Abstract
The Translational Chickpea Genomics Consortium (TCGC) was set up to increase the production and productivity of chickpea (Cicer arietinum L.). It represents research institutes from six major chickpea growing states (Madhya Pradesh, Maharashtra, Andhra Pradesh, Telangana, Karnataka and Uttar Pradesh) of India. The TCGC team has been engaged in deploying modern genomics approaches in breeding and popularizing improved varieties in farmers’ fields across the states. Using marker-assisted backcrossing, introgression lines with enhanced drought tolerance and fusarium wilt resistance have been developed in the genetic background of 10 elite varieties of chickpea. Multi-location evaluation of 100 improved lines (70 desi and 30 kabuli) during 2016–2017 and 2018–2019 enabled the identification of top performing desi and kabuli lines. In total, 909 Farmer Participatory Varietal Selection trials were conducted in 158 villages in 16 districts of the five states, during 2017–2018, 2018–2019, and 2019–2020, involving 16 improved varieties. New molecular breeding lines developed in different genetic backgrounds are potential candidates for national trials under the ICAR-All India Coordinated Research Project on Chickpea. The comprehensive efforts of TCGC resulted in the development and adoption of high-yielding varieties that will increase chickpea productivity and the profitability of chickpea growing farmers.
1. Introduction
Chickpea (Cicer arietinum L.; 2x = 2n = 16) is an important food legume crop cultivated on 13.72 M ha with a total production of 14.25 M t [1]. Although India is the largest producer of chickpea, it imports large quantities from Australia (83.5%), USA (3.8%), Myanmar (3.5%), Tanzania (3.3%), and Sudan (2.1%) to meet local demand (http://agricoop.nic.in/sites/default/files/Pulses%20profie_Mar%2C%202019_0.pdf; Last accessed on 25 July 2021). In terms of production in India, Madhya Pradesh ranks first, contributing 33.99% of the area and 40.92% of production, followed by Maharashtra, Rajasthan, Karnataka, Andhra Pradesh and Uttar Pradesh (http://dpd.gov.in/Annual%20Report%202017-18.pdf; Last accessed on 25 July 2021). Limited genetic diversity coupled with climate change during recent years has increased the frequency and severity of biotic and abiotic stresses and emerging diseases that are serious threats to chickpea production [2,3,4]. In order to achieve the crop’s actual yield potential, it is essential to enhance its genetic diversity and resistance/tolerance to biotic and abiotic stresses in the varieties grown by farmers.
Modern breeding technologies have proven useful in developing superior varieties in crops such as maize, rice, wheat, barley and soybean [5]. This was not the case in chickpea until recently, primarily due to limited information on genes and the ability to deploy genomics tools. In recent years, the tremendous progress made in developing novel genomic tools in chickpea, such as the draft genome sequence [6], several millions of SNP markers from whole genome sequence information on germplasm lines [7,8,9] and cost-effective genotyping platforms including low- to high-density SNP arrays [10,11]. Likewise, QTLs and markers associated with abiotic stresses like drought [12,13,14,15], heat [16] and salinity [17], and biotic stresses like fusarium wilt (FW) [18,19] and Ascochyta blight (AB) [18,20] are available for chickpea improvement. These have facilitated marker-assisted selection/introgression in chickpea breeding programs. Marker-assisted backcrossing (MABC) approach has been successfully deployed to develop superior varieties in the crop [21].
Once the chickpea genome sequence was available, the Department of Agriculture & Farmers Welfare (DA & FW) encouraged a consortium approach to translate this knowledge to enhance chickpea improvement through funding support. As a result, the Translational Chickpea Genomics Consortium (TCGC) was established in 2016, comprising six research institutions/agricultural universities from six major chickpea growing states of India: International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Hyderabad, Telangana; Regional Agricultural Research Station (RARS), Nandyal, Andhra Pradesh; Agricultural Research Station (ARS), Kalaburagi, University of Agricultural Sciences (UAS), Raichur, Karnataka; RAK College of Agriculture (RAKCA), Rajmata Vijayaraje Scindia Krishi Vishwa Vidyalaya (RVSKVV), Sehore, Madhya Pradesh; Mahatma Phule Krishi Vidyapeeth (MPKV), Rahuri, Maharashtra; and Indian Institute of Pulses Research (ICAR-IIPR), Kanpur, Uttar Pradesh. These research institutions are very well catering needs of the almost 80% of the chickpea area in India. The consortium’s major focus has been on deploying genomics information for chickpea improvement, developing/identifying improved varieties and enhancing the adoption of superior lines in farmers’ fields. During the past five years, TCGC has made significant progress that has involved: (a) deployment of genomics-assisted breeding in 10 chickpea varieties for drought tolerance and fusarium wilt resistance; (b) evaluation of 100 improved elite breeding lines (70 desi and 30 kabuli) for their performance in multi-location trials and (c) demonstration of improved crop varieties in 909 farmers’ fields across 158 villages spanning 16 districts in the states of Andhra Pradesh, Karnataka, Madhya Pradesh, Maharashtra and Uttar Pradesh. This paper reports on the significant achievements of the TCGC that can guide future chickpea improvement programs and lead to the development and popularization of improved varieties in important regions where the crop is grown in India.
