Assessment of the Origin and Diversity of Croatian Common Bean Germplasm Using Phaseolin Type, SSR and SNP Markers and Morphological Traits
Abstract
1. Introduction
2. Results
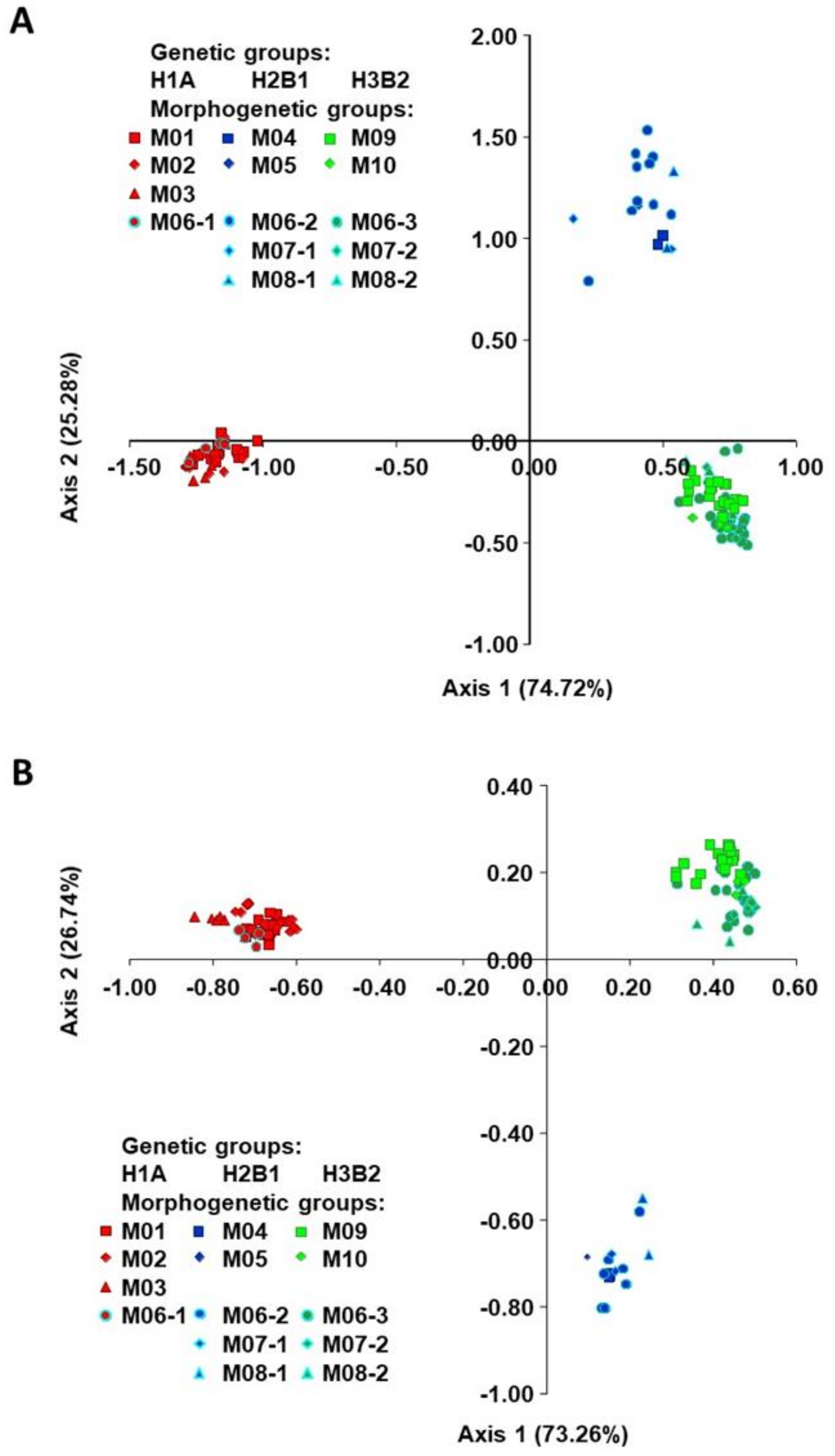
2.1. Origin, Genetic Diversity, and Structure
2.2. Classification of Accessions into Morphogenetic Groups (True-Types)
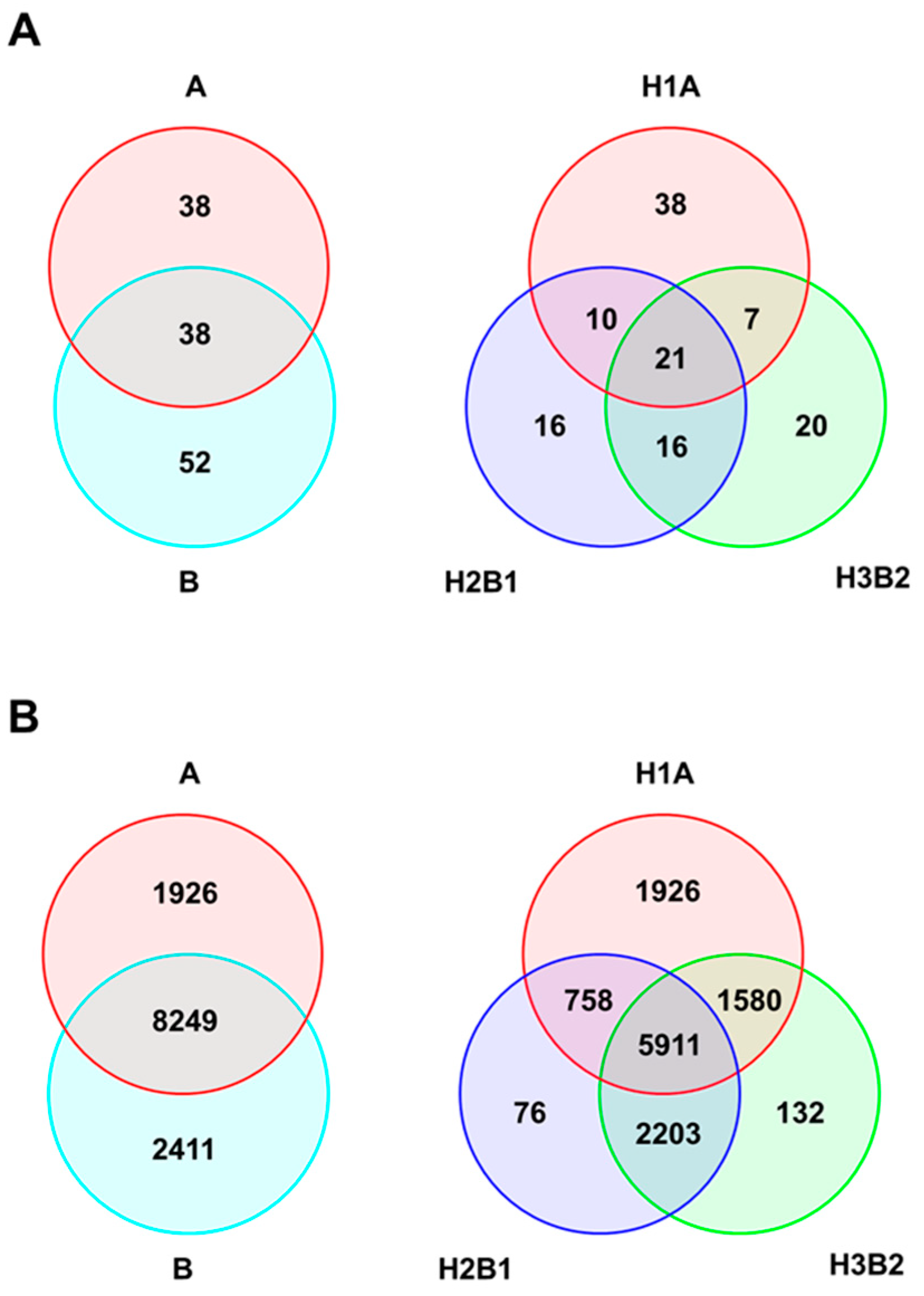
2.3. Genetic Diversity of Genetic Groups (SSR/SNP)
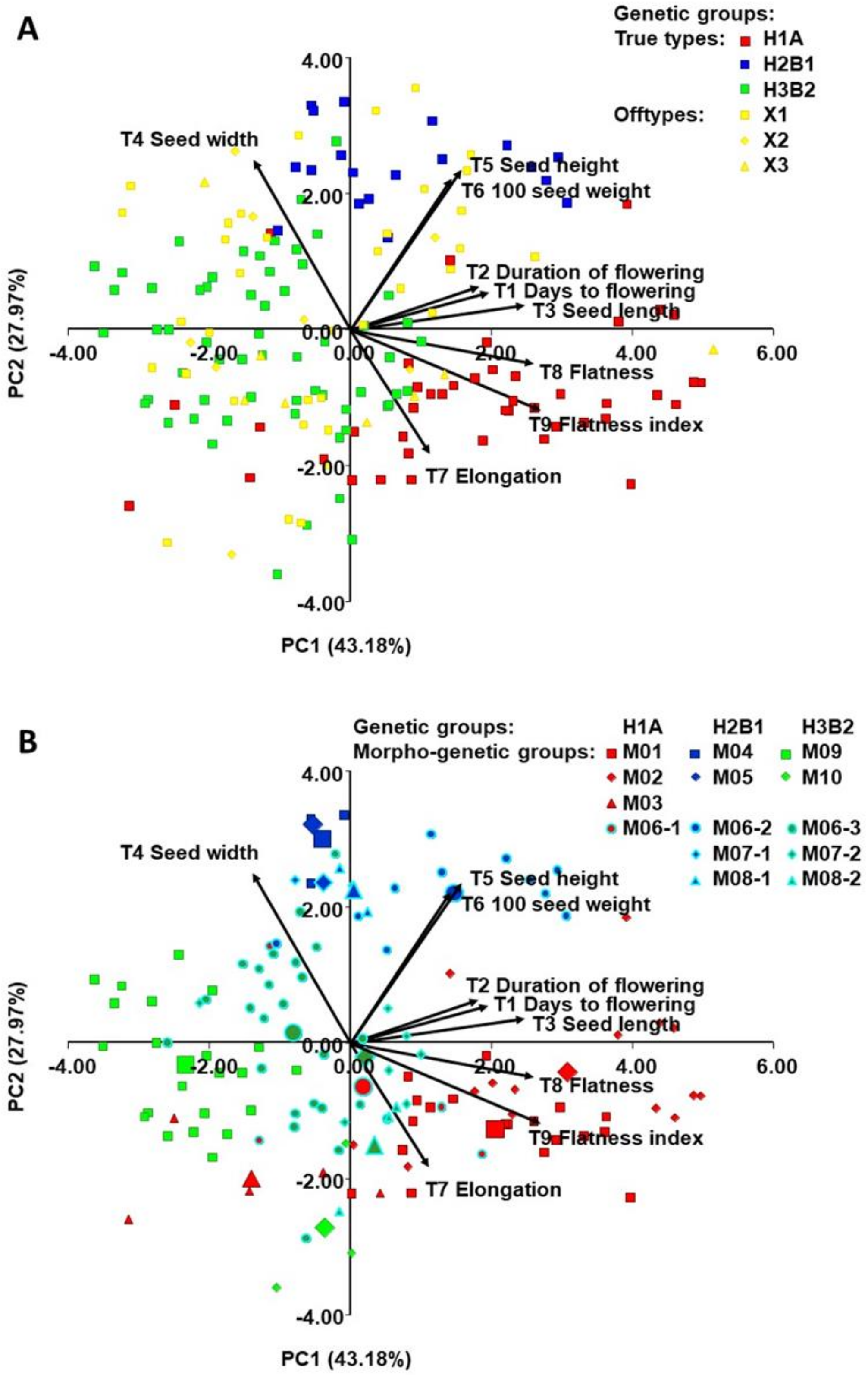
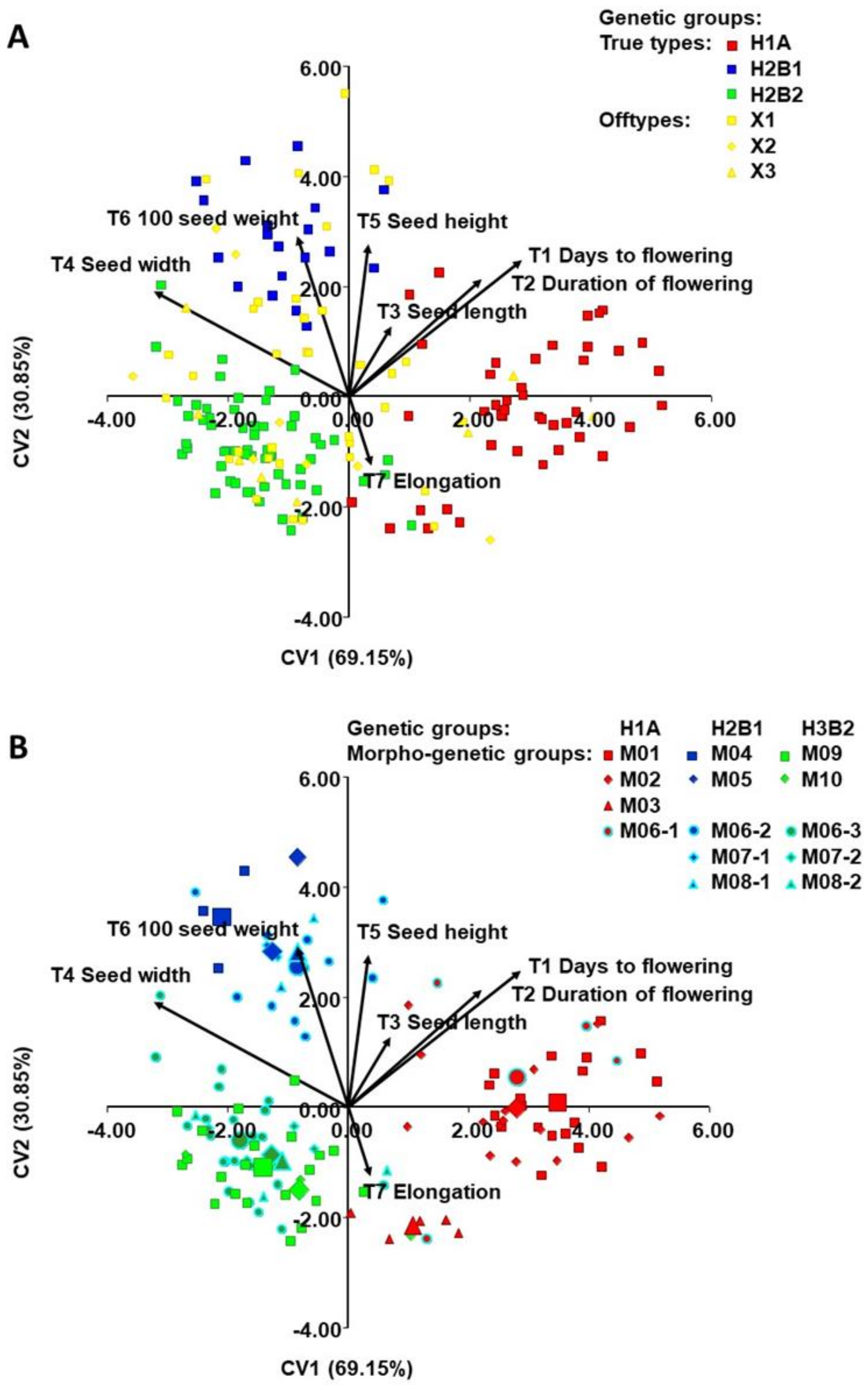
2.4. Morphological Diversity of Genetic Groups (T1-T9)
3. Discussion
3.1. Genetic Diversity and Structure
3.2. Establishment of Morphogenetic Groups
3.3. Morphological Diversity
3.4. Origin and Domestication
4. Materials and Methods
4.1. Plant Material
4.2. Phaseolin Type Methods
4.3. SSR Methods
4.4. SNP Methods
4.5. Morphological Analysis
4.6. Data Analysis
4.6.1. SSR
4.6.2. SNP
4.7. Classification into Morphogenetic Groups
4.8. Genetic Diversity
4.9. Morphological Diversity
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lopes, M.S.; El-Basyoni, I.; Baenziger, P.S.; Singh, S.; Royo, C.; Ozbek, K.; Aktas, H.; Ozer, E.; Ozdemir, F.; Manickavelu, A.; et al. Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot. 2015, 66, 3477–3486. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, S.L.; Ceccarelli, S.; Blair, M.W.; Upadhyaya, H.D.; Are, A.K.; Ortiz, R. Landrace Germplasm for Improving Yield and Abiotic Stress Adaptation. Trends Plant Sci. 2016, 21, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Pathania, A.; Sharma, S.K.; Sharma, P.N. Common bean. In Broadening the Genetic Base of Grain Legumes; Singh, M., Bisht, I.S., Dutta, M., Eds.; Springer: New Delhi, India, 2014; pp. 11–50. [Google Scholar]
- Azeez, M.A.; Adubi, A.O.; Durodola, F.A. Landraces and Crop Genetic Improvement. In Rediscovery of Landraces as a Resource for the Future; Grillo, O., Ed.; IntechOpen: London, UK, 2018; pp. 1–19. [Google Scholar]
- Anunda, H.N.; Nyaboga, E.N.; Amugune, N.O. Genetic diversity of common bean (Phaseolus vulgaris L.) landraces from South Western Kenya for resistance to Pythium root rot disease. Afr. J. Biotechnol. 2019, 18, 316–324. [Google Scholar]
