The CBL-Interacting Protein Kinase NtCIPK23 Positively Regulates Seed Germination and Early Seedling Development in Tobacco (Nicotiana tabacum L.)
Abstract
:1. Introduction
2. Results
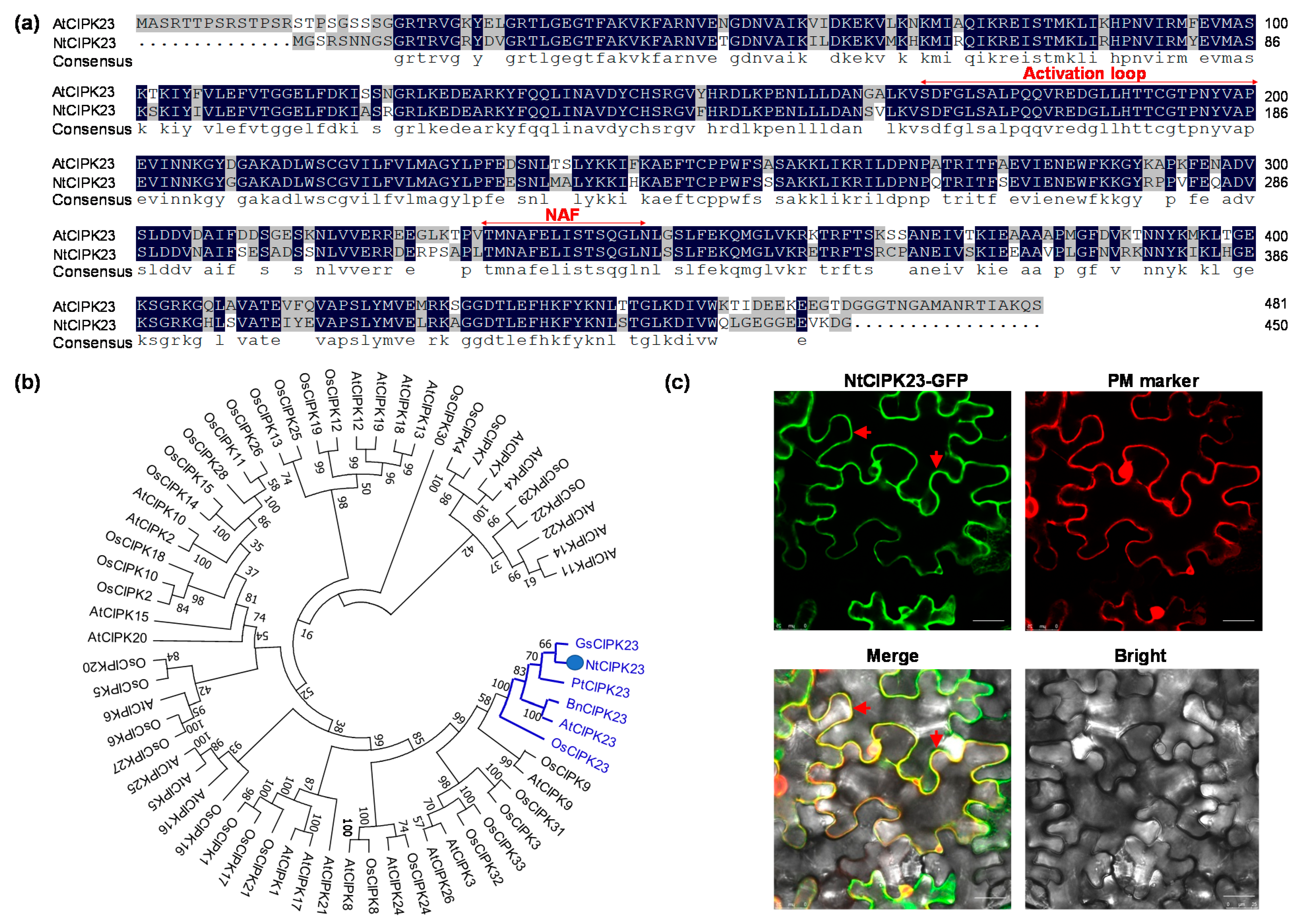
2.1. Sequence Analysis and the Subcellular Localization of NtCIPK23
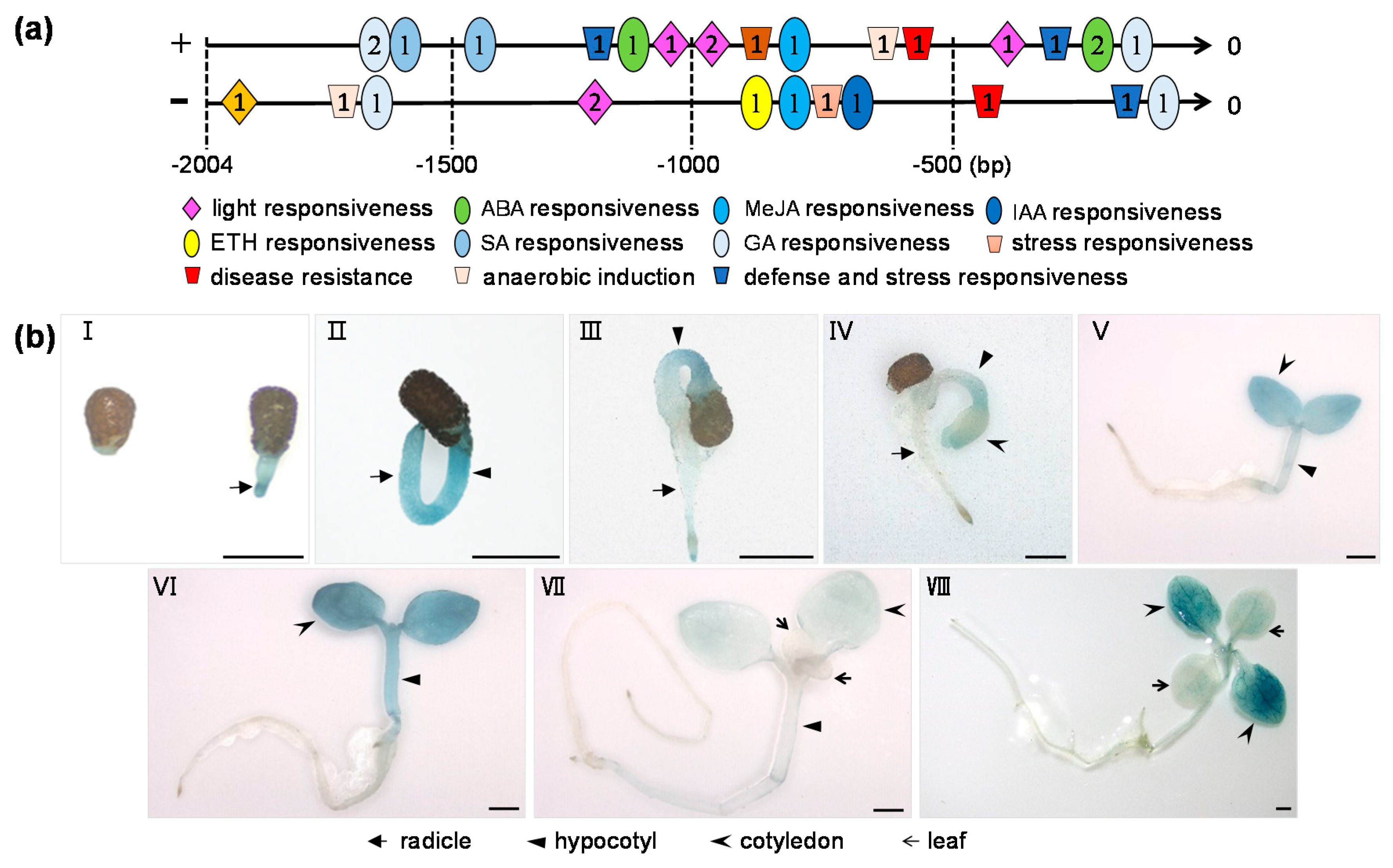
2.2. Expression Pattern of NtCIPK23 during Seed Germination and Early Seedling Growth
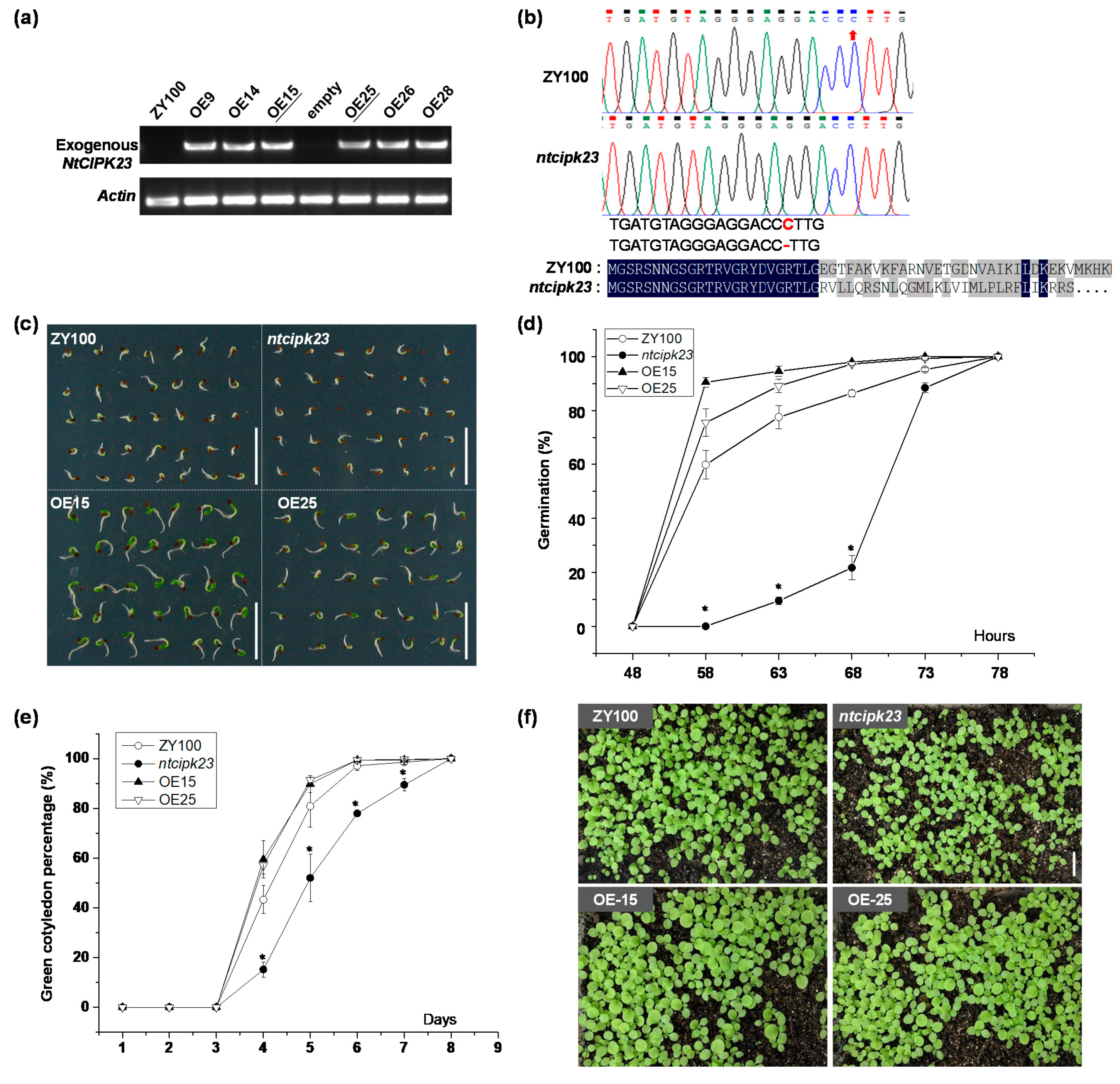
2.3. NtCIPK23 Plays a Positive Role in Seed Germination and Post-Germination Seedling Growth under Normal Conditions
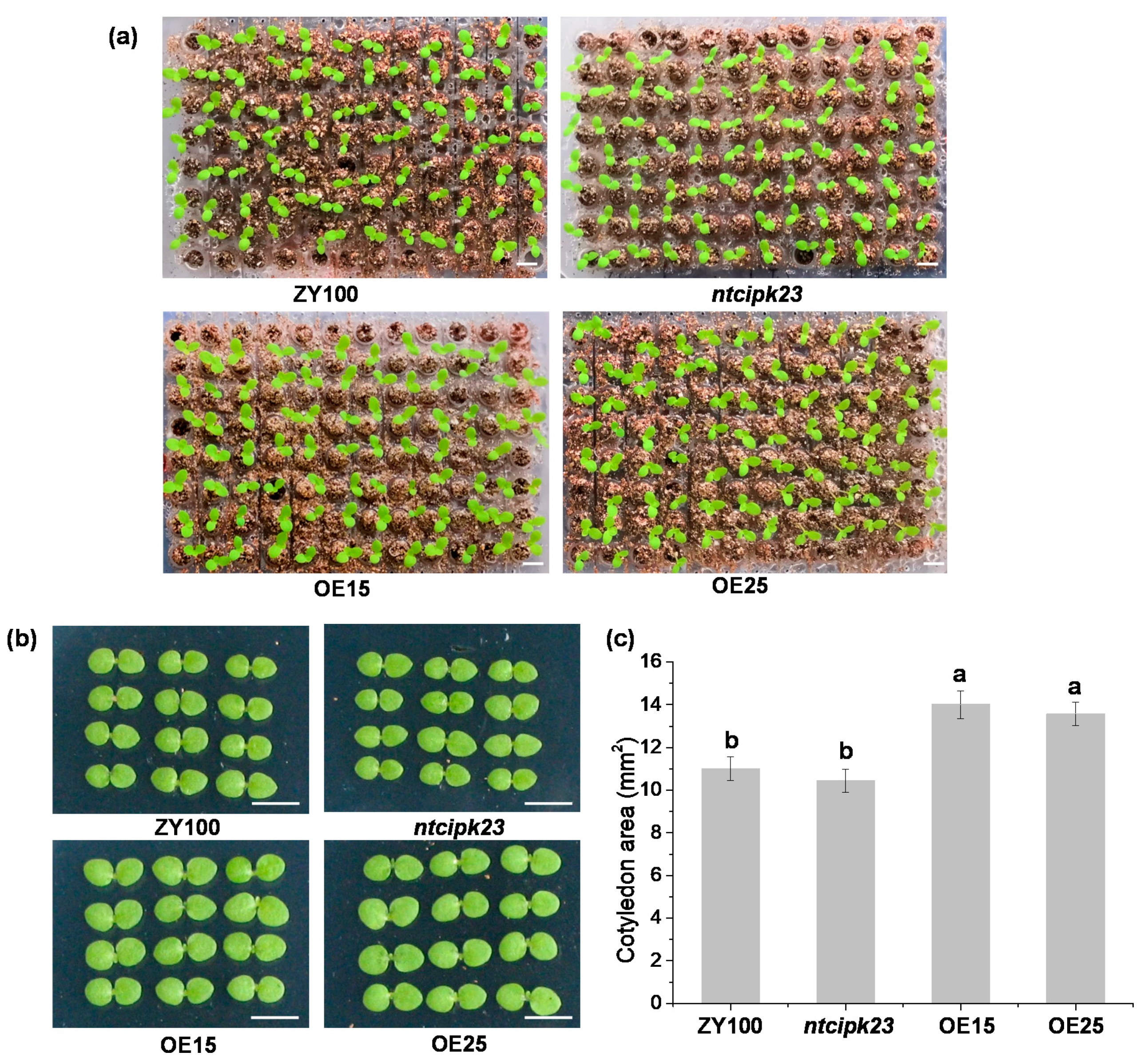
2.4. Overexpression of NtCIPK23 Promotes the Cotyledon Expansion of Young Tobacco Seedlings
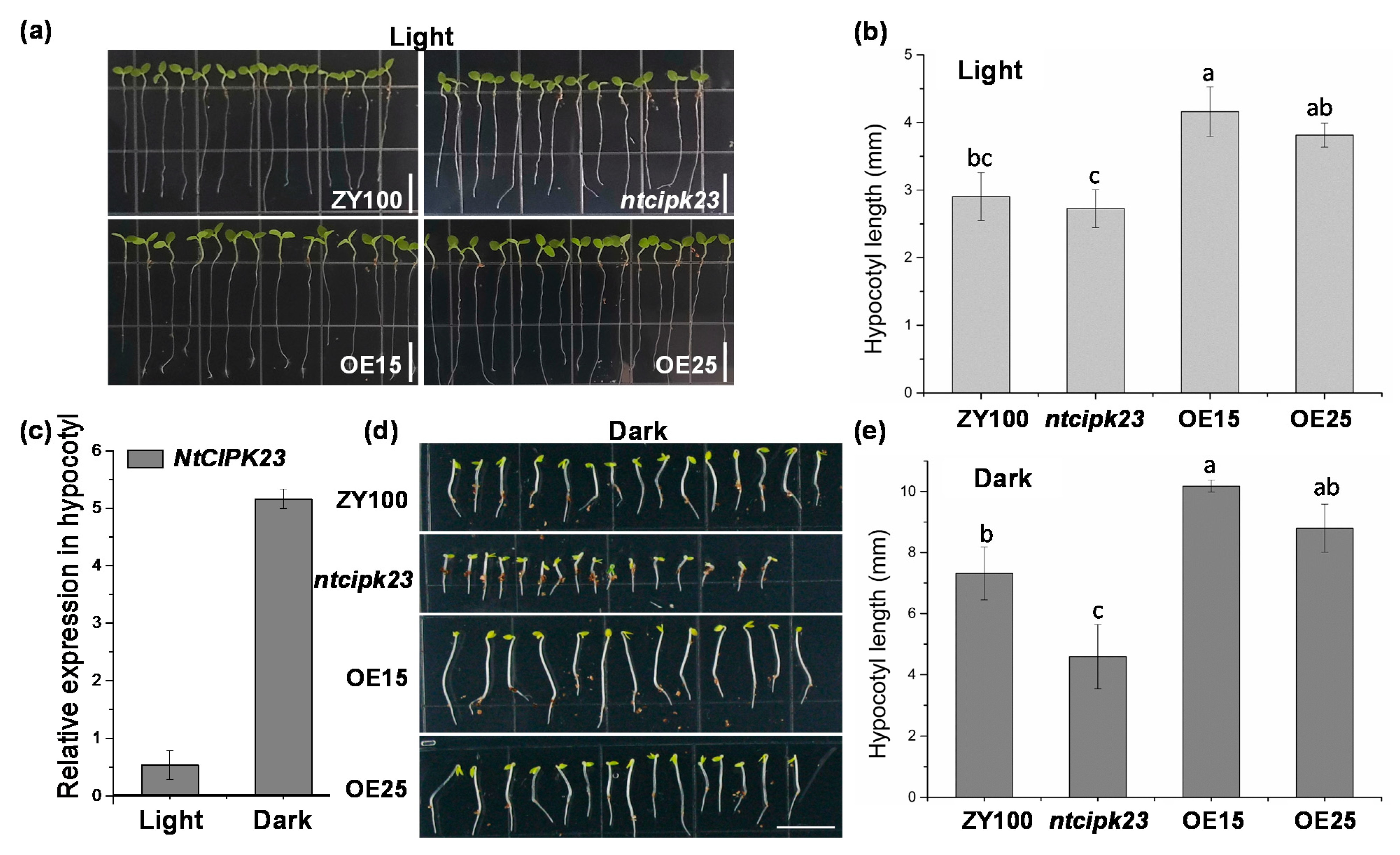
2.5. NtCIPK23 Positively Regulates the Hypocotyl Elongation of Young Tobacco Seedlings
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Gene Cloning and Plasmid Construction
4.3. RNA Extraction, RT-PCR, and Real-Time Quantitative PCR (RT-qPCR) Analyses
4.4. Generation of Transgenic Materials
4.5. GUS Histochemical Assay
4.6. Subcellular Localization Assay
4.7. Measurement and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reddy, A.S.N. Calcium: Silver bullet in signaling. Plant Sci. 2001, 160, 381–404. [Google Scholar] [CrossRef]
- Batistic, O.; Kudla, J. Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 2004, 219, 915–924. [Google Scholar] [CrossRef]
- Weinl, S.; Kudla, J. The CBL-CIPK Ca2+-decoding signaling network: Function and perspectives. New Phytol. 2009, 184, 517–528. [Google Scholar] [CrossRef]
