Application of UPLC-QTOF-MS Based Untargeted Metabolomics in Identification of Metabolites Induced in Pathogen-Infected Rice
Abstract
1. Introduction
2. Results and Discussion
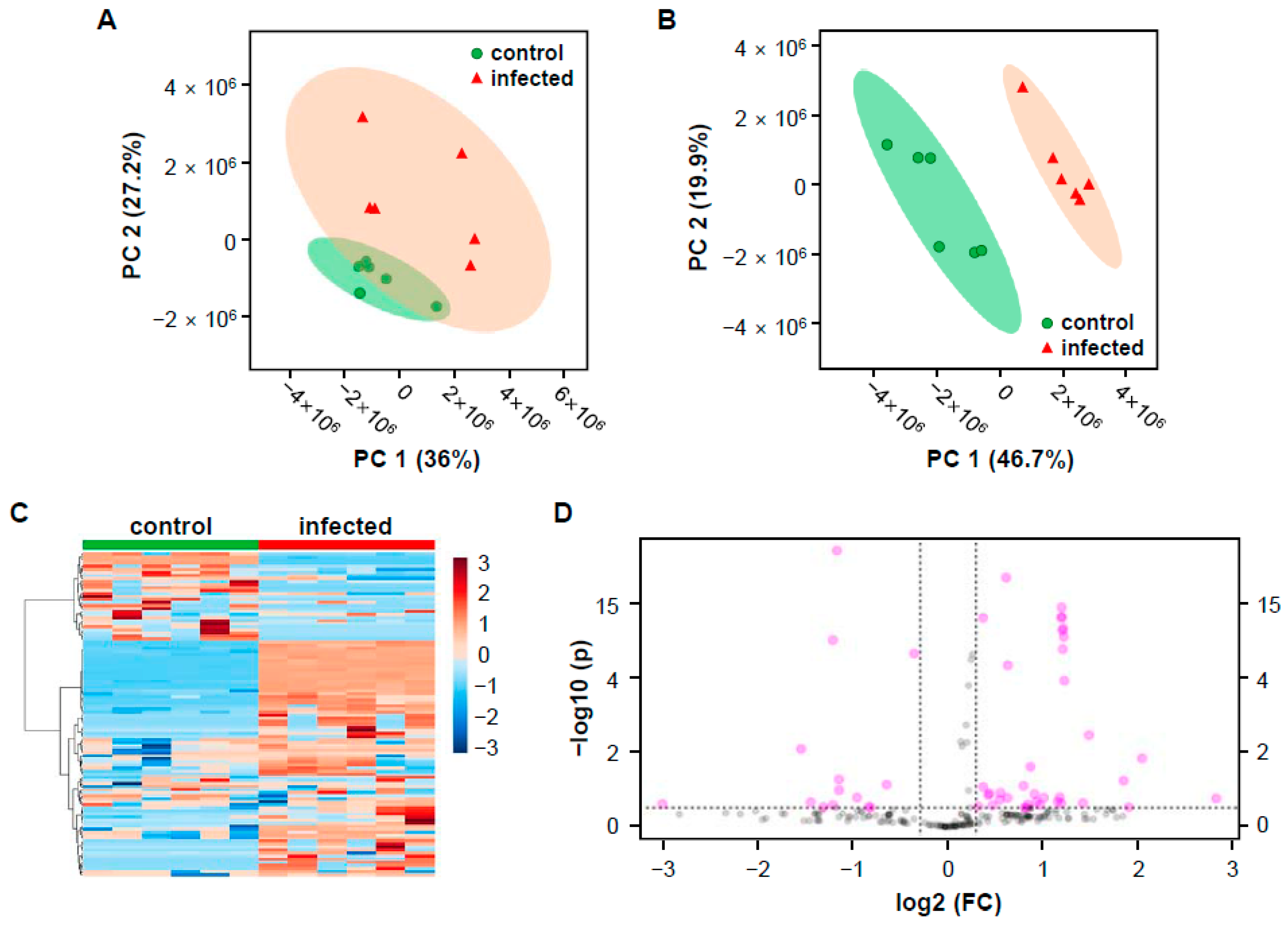
2.1. LC-MS Based Untargeted Metabolomics
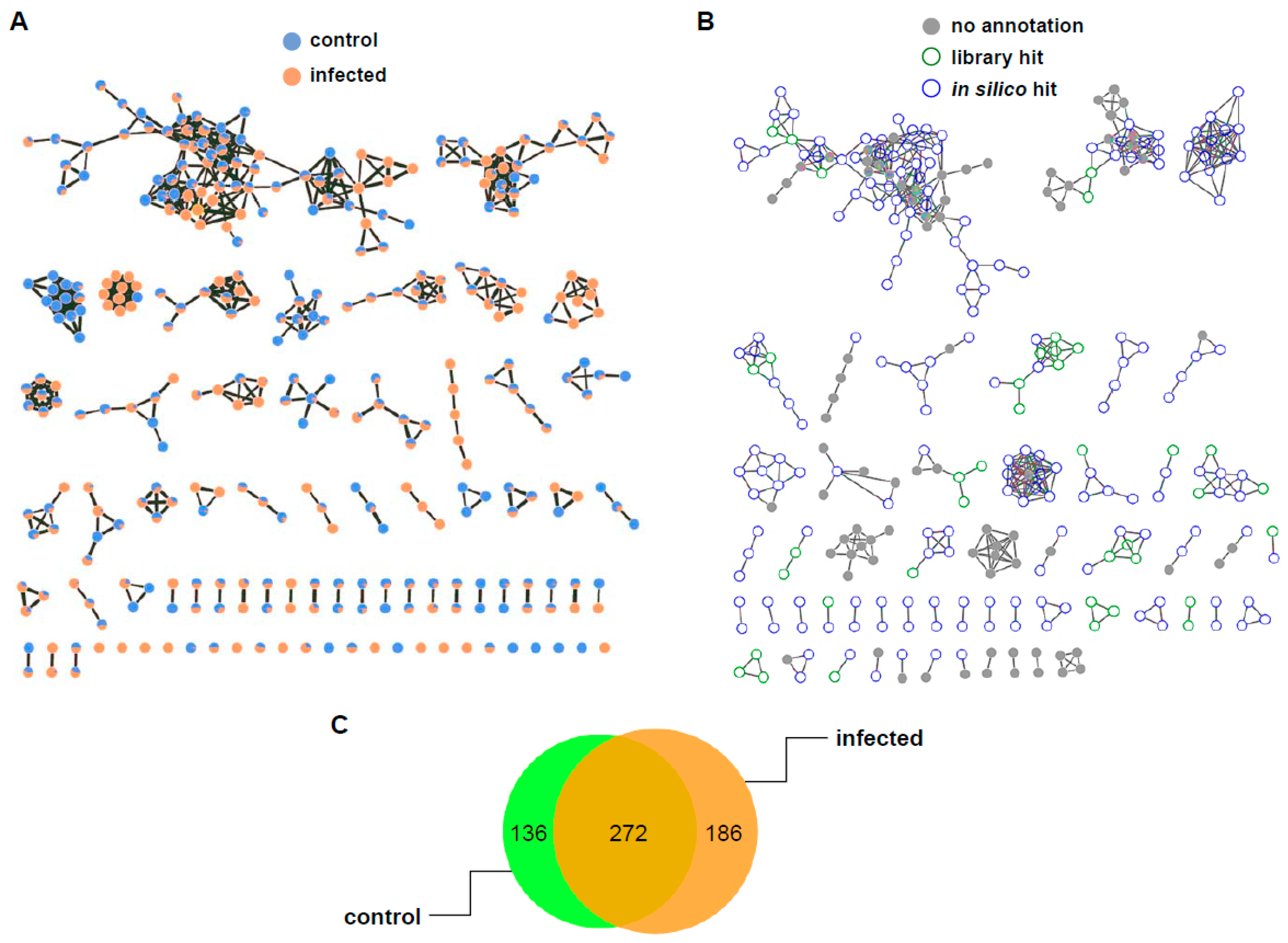
2.2. Mass Differences between Two Rice Groups
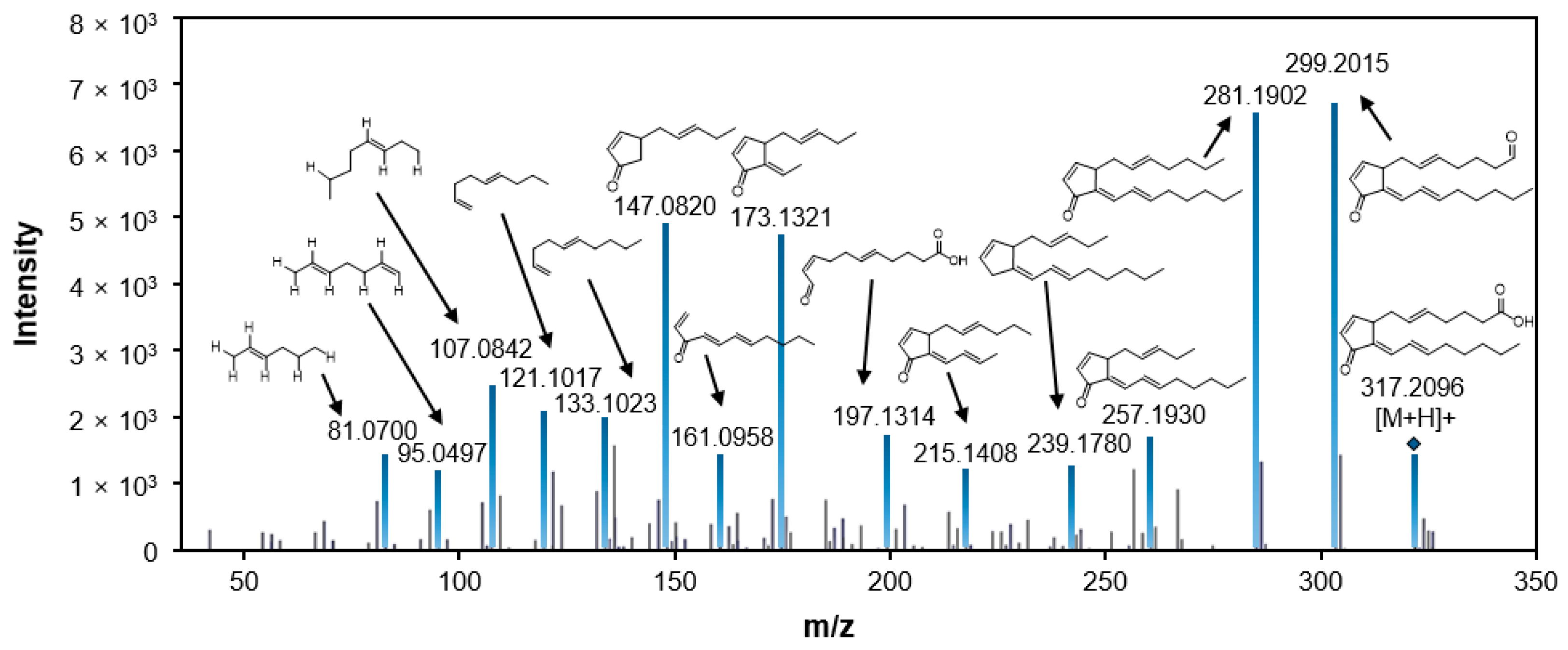
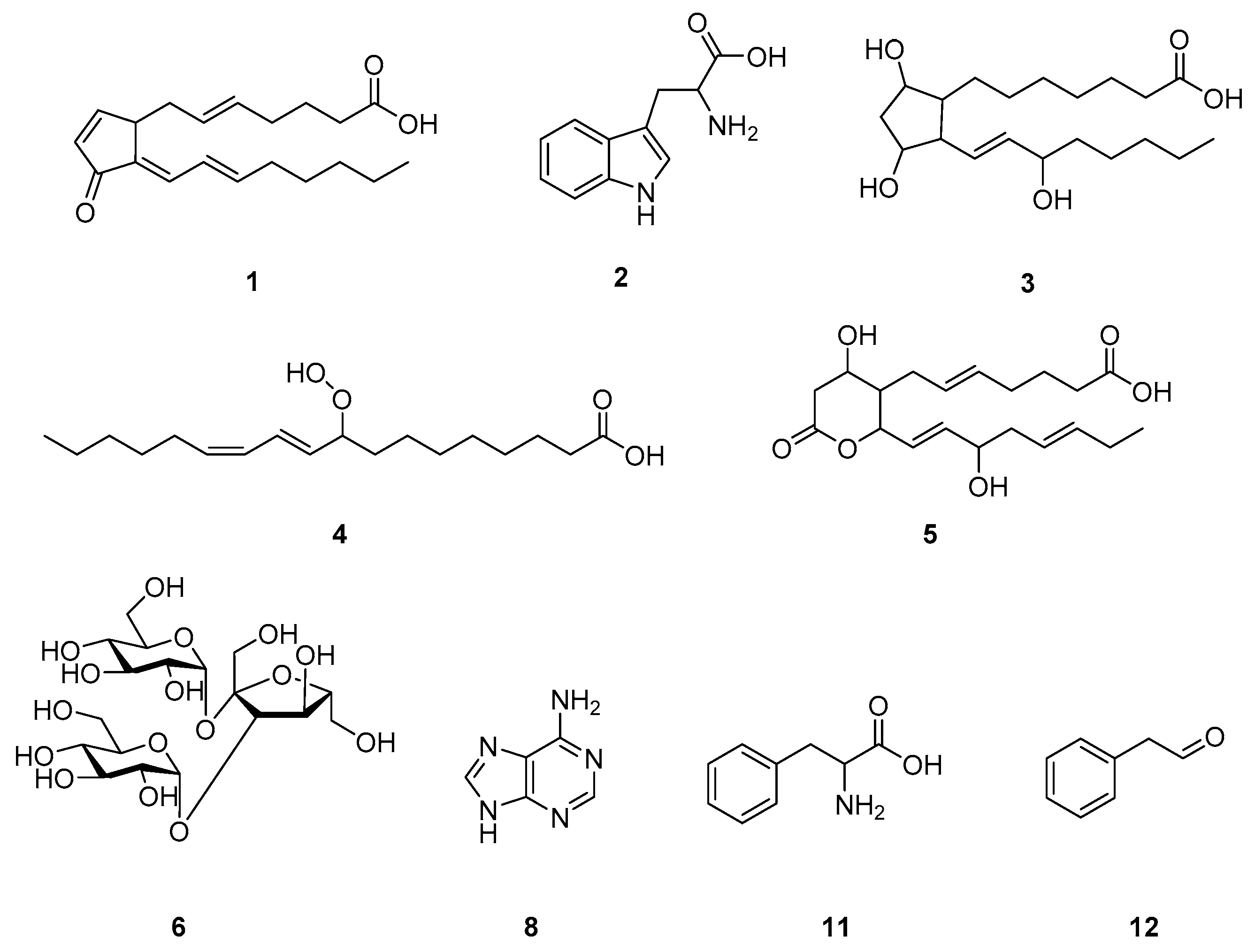
2.3. Putative Identification of Differentially Induced Metabolites
2.4. Metabolic Pathway Analysis
3. Materials and Methods
3.1. Host Plant and Fungal Pathogen
3.2. Sample Preparation
3.3. UPLC-QTOF-MS Analysis
3.4. Feature Finding
3.5. Statistical Data Analysis
3.6. Annotation of Metabolites and Metabolic Pathway Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jeandet, P.; Hébrard, C.; Deville, M.-A.; Cordelier, S.; Dorey, S.; Aziz, A.; Crouzet, J. Deciphering the role of phytoalexins in plant-microorganism interactions and human health. Molecules 2014, 19, 18033–18056. [Google Scholar] [CrossRef]
- López-Gresa, M.P.; Maltese, F.; Bellés, J.M.; Conejero, V.; Kim, H.K.; Choi, Y.H.; Verpoorte, R. Metabolic response of tomato leaves upon different plant-pathogen interactions. Phytochem. Anal. 2010, 21, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Jones, O.A.; Maguire, M.L.; Griffin, J.L.; Jung, Y.-H.; Shibato, J.; Rakwal, R.; Agrawal, G.K.; Jwa, N.-S. Using metabolic profiling to assess plant-pathogen interactions: An example using rice (Oryza sativa) and the blast pathogen Magnaporthe grisea. Eur. J. Plant Pathol. 2011, 129, 539–554. [Google Scholar] [CrossRef]