- Bertoldo, J.G.; Coimbra, J.L.M.; Guidolin, A.F.; de Andrade, L.R.B.; Nodari, R.O. Agronomic potential of genebank landrace elite accessions for common bean genetic breeding. Sci. Agric. 2014, 71, 120–125. [Google Scholar] [CrossRef]
- Cortés, A.J.; Blair, M.W. Lessons from Common Bean on How Wild Relatives and Landraces Can Make Tropical Crops More Resistant to Climate Change. In Rediscovery of Landraces as a Resource for the Future; Grillo, O., Ed.; IntechOpen: London, UK, 2018; pp. 113–127. [Google Scholar]
- Petropoulos, S.A.; Barros, L.; Ferreira, I.C.F.R. Editorial: Rediscovering local landraces: Shaping horticulture for the future. Front. Plant Sci. 2019, 10, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Hammer, K.; Teklu, Y. Plant genetic resources: Selected issues from genetic erosion to genetic engineering. J. Agric. Rural Dev. Trop. Subtrop. 2008, 109, 15–50. [Google Scholar]
- Negri, V.; Maxted, N.; Veteläinen, M. European landrace conservation: An introduction. In European Landraces: On-Farm Conservation, Management and Use; Veteläinen, M., Negri, V., Maxted, N., Eds.; Bioversity International: Rome, Italy, 2009; pp. 1–22. ISBN 9789290438052. [Google Scholar]
- Gepts, P. Phaseolus vulgaris (Beans). In Encyclopedia of Genetics; Brenner, S., Miller, J.H., Eds.; Elsevier Science Inc.: New York, NY, USA, 2001; pp. 1444–1445. [Google Scholar]
- Assefa, T.; Assibi Mahama, A.; Brown, A.V.; Cannon, E.K.S.; Rubyogo, J.C.; Rao, I.M.; Blair, M.W.; Cannon, S.B. A review of breeding objectives, genomic resources, and marker-assisted methods in common bean (Phaseolus vulgaris L.). Mol. Breed. 2019, 39, 1–23. [Google Scholar] [CrossRef]
- Carović-Stanko, K.; Liber, Z.; Vidak, M.; Barešić, A.; Grdiša, M.; Lazarević, B.; Šatović, Z. Genetic diversity of Croatian common bean landraces. Front. Plant Sci. 2017, 8, 604. [Google Scholar] [CrossRef]