2. Materials and Methods
2.1. Marker-Assisted Backcrossing for Drought Tolerance and Fusarium wilt Resistance
The MABC approach was adopted to enhance drought tolerance and FW resistance in two leading varieties from each of the consortium partners (Figure 1). In brief, 10 select recurrent parents, namely RSG 888, DCP 92-3, RVG 202, RVG 203, DigVijay, Phule Vikram, NBeG 47, NBeG 49, GBM 2 and BGD 103 were crossed with ICC 4958 as a donor (“QTL-hotspot”) for drought tolerance (Figure 1). Similarly, the genomic regions for FW resistance were also introgressed into the genetic background of 10 elite varieties (JAKI 9218, GNG 1581, RVG 202, RVG 203, Digvijay, Vijay, NBeG 47, NBeG 49, GBM 2 and BGD 103) using Super Annigeri 1 as a donor (Figure 1).
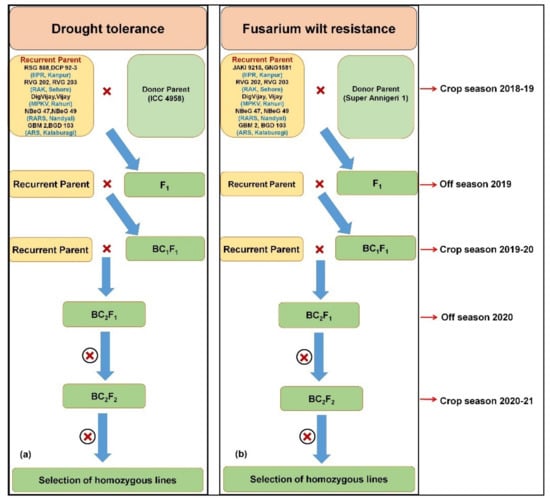
Figure 1.
Marker-assisted backcrossing was adopted for (a) drought tolerance and (b) fusarium wilt resistance using two recurrent parents from each center. ICC 4958 was used to introgress “QTL-hotspot” harbouring QTLs for drought tolerance related traits and Super Annigeri 1 was used as donor to introgress FW resistance in elite chickpea cultivars.
2.2. Development of Trait-Associated Markers
To develop trait associated markers, available genomic information on candidate regions for drought and FW was collected from previous studies and focused on the identification of major QTL regions [12,14,15,18,19,20]. For drought, a total of 26 genes identified in the “QTL-hotspot” region containing 24 SNPs were used to develop molecular markers for use in early generation selection. As a result, 10 SNP panel for drought was developed and tested for early generation selection for the backcross progenies (Table 1). Similarly, for the development of FW markers, genomic information on candidate regions of QTL associated with FW resistance was assembled from the studies [18,19,20] (Table 1 and Table S1). We used these markers on the backcross progenies and validated their usefulness in chickpea breeding programs. These findings have been considered important milestones for accelerated chickpea breeding programs.

Table 1.
The SNP panels used to select true hybrids.
2.3. Multi-Location Trials for Promising Chickpea Lines
To identify the best performing lines across locations, 70 desi and 30 kabuli lines (including molecular breeding lines) were evaluated for their yield performance (kg/ha) during 2016–2017 and 2018–2019. Based on the availability of seed, multi-location trials were conducted in three locations (Jabalpur, Nandyal and Sehore) during 2016–2017 and in five locations (Nandyal, Kalaburagi, Sehore, Rahuri and Kanpur) during 2018–2019. However, kabuli lines were not evaluated in Sehore and Kanpur locations during 2018–2019 owing to limited seed availability. Desi and kabuli lines were evaluated in an alpha lattice design with 14 entries per block. A spacing of 30 cm × 10 cm (intra-row) and 4 m (inter-row) was adopted to evaluate desi lines and a spacing of 45 cm × 10 cm (intra-row) and 4 m (inter-row) was adopted for kabuli lines. Combined analysis of variance (ANOVA) was carried out using SAS mixed procedure [22] to test the significance of main and interaction effects of environments and genotypes, considering environment, genotype, replication and block as random effects. Individual environmental variances were modelled into combined analysis with REPEATED statement using REML (Restricted Maximum Likelihood) method. Best Linear Unbiased Predictors (BLUPs) were calculated from the combined analysis. To identify the best test environment and superior genotype with high yield and stable performance across locations, GGE (genotype (G) + (genotype (G) × environment (E) interaction), a biplot analysis was done with the mean grain yield data obtained from multi-location trials. Using SREG (Site regression), GGE biplots [23] were performed for significant interaction effects of district and variety (FPSV trials) and environment and genotype (desi and kabuli multi-environment trials) to draw conclusions on genotype and environment evaluations.