- Schauer, N.; Fernie, A.R. Plant metabolomics: Towards biological function and mechanism. Trends Plant Sci. 2006, 11, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Mendes, P.; Dixon, R.A. Plant metabolomics: Large-scale phytochemistry in the functional genomics era. Phytochemistry 2003, 62, 817–836. [Google Scholar] [CrossRef]
- Weckwerth, W. Metabolomics in systems biology. Annu. Rev. Plant Biol. 2003, 54, 669–689. [Google Scholar] [CrossRef]
- Navarro-Reig, M.; Jaumot, J.; Piña, B.; Moyano, E.; Galceran, M.T.; Tauler, R. Metabolomic analysis of the effects of cadmium and copper treatment in Oryza sativa L. using untargeted liquid chromatography coupled to high resolution mass spectrometry and all-ion fragmentation. Metallomics 2017, 9, 660–675. [Google Scholar] [CrossRef]
- Andersen, M.-B.S.; Kristensen, M.; Manach, C.; Pujos-Guillot, E.; Poulsen, S.K.; Larsen, T.M.; Astrup, A.; Dragsted, L. Discovery and validation of urinary exposure markers for different plant foods by untargeted metabolomics. Anal. Bioanal. Chem. 2014, 406, 1829–1844. [Google Scholar] [CrossRef]
- Commisso, M.; Strazzer, P.; Toffali, K.; Stocchero, M.; Guzzo, F. Untargeted metabolomics: An emerging approach to determine the composition of herbal products. Comput. Struct. Biotechnol. J. 2013, 4, e201301007. [Google Scholar] [CrossRef]
- Sawada, Y.; Nakabayashi, R.; Yamada, Y.; Suzuki, M.; Sato, M.; Sakata, A.; Akiyama, K.; Sakurai, T.; Matsuda, F.; Aoki, T. RIKEN tandem mass spectral database (ReSpect) for phytochemicals: A plant-specific MS/MS-based data resource and database. Phytochemistry 2012, 82, 38–45. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Rahman, M.A.; Rathinasabapathi, B.; Babar, M.A. UPLC-HRMS-based untargeted metabolic profiling reveals changes in chickpea (Cicer arietinum) metabolome following long-term drought stress. Plant Cell Environ. 