- Čupić, T.; Gantner, R.; Popović, S.; Tucak, M.; Sudar, R.; Stjepanović, M. Widespread annual legumes in Croatia. In Proceedings of the 5th International Scientific/Professional Conference, Vukovar, Croatia, 4–6 June 2012; Stipešević, B., Sorić, R., Eds.; Glas Slavonije d.d.: Osijek, Croatia, 2012; pp. 220–225. [Google Scholar]
- Vidak, M.; Malešević, S.; Grdiša, M.; Šatović, Z.; Lazarević, B.; Carović-Stanko, K. Phenotypic diversity among Croatian common bean (Phaseolus vulgaris L.) landraces. Agric. Conspec. Sci. 2015, 80, 133–137. [Google Scholar]
- Šatović, Z.; Liber, Z.; Vidak, M.; Gunjača, J.; Grdiša, M.; Lazarević, B.; Carović-Stanko, K. Origin and Genetic Diversity of Croatian Common Bean Landraces. In Proceedings of the 3rd International Conference on Plant Biology, Beograd, Serbia, 9–12 June 2018; Uzelac, B., Ed.; Serbian Plant Physiology Society: Beograd, Serbia; Institute for Biological Research: Beograd, Serbia, 2018; p. 80. [Google Scholar]
- Gioia, T.; Logozzo, G.; Attene, G.; Bellucci, E.; Benedettelli, S.; Negri, V.; Papa, R.; Spagnoletti Zeuli, P. Evidence for Introduction Bottleneck and Extensive Inter-Gene Pool (Mesoamerica × Andes) Hybridization in the European Common Bean (Phaseolus vulgaris L.) Germplasm. PLoS ONE 2013, 8, e075974. [Google Scholar] [CrossRef] [PubMed]
- Díaz, L.M.; Blair, M.W. Race structure within the Mesoamerican gene pool of common bean (Phaseolus vulgaris L.) as determined by microsatellite markers. Theor. Appl. Genet. 2006, 114, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Chávez-Servia, J.L.; Heredia-García, E.; Mayek-Pérez, N.; Aquino-Bolaños, E.N.; Hernández-Delgado, S.; Carrillo-Rodríguez, J.C.; Gill-Langarica, H.R.; Vera-Guzmán, A.M. Diversity of Common Bean (Phaseolus vulgaris L.) Landraces and the Nutritional Value of their Grains. In Grain Legumes; Goyal, A.K., Ed.; IntechOpen: London, UK, 2016; pp. 1–33. [Google Scholar]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef]
- Papa, R.; Nanni, L.; Sicard, D.; Rau, D.; Attene, G. Evolution of genetic diversity in Phaseolus vulgaris L. In Darwin ’s Harvest: New Approaches to the Origins, Evolution, and Conservation of Crops; Motley, T.J., Zerega, N., Cross, H., Eds.; Columbia University Press: New York, NY, USA, 2006; pp. 121–142. [Google Scholar]