2.4. Varietal Adoption through Farmer Participatory Varietal Selection (FPVS) Trials
To enhance the adoption of improved varieties that are already available, FPVS trials were conducted in five major chickpea growing states (Andhra Pradesh, Karnataka, Madhya Pradesh, Maharashtra and Uttar Pradesh) in India during 2017–2018, 2018–2019 and 2019–2020 (Table 2). In total, 909 FPVS trials were conducted during three years. To date, the data from the three years has been compiled and analyzed. During the three years, two to five improved varieties in each state were distributed to farmers to conduct FPVS. A total of 909 FPVS trials were conducted and 16 improved chickpea varieties (two to five varieties in each state) were tested in 16 districts in the five states (Table 2).

Table 2.
Summary of FPVS trials conducted during 2017–2018, 2018–2019 and 2019–2020 seasons.
3. Results
3.1. Genomics-Assisted Breeding
Drought and FW are major abiotic and biotic stresses that hamper chickpea production. Some of the chickpea varieties with enhanced drought tolerance and FW resistance which were developed earlier using MABC approach and released (https://icar.org.in/content/development-two-superior-chickpea-varieties-genomics-assisted-breeding; https://icar.org.in/content/development-two-superior-chickpea-varieties-genomics-assisted-breeding; last accessed on 25 July 2021) [24,25,26,27] are already in the seed chain and have started paying dividends.
F1s were harvested at each center by crossing chosen recipient parents with ICC 4958 as a donor (“QTL-hotspot”) for drought tolerance and with Super Annigeri 1 as a donor for FW resistance during 2018–2019. True hybrids were confirmed using 10 SNP panel and two allele specific markers for the respective traits (Table 1 and Table S1 in Supplementary Materials). Subsequently, BC1F1 were obtained by crossing true F1s to their respective recurrent parents during 2019–2020. At all centers, heterozygous plants were used for subsequent backcrossing with respective recurrent parents, and BC2F1 seeds were harvested during the off season 2020. After foreground selection, positive BC2F1 plants were selfed to generate BC2F2 seeds during crop season 2020–2021 (Table S2). The back cross progenies after background selection needs further evaluation for their yield performance under drought.
3.2. Mean Performance and Stability of Elite Breeding Lines for Grain Yield in Multi-Location Trials
A large variation was observed in the mean performance of grain yields in elite desi and kabuli lines (including molecular breeding lines) in the states Andhra Pradesh, Karnataka, Madhya Pradesh, Maharashtra and Uttar Pradesh (Tables S3 and S4). Combined analysis of variance revealed significant differences among genotypes, environments, and genotype × environment interaction effects for grain yield in desi lines (Table 3). In the case of kabuli lines, genotype × environment and environment differed significantly. In the GGE biplot, desi and kabuli lines account for 50.18% and 65.58% of the total variation of the environment-centered genotype and GEI variation for grain yield, respectively (Figure 2a,b). The large variation among environmental means caused most of the variation in grain yield. Of 70 desi and 30 kabuli elite breeding lines, the high stability performance lines in two multi-location trials are presented in Table 4. In desi elite breeding lines across the locations in two multi-location trials, Jabalpur_2017 was identified as a highly discriminating environment. Other locations such as Nandyal_2018 and Sehore_2017 were identified as moderately discriminating environments and the rest of the environments were least discriminating for grain yield (Figure 2a). In kabuli lines, Sehore_2017 was identified as a highly discriminating environment for grain yield, while Jabalpur_2017, Nandyal_2017 and Nandyal_2018 were identified as moderately discriminating environments and Kalaburagi_2018 and Rahuri_2018 were identified as the least discriminating environments (Figure 2b).

Table 3.
Combined analysis of variance for grain yield based on multi-location trials of 100 elite lines (70 desi and 30 kabuli) conducted during cropping seasons 2016–2017 and 2018–2019.
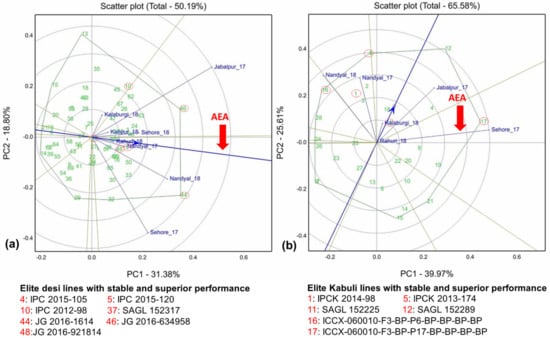
Figure 2.