- Luan, S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009, 14, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Manik, S.M.N.; Shi, S.; Chao, J.; Jin, Y.; Wang, Q.; Liu, H. Mechanisms and physiological roles of the CBL-CIPK networking system in Arabidopsis thaliana. Genes 2016, 7, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coello, P.; Hey, S.J.; Halford, N.G. The sucrose non-fermenting-1-related (SnRK) family of protein kinases: Potential for manipulation to improve stress tolerance and increase yield. J. Exp. Bot. 2011, 62, 883–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Barrena, M.J.; Martínez-Ripoll, M.; Albert, A. Structural biology of a major signaling network that regulates plant abiotic stress: The CBL-CIPK mediated pathway. Int. J. Mol. Sci. 2013, 14, 5734–5749. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Y.; Huang, Y.; Xiong, L. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 2007, 144, 1416–1428. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Gu, Z.; Xin, D.; Hao, L.; Liu, C.; Huang, J.; Ma, B.; Zhang, H. Identification and characterization of putative CIPK genes in maize. J. Genet. Genomics 2011, 38, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Wang, Y.; Wang, M.; Li, T.; Zhou, Y.; Wang, X.; Wei, S.; He, G.; Yang, G. Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 269. [Google Scholar] [CrossRef] [Green Version]
- Zhu, K.; Chen, F.; Liu, J.; Chen, X.; Hewezi, T.; Cheng, Z.M. Evolution of an intron-poor cluster of the CIPK gene family and expression in response to drought stress in soybean. Sci. Rep. 2016, 6, 28225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aleman, F.; Nieves-Cordones, M.; Martinez, V.; Rubio, F. Root K+ acquisition in plants: The Arabidopsis thaliana model. Plant Cell Physiol. 2011, 52, 1603–1612. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Kim, B.-G.; Cheong, Y.H.; Pandey, G.K.; Luan, S. A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 12625–12630. [Google Scholar] [CrossRef] [Green Version]
- Ragel, P.; Ródenas, R.; García-Martín, E.; Andrés, Z.; Villalta, I.; Nieves-Cordones, M.; Rivero, R.M.; Martínez, V.; Pardo, J.M.; Quintero, F.J. The CBL-interacting protein kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiol. 2015, 169, 2863–2873. [Google Scholar]
- Xu, J.; Li, H.; Chen, L.; Wang, Y.; Liu, L.; He, L.; Wu, W. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 2006, 125, 1347–1360. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Long, Y.; Qi, G.; Li, J.; Xu, Z.; Wu, W.; Wang, Y. The Os-AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell 2014, 26, 3387–4402. [Google Scholar] [CrossRef] [Green Version]
- Cuellar, T.; Pascaud, F.; Verdeil, J.L.; Torregrosa, L.; Adam-Blondon, A.F.; Thibaud, J.B.; Sentenac, H.; Gaillard, I. A grapevine shaker inward K+ channel activated by the calcineurin B-like calcium sensor 1-protein kinase CIPK23 network is expressed in grape berries under drought stress conditions. Plant J. 2010, 61, 58–69. [Google Scholar] [CrossRef]