- Angioi, S.A.; Rau, D.; Nanni, L.; Bellucci, E.; Papa, R.; Attene, G. The genetic make-up of the European landraces of the common bean. Plant Genet. Resour. Charact. Util. 2011, 9, 197–201. [Google Scholar] [CrossRef]
- Bitocchi, E.; Rau, D.; Bellucci, E.; Rodriguez, M.; Murgia, M.L.; Gioia, T.; Santo, D.; Nanni, L.; Attene, G.; Papa, R. Beans (Phaseolus ssp.) as a model for understanding crop evolution. Front. Plant Sci. 2017, 8, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Valdisser, P.A.M.R.; Pereira, W.J.; Almeida Filho, J.E.; Müller, B.S.F.; Coelho, G.R.C.; de Menezes, I.P.P.; Vianna, J.P.G.; Zucchi, M.I.; Lanna, A.C.; Coelho, A.S.G.; et al. In-depth genome characterization of a Brazilian common bean core collection using DArTseq high-density SNP genotyping. BMC Genom. 2017, 18, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Gaitán-Solís, E.; Duque, M.C.; Edwards, K.J.; Tohme, J. Microsatellite Repeats in Common Bean (Phaseolus vulgaris). Crop Sci. 2002, 42, 2128–2136. [Google Scholar] [CrossRef]
- De La Fuente, M.D.; López-Pedrouso, M.; Alonso, J.; Santalla, M.; De Ron, A.M.; Álvarez, G.; Zapata, C. In-depth characterization of the phaseolin protein diversity of common bean (Phaseolus vulgaris L.) based on two-dimensional electrophoresis and mass spectrometry. Food Technol. Biotechnol. 2012, 50, 315–325. [Google Scholar]
- Razvi, S.M.; Dar, M.H.; Groach, R.; Bhat, A. Molecular Diversity and Gene Pool Structure in Common Bean (Phaseolus vulgaris L.): A Review. Int. J. Curr. Trends Sci. Technol. 2017, 7, 20185–20202. [Google Scholar]
- Resende, R.T.; de Resende, M.D.V.; Azevedo, C.F.; Fonseca e Silva, F.; Melo, L.C.; Pereira, H.S.; Souza, T.L.P.O.; Valdisser, P.A.M.R.; Brondani, C.; Vianello, R.P. Genome-wide association and Regional Heritability Mapping of plant architecture, lodging and productivity in phaseolus vulgaris. G3 Genes Genomes Genet. 2018, 8, 2841–2854. [Google Scholar] [CrossRef]