GGE biplots of (a) 70 desi elite lines evaluated in five locations and (b) 30 kabuli elite breeding lines evaluated in three locations for yield (kg/ha) during crop seasons 2016–2017 and 2018–2019. The Average Environment Axis (AEA) or Average Environment Coordination (AEC) abscissa (in blue) is the single arrowed line, which passes through the origin of the biplot and through the hypothetical average environment, denoted by the circle near (Rahuri_18 and Nandyal_17 (Desi line biplot) and Kalaburgi_18 (Kabuli lines biplot). The direction of the arrowhead on the AEA points to higher mean values for grain yield. PC1 and PC2 are the first and second principal components, respectively. Desi and kabuli lines with stable yield performance are circled in red and their genotype names are indicated below the GGE plots.

Table 4.
High-yielding (kg/ha) desi and kabuli lines with stable performance in multi-location trials conducted during 2016–2017 and 2018–2019.
3.3. Promising Desi Lines for Grain Yield Identified through Multi-Location Trials
Desi lines Phule G 0919-4-8 at Nandyal and SAGL 152210 at Sehore revealed yield advantage (6.02% and 24.15%) over the local checks across the years (Table S2). Phule G 0914-8-14 at Rahuri and IPC 2008-11 at Kanpur recorded significant yield advantage (6.42% and 7.63%) compared to the local checks (Table S2). Importantly, JG 2016-634958 at Jabalpur and SAGL 152278 at Kalaburagi demonstrated yield advantage (73.44% and 109.48%) over the local checks. Across the locations and over the years, desi line JG 2016-1614 was identified as a superior line with high grain yield and high stability and a 2.6% yield advantage over the check (Table S3).
3.4. Promising Kabuli Lines for Grain Yield Identified through Multi-Location Trials
Kabuli lines IPCK 2013-152 at Nandyal exhibited a yield advantage (111%) over the local check across the years (Table S3). SAGL 152220 at Kalaburagi, ICCX-060010-F3-BP-P17-BP-BP-BP-BP at Sehore, IPCK 2012-129 at Rahuri and SAGL 152289 at Jabalpur also recorded higher grain yield and yield advantage (141.29%, 21.43%, 33.25% and 116.04%, respectively) over the local checks (Table S3). A total of 30 kabuli elite breeding lines and three lines (ICCX-060010-F3-BP-P6-BP-BP-BP-BP, IPCK 2013-174 and SAGL 152289) have been identified as superior with higher grain yield and high stability and a 12.11–11.11% yield advantage over the national check (Vihar) across the locations and over the years (Table S4).
3.5. Enhancing Varietal Adoption through FPVS Trials
The mean yields of different varieties during 2017–2018, 2018–2019 and 2019–2020 are presented in Table 5. The ANOVA revealed significant differences among genotypes, but non-significant genotype × environment interaction for grain yield at all the locations over the years (Andhra Pradesh, Madhya Pradesh, Maharashtra and Uttar Pradesh) except Karnataka in 2017–2018 (Table 6). In Karnataka in 2017–2018, a GGE biplot explained stability analysis for grain yield, which accounted for 100% of the total variation of the environment-centered G × E (Table 6). Due to significant GEI, the Dharwad and Kalaburagi were identified as the most discriminating environment and significantly differed with Bijapur for grain yield (Figure 3).

Table 5.
Mean grain yield (kg/ha) of select chickpea cultivars in FPVS trials conducted in five states of India during cropping seasons 2017–2018 2018–2019 and 2019–2020.

Table 6.
Combined analysis of variance (F-value) for grain yield based on FPVS trials in centers in the respective states.
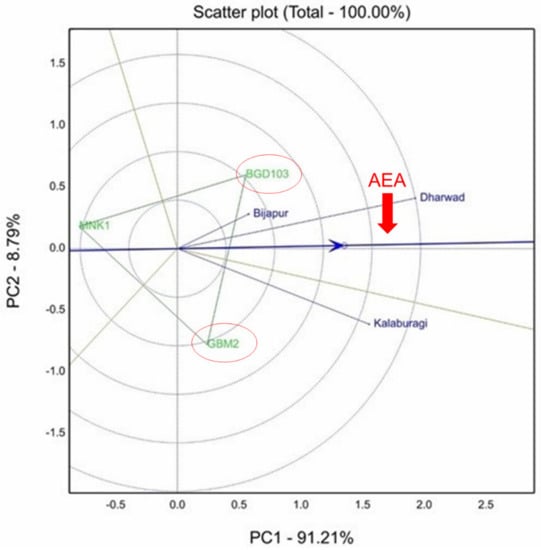
Figure 3.
GGE biplot showing significant genotype × environment interaction (GEI) for grain yield in Karnataka during 2017–2018. Dharwad and Kalaburagi were identified as the most discriminating environments.
