. Structural diversity of bacterial flagellar motors. EMBO J. 2011, 30, 2972–2981. [Google Scholar] [CrossRef]
- Tan, J.; Zhang, X.; Wang, X.; Xu, C.; Chang, S.; Wu, H.; Wang, T.; Liang, H.; Gao, H.; Zhou, Y.; et al. Structural basis of assembly and torque transmission of the bacterial flagellar motor. Cell 2021, 184, 2665–2679 e2619. [Google Scholar] [CrossRef]
- Thomas, D.R.; Morgan, D.G.; DeRosier, D.J. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc. Natl. Acad. Sci. USA 1999, 96, 10134–10139. [Google Scholar] [CrossRef]
- Kojima, S.; Imada, K.; Sakuma, M.; Sudo, Y.; Kojima, C.; Minamino, T.; Homma, M.; Namba, K. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol. Microbiol. 2009, 73, 710–718. [Google Scholar] [CrossRef]
- Leake, M.C.; Chandler, J.H.; Wadhams, G.H.; Bai, F.; Berry, R.M.; Armitage, J.P. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 2006, 443, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Reid, S.W.; Leake, M.C.; Chandler, J.H.; Lo, C.J.; Armitage, J.P.; Berry, R.M. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl. Acad. Sci. USA 2006, 103, 8066–8071. [Google Scholar] [CrossRef] [PubMed]
- Terashima, H.; Fukuoka, H.; Yakushi, T.; Kojima, S.; Homma, M. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na-driven flagella and required for stator formation. Mol. Microbiol. 2006, 62, 1170–1180. [Google Scholar] [CrossRef]
- Terashima, H.; Li, N.; Sakuma, M.; Koike, M.; Kojima, S.; Homma, M.; Imada, K. Insight into the assembly mechanism in the supramolecular rings of the sodium-driven Vibrio flagellar motor from the structure of FlgT. Proc. Natl. Acad. Sci. USA 2013, 110, 6133–6138. [Google Scholar] [CrossRef]
- Zhu, S.; Nishikino, T.; Takekawa, N.; Terashima, H.; Kojima, S.; Imada, K.; Homma, M.; Liu, J. In Situ Structure of the Vibrio Polar Flagellum Reveals a Distinct Outer Membrane Complex and Its Specific Interaction with the Stator. J. Bacteriol. 2020, 202. [Google Scholar] [CrossRef]
- Lloyd, S.A.; Whitby, F.G.; Blair, D.F.; Hill, C.P. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature 1999, 400, 472–475. [Google Scholar] [CrossRef]
- Zhou, J.; Lloyd, S.A.; Blair, D.F. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 1998, 95, 6436–6441. [Google Scholar] [CrossRef]
- Asai, Y.; Yakushi, T.; Kawagishi, I.; Homma, M. Ion-coupling determinants of Na+-driven and H+-driven flagellar motors. J. Mol. Biol. 2003, 327, 453–463. [Google Scholar] [CrossRef]
- Gosink, K.K.; Häse, C.C. Requirements for conversion of the Na+-driven flagellar motor of Vibrio cholerae to the H+-driven motor of Escherichia coli. J. Bacteriol. 2000, 182, 4234–4240. [Google Scholar] [CrossRef]
- Paulick, A.; Delalez, N.J.; Brenzinger, S.; Steel, B.C.; Berry, R.M.; Armitage, J.P.; Thormann, K.M. Dual stator dynamics in the Shewanella oneidensis MR-1 flagellar motor. Mol. Microbiol. 2015, 96, 993–1001. [Google Scholar] [CrossRef]
- Sowa, Y.; Homma, M.; Ishijima, A.; Berry, R.M. Hybrid-fuel bacterial flagellar motors in Escherichia coli. Proc. Natl. Acad. Sci. USA 2014, 111, 3436–3441. [Google Scholar] [CrossRef]