- Nemli, S.; Kaygisiz Aşçioğul, T.; Ateş, D.; Eşiyok, D.; Tanyolaç, M.B. Diversity and genetic analysis through DArTseq in common bean (Phaseolus vulgaris L.) germplasm from Turkey. Turk. J. Agric. For. 2017, 41, 389–404. [Google Scholar] [CrossRef]
- Jaccoud, D.; Peng, K.; Feinstein, D.; Kilian, A. Diversity Arrays: A solid state technology for sequence information independent genotyping. Nucleic Acids Res. 2001, 29, e25. [Google Scholar] [CrossRef]
- Nkhata, W.; Shimelis, H.; Melis, R.; Chirwa, R.; Mzengeza, T.; Mathew, I.; Shayanowako, A. Population structure and genetic diversity analyses of common bean germplasm collections of East and Southern Africa using morphological traits and high-density SNP markers. PLoS ONE 2020, 15, e0243238. [Google Scholar] [CrossRef]
- Angioi, S.A.; Rau, D.; Attene, G.; Nanni, L.; Bellucci, E.; Logozzo, G.; Negri, V.; Zeuli, P.L.S.; Papa, R. Beans in Europe: Origin and structure of the European landraces of Phaseolus vulgaris L. Theor. Appl. Genet. 2010, 121, 829–843. [Google Scholar] [CrossRef]
- Bellucci, E.; Bitocchi, E.; Rau, D.; Rodriguez, M.; Biagetti, E.; Giardini, A.; Attene, G.; Nanni, L.; Papa, R. Genomics of origin, domestication and evolution of Phaseolus vulgaris. In Genomics of Plant Genetic Resources: Managing, Sequencing and Mining Genetic Resources; Tuberosa, R., Graner, A., Frison, E., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 483–507. [Google Scholar]
- Maras, M.; Pipan, B.; Šuštar-Vozlič, J.; Todorović, V.; Ðurić, G.; Vasić, M.; Kratovalieva, S.; Ibusoska, A.; Agić, R.; Matotan, Z.; et al. Examination of genetic diversity of common bean from the western Balkans. J. Am. Soc. Hortic. Sci. 2015, 140, 308–316. [Google Scholar] [CrossRef]
- Tomlekova, N.; Sofkova-Bobcheva, S.; Sarsu, F.; Baudoin, J.P. Genetic diversity of Bulgarian Phaseolus vulgaris L. based on phaseolin type and seed-coat colour. Bulg. J. Agric. Sci. 2016, 22, 447–451. [Google Scholar]
- Leitão, S.T.; Dinis, M.; Veloso, M.M.; Šatović, Z.; Vaz Patto, M.C. Establishing the bases for introducing the unexplored Portuguese common bean germplasm into the breeding world. Front. Plant Sci. 2017, 8, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Caproni, L.; Raggi, L.; Ceccarelli, S.; Negri, V.; Carboni, A. In-depth characterisation of common bean diversity discloses its breeding potential for sustainable agriculture. Sustainability 2019, 11, 5443. [Google Scholar] [CrossRef]