3.6. Selection of High-Performing Varieties in Different States
In the FPVS trials conducted during 2017–2018 in Andhra Pradesh, NBeG 49 showed outstanding performance for grain yield and was the choice of farmers in all the three districts of Kurnool, Anantapur and Prakasam (Table 5 and Figure 4a). In the trials conducted in 2018–2019, NBeG 49 was found superior over varieties NBeG 47 and NBeG 3 tested in the region, with a 37.1% yield advantage over the local check. It is important to mention that in Kurnool district, NBeG 49 recorded a 41.9% yield advantage over the local check (Table 5). In Kurnool district, during 2019–2020, desi variety NBeG 49 and kabuli variety NBeG 119 showed 9.69% and 18.89% yield advantages, respectively over their respective local checks. Similarly, in Prakasam district, 9.37% and 13.4% yield advantage was recorded in NBeG 49 and NBeG 119, respectively. In Anantapur district, 10.77% yield advantage was recorded in NBeG 49 while it was only 1.56% in NBeG 119. Based on three years’ FPVS trials conducted in Andhra Pradesh, NBeG 49 was identified as the best performing line and was preferred by the farming community.
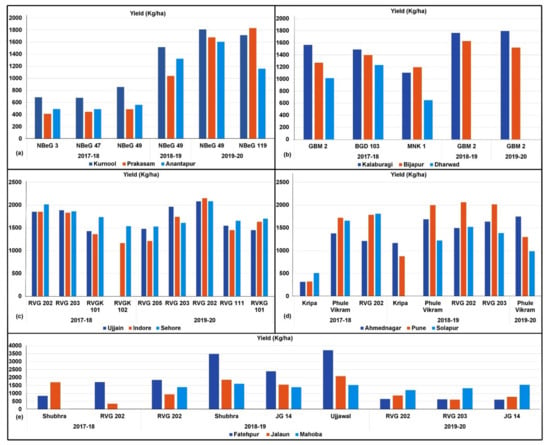
Figure 4.
Farmer preferred varieties identified through FPVS trials conducted in different chickpea growing states. The best performing lines were (a) NBeG 49 in Andhra Pradesh; (b) GBM 2 in Karnataka; (c) RVG 202 in Madhya Pradesh; (d) Phule Vikram and RVG 202 in Maharashtra and (e) RVG 202 and Shubhra (IPCK 2002-29) in Uttar Pradesh.
Based on FPVS trials conducted in Karnataka during 2017–2018, among the three elite chickpea varieties (GBM 2, BGD 103 and MNK 1), GBM 2 was found high yielding in Kalaburagi district, whereas, BGD 103 recorded high yield in Bijapur and Dharwad districts (Table 5 and Figure 4b). However, due to its traits that are amenable to mechanical harvesting, farmers from all the three districts preferred GBM 2 in 2018–2019 and 2019–2020 (Table 5 and Figure 4b).
Based on FPVS trials conducted in Madhya Pradesh during 2017–2018, RVG 202 was identified as high yielding in Indore and Sehore districts, whereas, RVG 203 was high yielding in Ujjain district (Table 5 and Figure 4c). During 2018–2019, farmers ranked the performance of different varieties. RVG 202 demonstrated superior performance in all three districts. No yield data was provided during 2018–2019 trials. In 2019–2020, FPVS trials were conducted with RVG 205, RVG 202, RVG 203, RVG 111 and RVKG 101. Of these, RVG 202 exhibited the highest grain yield of 2105 kg/ha followed by RVG 203 (1772 kg/ha) in three districts (Table 5). In Madhya Pradesh, RVG 202 followed by RVG 203 were identified as the best performing lines.
In Maharashtra, Phule Vikram was found high yielding in Ahmednagar district, whereas RVG 202 was identified as high yielding in Pune and Solapur districts in the 2017–2018 FPVS trials (Table 5 and Figure 4d). Similarly, during 2018–2019, Phule Vikram in Ahmednagar and RVG 202 in Pune and Solapur districts were identified as high-yielding varieties (Table 5 and Figure 4d). In the demonstrations in 2019–2020, the yield of Phule Vikram varied widely in all the three districts. For instance, in Ahmednagar district, it recorded the highest grain yield of 1746 kg/ha (Table 5) while yields recorded in Pune and Sholapur districts were 1278 kg/ha and 987 kg/ha yield, respectively (Table 5). Overall, Phule Vikram was widely preferred by farmers in Maharashtra. Under this study, Phule Vikram reached farmers of Pune and Solapur districts. Overall, the farmers expressed satisfaction about the varieties and showed preference for it. The productivity in Solapur district was less as compared to Ahmednagar and Pune districts due to the scarcity or nonavailability of water for irrigation during the crop seasons.