2019, 42, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.A.; Talbot, N.J.; Ebbole, D.J.; Farman, M.L.; Mitchell, T.K.; Orbach, M.J.; Thon, M.; Kulkarni, R.; Xu, J.-R.; Pan, H. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 2005, 434, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.A.; Zhou, X.; Chen, J.; Yang, Y. Role of ethylene, abscisic acid and MAP kinase pathways in rice blast resistance. In Advances in Genetics, Genomics and Control of Rice Blast Disease; Wang, G.L., Valent, B., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 185–190. [Google Scholar]
- Ohta, H.; Shida, K.; Peng, Y.-L.; Furusawa, I.; Shishiyama, J.; Aibara, S.; Morita, Y. A lipoxygenase pathway is activated in rice after infection with the rice blast fungus Magnaporthe grisea. Plant Physiol. 1991, 97, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, H.; Li, D.; Huang, L.; Hong, Y.; Ding, X.S.; Nelson, R.S.; Zhou, X.; Song, F. Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol. Biol. 2013, 81, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.-R.; Hamer, J.E. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 1996, 10, 2696–2706. [Google Scholar] [CrossRef]
- Azizi, P.; Osman, M.; Hanafi, M.M.; Sahebi, M.; Yusop, M.R.; Taheri, S. Adaptation of the metabolomics profile of rice after Pyricularia oryzae infection. Plant Physiol. Biochem. 2019, 144, 466–479. [Google Scholar] [CrossRef]
- Okazaki, Y.; Saito, K. Integrated metabolomics and phytochemical genomics approaches for studies on rice. GigaScience 2016, 5. [Google Scholar] [CrossRef]
- Allwood, J.W.; Ellis, D.I.; Goodacre, R. Metabolomic technologies and their application to the study of plants and plant-host interactions. Physiol. Plantarum 2008, 132, 117–135. [Google Scholar] [CrossRef]