- Raggi, L.; Tiranti, B.; Negri, V. Italian common bean landraces: Diversity and population structure. Genet. Resour. Crop Evol. 2013, 60, 1515–1530. [Google Scholar] [CrossRef]
- Campa, A.; Murube, E.; Ferreira, J.J. Genetic diversity, population structure, and linkage disequilibrium in a Spanish common bean diversity panel revealed through genotyping-by-sequencing. Genes 2018, 9, 518. [Google Scholar] [CrossRef] [PubMed]
- Savić, A.; Zorić, M.; Brdar-Jokanović, M.; Zdravković, M.; Dimitrijević, M.; Petrović, S.; Živanov, D.; Vasić, M. Origin and diversity study of local common bean (Phaseolus vulgaris L.) germplasm from Serbia: Phaseolin and phenotyping approach. Genet. Resour. Crop Evol. 2020, 67, 2195–2212. [Google Scholar] [CrossRef]
- Gepts, P.; Bliss, F.A. Dissemination Pathways of Common Bean (Phaseolus vulgaris, Fabaceae) Deduced from Phaseolin Electrophoretic Variability. II. Europe and Africa. Econ. Bot. 1988, 42, 86–104. [Google Scholar] [CrossRef]
- Lioi, L. Geographical variation of phaseolin patterns in an old world collection of Phaseolus vulgaris. Seed Sci. Technol. 1989, 17, 317–324. [Google Scholar]
- Rodiño, A.P.; Santalla, M.; De Ron, A.M.; Singh, S.P. A core collection of common bean from the Iberian Peninsula. Euphytica 2003, 131, 165–175. [Google Scholar] [CrossRef]
- Logozzo, G.; Donnoli, R.; Macaluso, L.; Papa, R.; Knüpffer, H.; Zeuli, P.S. Analysis of the contribution of Mesoamerican and Andean gene pools to European common bean (Phaseolus vulgaris L.) germplasm and strategies to establish a core collection. Genet. Resour. Crop Evol. 2007, 54, 1763–1779. [Google Scholar] [CrossRef]
- Savić, A.; Petrović, G.; Milošević, M.; Nikolić, Z.; Stojanović, A.; Gvozdanović-Varga, J.; Todorović, V.; Vasić, M. Morpho-chemical characterization of dry and snap bean (Phaseolus vulgaris L.) landraces collected on Fruška gora mt. Genetika 2014, 46, 303–313. [Google Scholar] [CrossRef]