In Uttar Pradesh, during the 2017–2018 FPVS trials, variety RVG 202 showed highest grain yield in Fatehpur district and variety Shubhra was found higher yielding in Jalaun district (Table 5 and Figure 4e). While Shubhra and Ujjawal gave higher yields compared to RVG 202 in Fatehpur and Jalaun districts in 2018–2019 trials (Table 5 and Figure 4e), Shubhra recorded the highest yield in Mahoba district. In the 2019–2020 FPVS trials, RVG 202 recorded the highest grain yield in Fatehpur and Jalaun districts (Table 5) while JG 14 recorded highest grain yield of 1548 kg/ha (Table 5) in Mahoba district. RVG 202 was preferred as the best performing line by farmers in the state.
4. Conclusions
The TCGC has been involved in the development of traits for drought and FW associated markers and conduct of FPVS trials to enable improved varieties to reach farmers. It has successfully developed trait (biotic and abiotic stress)-associated molecular markers and is currently working on developing drought tolerance and FW resistance through the MABC approach. Multi-location and FPVS trials were used to identify high grain yield and highly stable varieties on station and in farmers’ fields. Among the desi elite breeding lines, Phule G 0919-4-8 at Nandyal, SAGL 152210 at Sehore, JG 2016-634958 at Jabalpur, SAGL 152278 at Kalaburagi, PhuleG 0914-8-14 at Rahuri and IPC 2008-11 at Kanpur revealed a high yield advantage over the local checks. Across all the locations and over the years, JG 2016-1614 was identified as a superior line with high grain yield and high stability. Likewise, kabuli elite breeding lines IPCK 2013-152 at Nandyal, SAGL 152220 at Kalaburagi, ICCX-060010-F3-BP-P17-BP-BP-BP-BP at Sehore, IPCK 2012-129 at Rahuri and SAGL 152289 at Jabalpur recorded the highest grain yields over the local checks. Across locations and over the years, ICCX-060010-F3-BP-P6-BP-BP-BP-BP followed by IPCK 2013-174 and SAGL 152289 were identified as superior lines with high grain yield and high stability. In short, NBeG 49 and GBM 2 were identified as the best performing lines preferred by farmers in Andhra Pradesh and Karnataka. RVG 202 followed by RVG 203 were the choice of farmers in Madhya Pradesh while Phule Vikram and RVG 202 were identified as the best performing lines in Maharashtra. In Uttar Pradesh, RVG 202 and Shubhra (IPCK 2002-29) were preferred by the farmers as best performing desi and kabuli lines, respectively.
The advanced drought tolerance and FW resistance backcross lines will be evaluated for grain yield for varietal release. The top performing lines and new molecular breeding lines developed in different genetic backgrounds can be further evaluated for their performance at the national level in AICRP trials on chickpea for release as improved varieties. The development of these high-yielding chickpea varieties and enhancing their adoption will increase the productivity and profitability of smallholder farmers.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/plants10122583/s1, Table S1: Details of allele specific markers for Fusarium wilt; Table S2: Details of marker-assisted backcrossing to improve elite lines for drought tolerance and resistance to fusarium wilt, Table S3: Mean performance of 70 elite desi lines evaluated in different locations during 2016–2017 and 2018–2019, Table S4: Mean performance of 30 elite kabuli lines evaluated in different locations during 2016–2017 and 2018–2019.
Author Contributions
Conceptualization R.K.V., N.P.S., MT.; Methodology: R.P., V.J., Y.K., P.K., M.Y., N.S.K., C.L., S.Y., S.S., K.R.S., B.M., G.P.D., C.B., S.K.C., P.M.G., M.R.; formal analysis: R.P., A.R., A.V.; data curation: A.R., A.V.; writing—original draft preparation: R.P., M.T.; writing—review and editing: M.T., R.K.V., M.R.;: R.K.V., MT., N.P.S.; project administration: R.K.V., N.P.S., M.T.; funding acquisition: R.K.V., N.P.S.; All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Department of Agriculture & Farmers Welfare (DA & FW; Grant ID: F.NO CPS 18-5/2017 NFSM), Ministry of Agriculture and Farmers Welfare, Government of India and partially funded by the Bill & Melinda Gates Foundation (BMGF; Grant ID: OPP1114827). This work was undertaken as part of the CGIAR Research Program on Grain Legumes and Dryland Cereals (CRP-GLDC).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The technical support rendered by Praveen Kumar Manchikatla is greatly appreciated.
Conflicts of Interest
The authors declare no conflict of interest.
References
- FAOSTAT. 2020. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 25 March 2021).