- Berger, S.; Sinha, A.K.; Roitsch, T. Plant physiology meets phytopathology: Plant primary metabolism and plant-pathogen interactions. J. Exp. Bot. 2007, 58, 4019–4026. [Google Scholar] [CrossRef]
- Parker, D.; Beckmann, M.; Zubair, H.; Enot, D.P.; Caracuel-Rios, Z.; Overy, D.P.; Snowdon, S.; Talbot, N.J.; Draper, J. Metabolomic analysis reveals a common pattern of metabolic re-programming during invasion of three host plant species by Magnaporthe grisea. Plant J. 2009, 59, 723–737. [Google Scholar] [CrossRef]
- Nebbioso, A.; De Martino, A.; Eltlbany, N.; Smalla, K.; Piccolo, A. Phytochemical profiling of tomato roots following treatments with different microbial inoculants as revealed by IT-TOF mass spectrometry. Chem. Biol. Technol. Agric. 2016, 3, 12. [Google Scholar] [CrossRef]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A tutorial review: Metabolomics and partial least squares-discriminant analysis-a marriage of convenience or a shotgun wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef]
- Austin, P.C.; Hux, J.E. A brief note on overlapping confidence intervals. J. Vasc. Surg. 2002, 36, 194–195. [Google Scholar] [CrossRef] [PubMed]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.-F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 1–38. [Google Scholar]
- da Silva, R.R.; Wang, M.; Nothias, L.-F.; van der Hooft, J.J.; Caraballo-Rodríguez, A.M.; Fox, E.; Balunas, M.J.; Klassen, J.L.; Lopes, N.P.; Dorrestein, P.C. Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol. 2018, 14, e1006089. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; da Silva, R.R.; Jang, H.S.; Kim, H.W.; Kwon, Y.S.; Kim, J.-H.; Yang, H. In silico annotation of discriminative markers of three Zanthoxylum species using molecular network derived annotation propagation. Food Chem. 2019, 295, 368–376. [Google Scholar] [CrossRef]