- Ho, C.H.; Lin, S.H.; Hu, H.C.; Tsay, Y.F. CHL1 functions as a nitrate sensor in plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leran, S.; Edel, K.H.; Pervent, M.; Hashimoto, K.; Corratge-Faillie, C.; Offenborn, J.N.; Tillard, P.; Gojon, A.; Kudla, J.; Lacombe, B. Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Science Signalling 2015, 8, ra43. [Google Scholar] [CrossRef]
- Straub, T.; Ludewig, U.; Neuhäuser, B. The kinase CIPK23 inhibits ammonium transport in Arabidopsis thaliana. Plant Cell 2017, 29, 409–422. [Google Scholar] [CrossRef] [Green Version]
- Shi, S.; Xu, F.; Ge, Y.; Mao, J.; An, L.; Deng, S.; Ullah, Z.; Yuan, X.; Liu, G.; Liu, H.; et al. NH4+ toxicity, which is mainly determined by the high NH4+/K+ ratio, is alleviated by CIPK23 in Arabidopsis. Plants 2020, 9, 501. [Google Scholar] [CrossRef] [Green Version]
- Tang, R.J.; Zhao, F.G.; Garcia, V.J.; Kleist, T.J.; Yang, L.; Zhang, H.X.; Luan, S. Tonoplast CBL-CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, 3134–3139. [Google Scholar] [CrossRef] [Green Version]
- Hedrich, R.; Kudla, J. Calcium signaling networks channel plant K+ uptake. Cell 2006, 125, 1221–1223. [Google Scholar] [CrossRef] [Green Version]
- Negi, J.; Matsuda, O.; Nagasawa, T.; Oba, Y.; Takahashi, H.; Kawai-Yamada, M.; Uchimiya, H.; Hashimoto, M.; Iba, K. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 2008, 452, 483–486. [Google Scholar] [CrossRef]