- Sharma, M.; Ghosh, R.; Pande, S. Dry root rot (Rhizoctonia bataticola (Taub.) Butler): An emerging disease of chickpea—Where do we stand. Arch. Phytopathol. Plant Prot. 2015, 48, 797–812. [Google Scholar] [CrossRef]
- Kadiyala, M.D.M.; Kumara, C.D.; Nedumaran, S.D.; Shyam, M.; Gumma, M.K. Agronomic management options for sustaining chickpea yield under climate change scenario. J. Agrometeorol. 2016, 18, 41–47. [Google Scholar]
- Khaliq, A.; Alam, S.; Khan, I.U.; Khan, D.; Naz, S.; Zhang, Y.; Shah, A.A. Integrated control of dry root rot of chickpea caused by Rhizoctonia bataticola under the natural field condition. Biotechnol. Rep. 2020, 25, e00423. [Google Scholar] [CrossRef]
- Thudi, M.; Palakurthi, R.; Schnable, J.C.; Chitikineni, A.; Dreisigacker, S.; Mace, E.; Srivastava, R.K.; Satyavathi, C.T.; Odeny, D.; Tiwari, V.K.; et al. Genomic resources in plant breeding for sustainable agriculture. J. Plant Physiol. 2020, 153351. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Song, C.; Saxena, R.K.; Azam, S.; Yu, S.; Sharpe, A.G.; Cannon, S.; Baek, J.; Rosen, B.D.; Tar’an, B.; et al. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 2013, 31, 240–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thudi, M.; Chitikineni, A.; Liu, X.; He, W.; Roorkiwal, M.; Yang, W.; Jian, J.; Doddamani, D.; Gaur, P.M.; Rathore, A. Recent breeding programs enhanced genetic diversity in both desi and kabuli varieties of chickpea (Cicer arietinum L.). Sci. Rep. 2016, 6, 38636. [Google Scholar] [CrossRef] [Green Version]
- Thudi, M.; Khan, A.W.; Kumar, V.; Gaur, P.M.; Katta, K.; Garg, V.; Roorkiwal, M.; Samineni, S.; Varshney, R.K. Whole genome re-sequencing reveals genome-wide variations among parental lines of 16 mapping populations in chickpea (Cicer arietinum L.). BMC Plant Biol. 2016, 16, 10. [Google Scholar] [CrossRef] [Green Version]
- Varshney, R.K.; Thudi, M.; Roorkiwal, M.; He, W.; Upadhyaya, H.D.; Yang, W.; Bajaj, P.; Cubry, P.; Rathore, A.; Jian, J.; et al. Resequencing of 429 chickpea accessions from 45 countries provides insights into genome diversity, domestication and agronomic traits. Nat. Genet. 2019, 51, 857–864. [Google Scholar] [CrossRef] [Green Version]
- Roorkiwal, M.; Sawargaonkar, S.L.; Chitikineni, A.; Thudi, M.; Saxena, R.K.; Upadhyaya, H.D.; Vales, M.I.; Riera-Lizarazu, O.; Varshney, R.K. Single nucleotide polymorphism genotyping for breeding and genetics applications in chickpea and pigeonpea using the BeadXpress platform. Plant Genome 2013, 6, plantgenome2013.05.0017. [Google Scholar] [CrossRef] [Green Version]
- Roorkiwal, M.; Jain, A.; Kale, S.M.; Doddamani, D.; Chitikineni, A.; Thudi, M.; Varshney, R.K. Development and evaluation of high-density Axiom® Cicer SNP Array for high-resolution genetic mapping and breeding applications in chickpea. Plant Biotechnol. J. 2018, 16, 890–901. [Google Scholar] [CrossRef] [Green Version]
- Varshney, R.K.; Thudi, M.; Nayak, S.N.; Gaur, P.M.; Kashiwagi, J.; Krishnamurthy, L.; Jaganathan, D.; Koppolu, J.; Bohra, A.; Tripathi, S.; et al. Genetic dissection of drought tolerance in chickpea (Cicer arietinum L.). Theor. Appl. Genet. 2014, 127, 445–462. [Google Scholar] [CrossRef]
- Thudi, M.; Upadhyaya, H.D.; Rathore, A.; Gaur, P.M.; Krishnamurthy, L.; Roorkiwal, M.; Nayak, S.N.; Chaturvedi, S.K.; Basu, P.S.; Gangarao, N.V.P.R.; et al. Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. PLoS ONE 2014, 9, e96758. [Google Scholar]
- Jaganathan, D.; Thudi, M.; Kale, S.; Azam, S.; Roorkiwal, R.; Gaur, P.M.; Kishor, P.K.; Nguyen, H.; Sutton, T.; Varshney, R.K. Genotyping-by-sequencing based intra-specific genetic map refines a “QTL-hotspot” region for drought tolerance in chickpea. Mol. Genet. 2015, 290, 559–571. [Google Scholar] [CrossRef]