- Wakasa, K.; Ishihara, A. Metabolic engineering of the tryptophan and phenylalanine biosynthetic pathways in rice. Plant Biotechnol. 2009, 26, 523–533. [Google Scholar] [CrossRef]
- Ishihara, A.; Nakao, T.; Mashimo, Y.; Murai, M.; Ichimaru, N.; Tanaka, C.; Nakajima, H.; Wakasa, K.; Miyagawa, H. Probing the role of tryptophan-derived secondary metabolism in defense responses against Bipolaris oryzae infection in rice leaves by a suicide substrate of tryptophan decarboxylase. Phytochemistry 2011, 72, 7–13. [Google Scholar] [CrossRef]
- Hull, A.K.; Vij, R.; Celenza, J.L. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 2379–2384. [Google Scholar] [CrossRef]
- Ueno, M.; Kumura, Y.; Ueda, K.; Kihara, J.; Arase, S. Indole derivatives enhance resistance against the rice blast fungus Magnaporthe oryzae. J. Gen. Plant Pathol. 2011, 77, 209–213. [Google Scholar] [CrossRef]
- Nicholson, R.L.; Hammerschmidt, R. Phenolic compounds and their role in disease resistance. Annu. Rev. Phytopathol. 1992, 30, 369–389. [Google Scholar] [CrossRef]
- Großkinsky, D.; Edelsbrunner, K.; Pfeifhofer, H.; Van der Graaff, E.; Roitsch, T. Cis-and trans-zeatin differentially modulate plant immunity. Plant Signal. Behav. 2013, 8, e24798. [Google Scholar] [CrossRef]
- Barna, B.; Smigocki, A.; Baker, J. Transgenic production of cytokinin suppresses bacterially induced hypersensitive response symptoms and increases antioxidative enzyme levels in Nicotiana spp. Phytopathology 2008, 98, 1242–1247. [Google Scholar] [CrossRef]
- Prost, I.; Dhondt, S.; Rothe, G.; Vicente, J.; Rodriguez, M.J.; Kift, N.; Carbonne, F.; Griffiths, G.; Esquerré-Tugayé, M.-T.; Rosahl, S. Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol. 2005, 139, 1902–1913. [Google Scholar] [CrossRef]