- Yan, Y.; He, X.; Hu, W.; Liu, G.; Wang, P.; He, C.; Shi, H. Functional analysis of MeCIPK23 and MeCBL1/9 in cassava defense response against Xanthomonas axonopodis pv. manihotis. Plant Cell Rep. 2018, 37, 887–900. [Google Scholar] [CrossRef]
- Yang, W.; Kong, Z.; Omo-Ikerodah, E.; Xu, W.; Li, Q.; Xue, Y. Calcineurin B-like interacting protein kinase OsCIPK23 functions in pollination and drought stress responses in rice (Oryza sativa L.). J. Genet. Genom. 2008, 35, 531–543. [Google Scholar] [CrossRef]
- Footitt, S.; Olcer-Footitt, H.; Hambidge, A.J.; Finch-Savage, W.E. A laboratory simulation of Arabidopsis seed dormancy cycling provides new insight into its regulation by clock genes and the dormancy-related genes DOG1, MFT, CIPK23 and PHYA. Plant Cell Environ. 2017, 40, 1474–1486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, S.; Kaiserli, E.; Zhao, X.; Waksman, T.; Takemiya, A.; Okumura, M.; Takahashi, H.; Seki, M.; Shinozaki, K.; Endo, Y.; et al. CIPK23 regulates blue light-dependent stomatal opening in Arabidopsis thaliana. Plant J. 2020, 104, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.K.; Cai, X.; Nebenfuhr, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef]
- Batistic, O.; Waadt, R.; Steinhorst, L.; Held, K.; Kudla, J. CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 2010, 61, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Ródenas, R.; Vert, G. Regulation of root nutrient transporters by CIPK23: “one kinase to rule them all”. Plant Cell Physiol. 2020, pcaa 156. [Google Scholar] [CrossRef]