- Kale, S.M.; Jaganathan, D.; Ruperao, P.; Chen, C.; Punna, R.; Kudapa, H.; Thudi, M.; Roorkiwal, M.; Katta, M.A.; Doddamani, D.; et al. Prioritization of candidate genes in “QTL-hotspot” region for drought tolerance in chickpea (Cicer arietinum L.). Sci. Rep. 2015, 5, 15296. [Google Scholar] [CrossRef] [Green Version]
- Soren, K.R.; Madugula, P.; Kumar, N.; Barmukh, R.; Sengar, M.S.; Bharadwaj, C.; Sharma, P.C.; Singh, S.; Bhandari, A.; Singh, J.; et al. Genetic dissection and identification of candidate genes for salinity tolerance using Axiom® CicerSNP Array in chickpea. Int. J. Mol. Sci. 2020, 21, 5058. [Google Scholar] [CrossRef] [PubMed]
- Sabbavarapu, M.M.; Sharma, M.; Chamarthi, S.K.; Swapna, N.; Rathore, A.; Thudi, M.; Gaur, P.M.; Pande, S.; Singh, S.; Kaur, L.; et al. Molecular mapping of QTLs for resistance to fusarium wilt (race 1) and ascochyta blight in chickpea (Cicer arietinum L.). Euphytica 2013, 193, 121–133. [Google Scholar] [CrossRef] [Green Version]
- Garg, T.; Mallikarjuna, B.P.; Thudi, M.; Samineni, S.; Singh, S.; Sandhu, J.S.; Kaur, L.; Singh, I.; Sirari, A.; Basandrai, A.K. Identification of QTLs for resistance to fusarium wilt and ascochyta blight in a recombinant inbred population of chickpea (Cicer arietinum L.). Euphytica 2018, 214, 45. [Google Scholar] [CrossRef] [Green Version]
- Jha, U.C.; Nayyar, H.; Palakurthi, R.; Jha, R.; Valluri, V.; Bajaj, P.; Chitikineni, A.; Singh, N.P.; Varshney, R.K.; Thudi, M. Major QTLs and potential candidate genes for heat stress tolerance identified in chickpea (Cicer arietinum L.). Front. Plant Sci. 2021, 12, 655103. [Google Scholar] [CrossRef] [PubMed]
- Deokar, A.; Sagi, M.; Tar’an, B. Genome-wide SNP discovery for development of high-density genetic map and QTL mapping of ascochyta blight resistance in chickpea (Cicer arietinum L.). Theor. Appl. Genet. 2019, 132, 1861–1872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roorkiwal, M.; Bharadwaj, C.; Barmukh, R.; Dixit, G.P.; Thudi, M.; Gaur, P.M.; Chaturvedi, S.K.; Fikre, A.; Hamwieh, A.; Kumar, S.; et al. Integrating genomics for chickpea improvement: Achievements and opportunities. Theor. Appl. Genet. 2020, 13, 1703–1720. [Google Scholar] [CrossRef] [Green Version]
- SAS Institute Inc. SAS/STAT® 15.1 User’s Guide; SAS Institute Inc.: Cary, NC, USA, 2008. [Google Scholar]
- Yan, W.; Tinker, N.A. Biplot analysis of multi-environment trial data: Principles and applications. Can. J. Plant Sci. 2006, 86, 623–645. [Google Scholar] [CrossRef] [Green Version]
- Varshney, R.K.; Mohan, S.M.; Gaur, P.M.; Chamarthi, S.K.; Singh, V.K.; Srinivasan, S.; Swapna, N.; Sharma, M.; Pande, S.; Singh, S.; et al. Marker-assisted backcrossing to introgress resistance to fusarium wilt race 1 and ascochyta blight in C 214, an elite cultivar of chickpea. Plant Genome 2014, 7, plantgenome2013.10.0035. [Google Scholar] [CrossRef] [Green Version]
- Pratap, A.; Chaturvedi, S.K.; Tomar, R.; Rajan, N.; Malviya, N.; Thudi, M.; Saabale, P.R.; Prajapati, U.; Varshney, R.K.; Singh, N.P. Marker-assisted introgression of resistance to fusarium wilt race 2 in Pusa 256, an elite cultivar of desi chickpea. Mol. Genet. Genom. 2017, 292, 1237–1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bharadwaj, C.; Tripathi, S.; Soren, K.R.; Thudi, M.; Singh, R.K.; Sheoran, S.; Roorkiwal, M.; Patil, B.S.; Chitikineni, A.; Palakurthi, R.; et al. Introgression of “QTL-hotspot” region enhances drought tolerance and grain yield in three elite chickpea cultivars. Plant Genome 2021, 20076. [Google Scholar] [CrossRef] [PubMed]
- Mannur, D.M.; Babbar, A.; Thudi, M.; Sabbavarapu, M.M.; Roorkiwal, M.; Sharanabasappa, B.Y.; Bansal, V.P.; Jayalakshmi, S.K.; Yadav, S.S.; Rathore, A.; et al. Super Annigeri 1 and improved JG 74: Two fusarium wilt-resistant introgression lines developed using marker-assisted backcrossing approach in chickpea (Cicer arietinum L.). Mol. Breed. 2019, 39, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).