- De León, I.P.; Sanz, A.; Hamberg, M.; Castresana, C. Involvement of the Arabidopsisα-DOX1 fatty acid dioxygenase in protection against oxidative stress and cell death. Plant J. 2002, 29, 61–72. [Google Scholar] [CrossRef]
- Weber, H.; Chételat, A.; Caldelari, D.; Farmer, E.E. Divinyl ether fatty acid synthesis in late blight-diseased potato leaves. Plant Cell 1999, 11, 485–493. [Google Scholar]
- Rustérucci, C.; Montillet, J.-L.; Agnel, J.-P.; Battesti, C.; Alonso, B.; Knoll, A.; Bessoule, J.-J.; Etienne, P.; Suty, L.; Blein, J.-P. Involvement of lipoxygenase-dependent production of fatty acid hydroperoxides in the development of the hypersensitive cell death induced by cryptogein on tobacco leaves. J. Biol. Chem. 1999, 274, 36446–36455. [Google Scholar] [CrossRef]
- Porta, H.; Rocha-Sosa, M. Plant lipoxygenases. Physiological and molecular features. Plant Physiol. 2002, 130, 15–21. [Google Scholar] [CrossRef]
- Smith, W.L.; Urade, Y.; Jakobsson, P.-J. Enzymes of the cyclooxygenase pathways of prostanoid biosynthesis. Chem. Rev. 2011, 111, 5821–5865. [Google Scholar] [CrossRef]
- Kim, J.C.; Choi, G.J.; Park, J.H.; Kim, H.T.; Cho, K.Y. Activity against plant pathogenic fungi of phomalactone isolated from Nigrospora sphaerica. Pest Manag. Sci. 2001, 57, 554–559. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]


| No | m/z | RT a (min) | log2(FC) | −log10(p) | Representative MS/MS Fragments | Compound ID | Class | Adduct Ions |
|---|---|---|---|---|---|---|---|---|
| 1 | 317.2096 | 13.64 | 20.503 | 15.026 | 107.0842 121.1017 147.0820 173.1321 299.2015 | 15-deoxy-Δ12,14-prostaglandin J2 | prostaglandins | [M + Na]+ |
| 2 | 188.0696 | 3.39 | 19.939 | 14.257 | 118.0642 143.0719 | tryptophan | amino acids | [M + H−NH3]+ |