- Deme, J.C.; Johnson, S.; Vickery, O.; Aron, A.; Monkhouse, H.; Griffiths, T.; James, R.H.; Berks, B.C.; Coulton, J.W.; Stansfeld, P.J.; et al. Structures of the stator complex that drives rotation of the bacterial flagellum. Nat. Microbiol. 2020, 5, 1553–1564. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Popp, P.F.; Santiveri, M.; Roa-Eguiara, A.; Yan, Y.; Martin, F.J.O.; Liu, Z.; Wadhwa, N.; Wang, Y.; Erhardt, M.; et al. Ion selectivity and rotor coupling of the Vibrio flagellar sodium-driven stator unit. Nat. Commun. 2023, 14, 4411. [Google Scholar] [CrossRef]
- Santiveri, M.; Roa-Eguiara, A.; Kuhne, C.; Wadhwa, N.; Hu, H.; Berg, H.C.; Erhardt, M.; Taylor, N.M.I. Structure and Function of Stator Units of the Bacterial Flagellar Motor. Cell 2020, 183, 244–257 e216. [Google Scholar] [CrossRef]
- Chang, Y.; Zhang, K.; Carroll, B.L.; Zhao, X.; Charon, N.W.; Norris, S.J.; Motaleb, M.A.; Li, C.; Liu, J. Molecular mechanism for rotational switching of the bacterial flagellar motor. Nat. Struct. Mol. Biol. 2020, 27, 1041–1047. [Google Scholar] [CrossRef]
- Falke, J.J.; Koshland, D.E., Jr. Global flexibility in a sensory receptor: A site-directed cross-linking approach. Science 1987, 237, 1596–1600. [Google Scholar] [CrossRef]
- Milligan, D.L.; Koshland, D.E., Jr. Site-directed cross-linking. Establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J. Biol. Chem. 1988, 263, 6268–6275. [Google Scholar] [CrossRef]
- Tajima, H.; Imada, K.; Sakuma, M.; Hattori, F.; Nara, T.; Kamo, N.; Homma, M.; Kawagishi, I. Ligand specificity determined by differentially arranged common ligand-binding residues in bacterial amino acid chemoreceptors Tsr and Tar. J. Biol. Chem. 2011, 286, 42200–42210. [Google Scholar] [CrossRef]
- Nishiyama, S.; Takahashi, Y.; Yamamoto, K.; Suzuki, D.; Itoh, Y.; Sumita, K.; Uchida, Y.; Homma, M.; Imada, K.; Kawagishi, I. Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants. Sci. Rep. 2016, 6, 20866. [Google Scholar] [CrossRef]
- Alexander, R.P.; Zhulin, I.B. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. USA 2007, 104, 2885–2890. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Liu, J.; Hu, B.; Morado, D.R.; Jani, S.; Manson, M.D.; Margolin, W. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc. Natl. Acad. Sci. USA 2012, 109, E1481–E1488. [Google Scholar] [CrossRef] [PubMed]
- Gestwicki, J.E.; Kiessling, L.L. Inter-receptor communication through arrays of bacterial chemoreceptors. Nature 2002, 415, 81–84. [Google Scholar] [CrossRef]
- Briegel, A.; Li, X.; Bilwes, A.M.; Hughes, K.T.; Jensen, G.J.; Crane, B.R. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc. Natl. Acad. Sci. USA 2012, 109, 3766–3771. [Google Scholar] [CrossRef]
- Ortega, D.R.; Kjaer, A.; Briegel, A. The chemosensory systems of Vibrio cholerae. Mol. Microbiol. 2020, 114, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Ringgaard, S.; Schirner, K.; Davis, B.M.; Waldor, M.K. A family of ParA-like ATPases promotes cell pole maturation by facilitating polar localization of chemotaxis proteins. Genes. Dev. 2011, 25, 1544–1555. [Google Scholar] [CrossRef]
- Anantharaman, V.; Aravind, L. Cache—A signaling domain common to animal Ca2+-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem. Sci. 2000, 25, 535–537. [Google Scholar] [CrossRef]