- Madakbaş, S.Y.; Awale, H.; Kelly, J. Determination of Phaseolin Types in Common Bean (Phaseolus vulgaris) Varieties from Turkey. Greener J. Agric. Sci. 2014, 4, 39–45. [Google Scholar]
- Šustar-Vozlič, J.; Maras, M.; Javornik, B.; Meglič, V. Genetic diversity and origin of Slovene common bean (Phaseolus vulgaris L.) germplasm as revealed by AFLP markers and phaseolin analysis. J. Am. Soc. Hortic. Sci. 2006, 131, 242–249. [Google Scholar] [CrossRef]
- Svetleva, D.; Pereira, G.; Carlier, J.; Cabrita, L.; Leitão, J.; Genchev, D. Molecular characterization of Phaseolus vulgaris L. genotypes included in Bulgarian collection by ISSR and AFLP™ analyses. Sci. Hortic. (Amst.) 2006, 109, 198–206. [Google Scholar] [CrossRef]
- da Silva, E.M.; Correia, A.S.A.; Lopes, N.A.A.; Nóbrega, H.G.M.; Ganança, J.F.T.; Domingues, A.M.; Khadem, M.; Slaski, J.J.; Carvalho, M.Â.A.P. de Phytogeographical origin of Madeiran common beans based on phaseolin patterns. Pesqui. Agropecuária Bras. 2010, 45, 863–871. [Google Scholar] [CrossRef]
- Cortés, A.J.; Chavarro, M.C.; Blair, M.W. SNP marker diversity in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2011, 123, 827–845. [Google Scholar] [CrossRef]
- Gaut, B.S. The complex domestication history of the common bean. Nat. Genet. 2014, 46, 663–664. [Google Scholar] [CrossRef]
- Croatian Agency for Agriculture and Food (CAAF). Guide to Plant Variety Protection; Croatian Agency for Agriculture and Food (CAAF), Center for Seed and Seedlings, Department for Plant: Osijek, Croatia, 2019. [Google Scholar]
- Singh, S.P.; Gutiérrez, J.A.; Molina, A.; Urrea, C.; Gepts, P. Genetic Diversity in Cultivated Common Bean: II. Marker-Based Analysis of Morphological and Agronomic Traits. Crop Sci. 1991, 31, 23–29. [Google Scholar] [CrossRef]
- Lioi, L.; Piergiovanni, A.R. Genetic diversity and seed quality of the Badda common Bean from Sicily (Italy). Diversity 2013, 5, 843–855. [Google Scholar] [CrossRef]