| 3 | 357.2580 | 15.43 | 19.457 | 14.129 | 102.0867 176.9264 216.9466 256.9701 340.3596 | prostaglandin F1α | prostaglandins | [M + H]+ |
| 4 | 313.2325 | 15.51 | 19.376 | 13.676 | 154.9296 196.9310 214.9464 256.9654 | 9-hydroperoxy-10Z,12E-octadecadienoic acid | octadecanoids | [M + H]+ |
| 5 | 330.2976 | 13.46 | 18.772 | 1.784 | 107.0844 121.1000 130.0855 264.2675 282.2745 | 11-dehydro-thromboxane B3 | thromboxanes | [M + H−2H2O]+ |
| 6 | 527.1528 | 1.40 | 17.835 | 1.955 | 185.0411 203.0513 305.0828 365.1016 | melezitose | trisaccharides | [M + Na]+ |
| 7 | 383.2025 | 16.89 | 16.834 | 1.673 | 104.9929 141.0741 156.0904 207.0625 267.1209 | Unknown 1 | oxadiazoles | - |
| 8 | 136.0610 | 1.40 | 2.366 | 1.408 | 119.0338 109.0500 | adenine | nucleobases | [M + H]+ |
| 9 | 303.1402 | 4.73 | 2.210 | 3.713 | 117.0324 127.0968 145.0266 177.0533 | Unknown 2 | - | - |
| 10 | 699.3502 | 12.67 | 1.660 | 1.124 | 185.0405 294.9074 347.0933 537.2977 699.3497 | Unknown 3 | - | - |
| 11 | 166.0854 | 2.37 | 1.252 | 10.862 | 103.0533 120.0795 | phenylalanine | amino acids | [M + H]+ |
| 12 | 120.0802 | 2.37 | 1.219 | 16.785 | 103.0536 | phenylacetaldehyde | phenylpropanoids | [M + H]+ |
| KEGG Pathway | Total a | Hits b | p-Value |
|---|---|---|---|
| Phenylalanine metabolism | 12 | 2 | 1.83 × 10−3 |
| Phenylalanine, tyrosine, and tryptophan biosynthesis | 22 | 2 | 6.21 × 10−3 |
| Aminoacyl-tRNA biosynthesis | 46 | 2 | 2.60 × 10−2 |
| Linoleic acid metabolism | 4 | 1 | 2.26 × 10−2 |
| Tropane, piperidine, and pyridine alkaloid biosynthesis | 8 | 1 | 4.48 × 10−2 |
| Zeatin biosynthesis | 21 | 1 | 1.14 × 10−1 |
| Tryptophan metabolism | 23 | 1 | 1.24 × 10−1 |
| Glycine, serine, and threonine metabolism | 33 | 1 | 1.74 × 10−1 |