- Wang, P.; Hsu, C.; Du, Y.; Zhu, P.; Zhao, C.; Fu, X.; Zhang, C.; Paez, J.; Macho, A.; Tao, W.; et al. Mapping proteome-wide targets of protein kinases in plant stress responses. Proc. Natl. Acad. Sci. USA 2020, 117, 3270–3280. [Google Scholar] [CrossRef]
- Sadhukhan, A.; Enomoto, T.; Kobayashi, Y.; Watanabe, T.; Iuchi, S.; Kobayashi, M.; Sahoo, L.; Yamamoto, Y.; Koyama, H. Sensitive to proton rhizotoxicity 1 regulates salt and drought tolerance of Arabidopsis thaliana through transcriptional regulation of CIPK23. Plant Cell Physiol. 2019, 60, 2113–2126. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Cui, X.; Su, L.; Fang, S.; Chu, J.; Gong, Q.; Yang, J.; Zhu, Z. Jasmonate inhibits COP1 activity to suppress hypocotyl elongation and promote cotyledon opening in etiolated Arabidopsis seedlings. Plant J. 2017, 90, 1144–1155. [Google Scholar] [CrossRef] [Green Version]
- Folta, K.M.; Spalding, E.P. Unexpected roles for cryptochrome 2 and phototropin revealed by high-resolution analysis of blue light-mediated hypocotyl growth inhibition. Plant J. 2001, 26, 471–478. [Google Scholar] [CrossRef]
- Sánchez-Barrena, M.; Chaves-Sanjuan, A.; Raddatz, N.; Mendoza, I.; Cortés, Á.; Gago, F.; González-Rubio, J.; Benavente, J.; Quintero, F.J.; Pardo, J.M.; et al. Recognition and activation of the plant AKT1 potassium channel by the kinase CIPK23. Plant Physiol. 2020, 182, 2143–2153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, J.E.F.; Kadonaga, J.T. The RNA polymerase II core promoter: A key component in the regulation of gene expression. Genes Dev. 2002, 16, 2583–2592. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Chen, Y.F.; Wu, W.H. Potassium and phosphorus transport and signaling in plants. J. Integr. Plant Biol. 2020. [Google Scholar] [CrossRef]
- Gendreau, E.; Jraas, T.; Desnos, T.; Grandjean, O.; Caboche, M.; Höfte, H. Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 1997, 114, 295–305. [Google Scholar] [CrossRef] [Green Version]
- Zhong, S.; Shi, H.; Xue, C.; Wei, N.; Guo, H.; Deng, X.W. Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc. Natl. Acad. Sci. USA 2014, 111, 3913–3920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forcella, F.; Arnold, R.L.B.; Sanchez, R.; Ghersa, C.M. Modeling seedling emergence. Field Crops Res. 2000, 67, 123–139. [Google Scholar] [CrossRef]
- Gao, J.; Wang, G.; Ma, S.; Xie, X.; Wu, X.; Zhang, X.; Wu, Y.; Zhao, P.; Xia, Q. CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol. Biol. 2015, 87, 99–110. [Google Scholar] [CrossRef]
- Schmidt, G.W.; Delaney, S.K. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genom. 2010, 283, 233–241. [Google Scholar] [CrossRef]
- Wu, M.L.; Cui, Y.C.; Ge, L.; Cui, L.P.; Xu, Z.C.; Zhang, H.Y.; Wang, Z.J.; Zhou, D.; Wu, S.; Chen, L.; et al. NbCycB2 represses Nbwo activity via a negative feedback loop in tobacco trichome development. J. Exp. Bot. 2020, 71, 1815–1827. [Google Scholar] [CrossRef]