- Kuroda, A.; Kumano, T.; Taguchi, K.; Nikata, T.; Kato, J.; Ohtake, H. Molecular cloning and characterization of a chemotactic transducer gene in Pseudomonas aeruginosa. J. Bacteriol. 1995, 177, 7019–7025. [Google Scholar] [CrossRef]
- Taguchi, K.; Fukutomi, H.; Kuroda, A.; Kato, J.; Ohtake, H. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology 1997, 143, 3223–3229. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, D.W.; Ordal, G.W. Cloning and characterization of genes encoding methyl-accepting chemotaxis proteins in Bacillus subtilis. J. Biol. Chem. 1994, 269, 14038–14046. [Google Scholar] [CrossRef]
- Muller, J.; Schiel, S.; Ordal, G.W.; Saxild, H.H. Functional and genetic characterization of mcpC, which encodes a third methyl-accepting chemotaxis protein in Bacillus subtilis. Microbiology 1997, 143, 3231–3240. [Google Scholar] [CrossRef]
- Takahashi, Y.; Nishiyama, S.; Kawagishi, I.; Imada, K. Structural basis of the binding affinity of chemoreceptors Mlp24p and Mlp37p for various amino acids. Biochem. Biophys. Res. Commun. 2020, 523, 233–238. [Google Scholar] [CrossRef]
- Takahashi, Y.; Nishiyama, S.; Sumita, K.; Kawagishi, I.; Imada, K. Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 of Vibrio cholerae. J. Bacteriol. 2019, 201, 10–1128. [Google Scholar] [CrossRef]
- Goers Sweeney, E.; Henderson, J.N.; Goers, J.; Wreden, C.; Hicks, K.G.; Foster, J.K.; Parthasarathy, R.; Remington, S.J.; Guillemin, K. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure 2012, 20, 1177–1188. [Google Scholar] [CrossRef]
- Ghosh, S.; Rao, K.H.; Sengupta, M.; Bhattacharya, S.K.; Datta, A. Two gene clusters co-ordinate for a functional N-acetylglucosamine catabolic pathway in Vibrio cholerae. Mol. Microbiol. 2011, 80, 1549–1560. [Google Scholar] [CrossRef]
- Taylor, R.K.; Manoil, C.; Mekalanos, J.J. Broad-host-range vectors for delivery of TnphoA: Use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J. Bacteriol. 1989, 171, 1870–1878. [Google Scholar] [CrossRef]
- Selvaraj, P.; Gupta, R.; Peterson, K.M. The Vibrio cholerae ToxR Regulon Encodes Host-Specific Chemotaxis Proteins that Function in Intestinal Colonization. SOJ Microbiol. Infect. Dis. 2015, 3, 1–5. [Google Scholar] [CrossRef]
- Valiente, E.; Davies, C.; Mills, D.C.; Getino, M.; Ritchie, J.M.; Wren, B.W. Vibrio cholerae accessory colonisation factor AcfC: A chemotactic protein with a role in hyperinfectivity. Sci. Rep. 2018, 8, 8390. [Google Scholar] [CrossRef]
- Boin, M.A.; Häse, C.C. Characterization of Vibrio cholerae aerotaxis. FEMS Microbiol. Lett. 2007, 276, 193–201. [Google Scholar] [CrossRef]
- Homma, M.; Oota, H.; Kojima, S.; Kawagishi, I.; Imae, Y. Chemotactic responses to an attractant and a repellent by the polar and lateral flagellar systems of Vibrio alginolyticus. Microbiology 1996, 142, 2777–2783. [Google Scholar] [CrossRef]
- Chaparro, A.P.; Ali, S.K.; Klose, K.E. The ToxT-dependent methyl-accepting chemoreceptors AcfB and TcpI contribute to Vibrio cholerae intestinal colonization. FEMS Microbiol. Lett. 2010, 302, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Harkey, C.W.; Everiss, K.D.; Peterson, K.M. The Vibrio cholerae toxin-coregulated-pilus gene tcpI encodes a homolog of methyl-accepting chemotaxis proteins. Infect. Immun. 1994, 62, 2669–2678. [Google Scholar] [CrossRef]