| Phenylpropanoid biosynthesis | 35 | 1 | 1.83 × 10−1 |
| Purine metabolism | 63 | 1 | 3.08 × 10−1 |
| Pathway Module | KEGG Pathway |
|---|---|
| Amino acid metabolism | Phenylalanine metabolism |
| Phenylalanine, tyrosine, and tryptophan biosynthesis | |
| Tryptophan metabolism | |
| Glycine, serine, and threonine metabolism | |
| Biosynthesis of other secondary metabolites | Tropane, piperidine, and pyridine alkaloid biosynthesis |
| Phenylpropanoid biosynthesis | |
| Lipid metabolism | Linoleic acid metabolism |
| Biosynthesis of terpenoids and polyketides | Zeatin biosynthesis |
| Nucleotide metabolism | Purine metabolism |
| Genetic transcription | Aminoacyl-tRNA biosynthesis |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, M.; Park, S.; Kim, H.; Choi, G.J.; Kim, S.H. Application of UPLC-QTOF-MS Based Untargeted Metabolomics in Identification of Metabolites Induced in Pathogen-Infected Rice. Plants 2021, 10, 213. https://doi.org/10.3390/plants10020213
Oh M, Park S, Kim H, Choi GJ, Kim SH. Application of UPLC-QTOF-MS Based Untargeted Metabolomics in Identification of Metabolites Induced in Pathogen-Infected Rice. Plants. 2021; 10(2):213. https://doi.org/10.3390/plants10020213
Chicago/Turabian StyleOh, Mira, SeonJu Park, Hun Kim, Gyung Ja Choi, and Seung Hyun Kim. 2021. "Application of UPLC-QTOF-MS Based Untargeted Metabolomics in Identification of Metabolites Induced in Pathogen-Infected Rice" Plants 10, no. 2: 213. https://doi.org/10.3390/plants10020213
APA StyleOh, M., Park, S., Kim, H., Choi, G. J., & Kim, S. H. (2021). Application of UPLC-QTOF-MS Based Untargeted Metabolomics in Identification of Metabolites Induced in Pathogen-Infected Rice. Plants, 10(2), 213. https://doi.org/10.3390/plants10020213