- Trolet, A.; Baldrich, P.; Criqui, M.C.; Dubois, M.; Clavel, M.; Meyers, B.C.; Genschik, P. Cell cycle-dependent regulation and function of ARGONAUTE1 in plants. Plant Cell 2019, 31, 1734–1750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horsch, R.; Fry, J.; Hoffmann, N.; Eichholtz, D.; Rogers, S. A simple and general method for transferring genes into plants. Science 1985, 227, 1229. [Google Scholar]
- Dong, L.; Wang, Q.; Manik, S.M.N.; Song, Y.; Shi, S.; Su, Y.; Liu, G.; Liu, H. Nicotiana sylvestris calcineurin B-like protein NsylCBL10 enhances salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 2015, 34, 2053–2063. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, S.; An, L.; Mao, J.; Aluko, O.O.; Ullah, Z.; Xu, F.; Liu, G.; Liu, H.; Wang, Q. The CBL-Interacting Protein Kinase NtCIPK23 Positively Regulates Seed Germination and Early Seedling Development in Tobacco (Nicotiana tabacum L.). Plants 2021, 10, 323. https://doi.org/10.3390/plants10020323
Shi S, An L, Mao J, Aluko OO, Ullah Z, Xu F, Liu G, Liu H, Wang Q. The CBL-Interacting Protein Kinase NtCIPK23 Positively Regulates Seed Germination and Early Seedling Development in Tobacco (Nicotiana tabacum L.). Plants. 2021; 10(2):323. https://doi.org/10.3390/plants10020323
Chicago/Turabian StyleShi, Sujuan, Lulu An, Jingjing Mao, Oluwaseun Olayemi Aluko, Zia Ullah, Fangzheng Xu, Guanshan Liu, Haobao Liu, and Qian Wang. 2021. "The CBL-Interacting Protein Kinase NtCIPK23 Positively Regulates Seed Germination and Early Seedling Development in Tobacco (Nicotiana tabacum L.)" Plants 10, no. 2: 323. https://doi.org/10.3390/plants10020323