- Everiss, K.D.; Hughes, K.J.; Kovach, M.E.; Peterson, K.M. The Vibrio cholerae acfB colonization determinant encodes an inner membrane protein that is related to a family of signal-transducing proteins. Infect. Immun. 1994, 62, 3289–3298. [Google Scholar] [CrossRef]
- Jeffery, C.J.; Koshland, D.E., Jr. Vibrio cholerae hlyB is a member of the chemotaxis receptor gene family. Protein Sci. 1993, 2, 1532–1535. [Google Scholar] [CrossRef]
- Rollins, S.M.; Peppercorn, A.; Hang, L.; Hillman, J.D.; Calderwood, S.B.; Handfield, M.; Ryan, E.T. In vivo induced antigen technology (IVIAT). Cell Microbiol. 2005, 7, 1–9. [Google Scholar] [CrossRef]
- Jugder, B.E.; Batista, J.H.; Gibson, J.A.; Cunningham, P.M.; Asara, J.M.; Watnick, P.I. Vibrio cholerae high cell density quorum sensing activates the host intestinal innate immune response. Cell Rep. 2022, 40, 111368. [Google Scholar] [CrossRef]
- Monteagudo-Cascales, E.; Lozano-Montoya, A.; Krell, T. Pseudomonas aeruginosa performs chemotaxis to serotonin, dopamine, epinephrine, and norepinephrine. bioRxiv 2024. bioRxiv:2024.2012.2005.626837. [Google Scholar] [CrossRef]
- Moorthy, S.; Watnick, P.I. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol. Microbiol. 2005, 57, 1623–1635. [Google Scholar] [CrossRef]
- Murphy, S.G.; Johnson, B.A.; Ledoux, C.M.; Dorr, T. Vibrio cholerae's mysterious Seventh Pandemic island (VSP-II) encodes novel Zur-regulated zinc starvation genes involved in chemotaxis and cell congregation. PLoS Genet 2021, 17, e1009624. [Google Scholar] [CrossRef] [PubMed]
- Irazoki, O.; Ter Beek, J.; Alvarez, L.; Mateus, A.; Colin, R.; Typas, A.; Savitski, M.M.; Sourjik, V.; Berntsson, R.P.; Cava, F. D-amino acids signal a stress-dependent run-away response in Vibrio cholerae. Nat. Microbiol. 2023, 8, 1549–1560. [Google Scholar] [CrossRef] [PubMed]
- Briegel, A.; Ortega, D.R.; Mann, P.; Kjaer, A.; Ringgaard, S.; Jensen, G.J. Chemotaxis cluster 1 proteins form cytoplasmic arrays in Vibrio cholerae and are stabilized by a double signaling domain receptor DosM. Proc. Natl. Acad. Sci. USA 2016, 113, 10412–10417. [Google Scholar] [CrossRef] [PubMed]
- Shiomi, D.; Zhulin, I.B.; Homma, M.; Kawagishi, I. Dual recognition of the bacterial chemoreceptor by chemotaxis-specific domains of the CheR methyltransferase. J. Biol. Chem. 2002, 277, 42325–42333. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Omori, F.; Tajima, H.; Asaoka, S.; Nishiyama, S.-i.; Sowa, Y.; Kawagishi, I. Chemotaxis and Related Signaling Systems in Vibrio cholerae. Biomolecules 2025, 15, 434. https://doi.org/10.3390/biom15030434
Omori F, Tajima H, Asaoka S, Nishiyama S-i, Sowa Y, Kawagishi I. Chemotaxis and Related Signaling Systems in Vibrio cholerae. Biomolecules. 2025; 15(3):434. https://doi.org/10.3390/biom15030434
Chicago/Turabian StyleOmori, Fuga, Hirotaka Tajima, Sotaro Asaoka, So-ichiro Nishiyama, Yoshiyuki Sowa, and Ikuro Kawagishi. 2025. "Chemotaxis and Related Signaling Systems in Vibrio cholerae" Biomolecules 15, no. 3: 434. https://doi.org/10.3390/biom15030434
APA StyleOmori, F., Tajima, H., Asaoka, S., Nishiyama, S.-i., Sowa, Y., & Kawagishi, I. (2025). Chemotaxis and Related Signaling Systems in Vibrio cholerae. Biomolecules, 15(3), 434. https://doi.org/10.3390/biom15030434