- Gepts, P.; Debouck, D. Origin, domestication and evolution of the common bean, Phaseolus vulgaris. In Common Beans: Research for Crop Improvement Research for Crop Improvement; Schoonhoven, A., Voysest, O., Eds.; CAB International: Wallingford, UK, 1991; pp. 7–53. [Google Scholar]
- Zeven, A.C.; Waninge, J.; Van Hintum, T.; Singh, S.P. Phenotypic variation in a core collection of common bean (Phaseolus vulgaris L.) in The Netherlands. Euphytica 1999, 109, 93–106. [Google Scholar] [CrossRef]
- Rodiño, A.P.; Santalla, M.; González, A.M.; De Ron, A.M.; Singh, S.P. Novel genetic variation in common bean from the Iberian Peninsula. Crop Sci. 2006, 46, 2540–2546. [Google Scholar] [CrossRef]
- Santalla, M.; Rodiño, A.P.; De Ron, A.M. Allozyme evidence supporting southwestern Europe as a secondary center of genetic diversity for the common bean. Theor. Appl. Genet. 2002, 104, 934–944. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.; Gepts, P. Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae). Theor. Appl. Genet. 2009, 118, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P. Patterns of variation in cultivated common bean (Phaseolus vulgaris, Fabaceae). Econ. Bot. 1989, 43, 39–57. [Google Scholar] [CrossRef]
- Kwak, M.; Toro, O.; Debouck, D.G.; Gepts, P. Multiple origins of the determinate growth habit in domesticated common bean (Phaseolus vulgaris). Ann. Bot. 2012, 110, 1573–1580. [Google Scholar] [CrossRef] [PubMed]
- Lithourgidis, A.S.; Dordas, C.A.; Damalas, C.A.; Vlachostergios, D.N. Annual intercrops: An alternative pathway for sustainable agriculture. Aust. J. Crop Sci. 2011, 5, 396–410. [Google Scholar]
- Kami, J.; Velásquez, V.B.; Debouck, D.G.; Gepts, P. Identification of presumed ancestral DNA sequences of phaseolin in Phaseolus vulgaris. Proc. Natl. Acad. Sci. USA 1995, 92, 1101–1104. [Google Scholar] [CrossRef]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Blair, M.W.; Pedraza, F.; Buendia, H.F.; Gaitán-Solís, E.; Beebe, S.E.; Gepts, P.; Tohme, J. Development of a genome-wide anchored microsatellite map for common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2003, 107, 1362–1374. [Google Scholar] [CrossRef] [PubMed]
- Córdoba, J.M.; Chavarro, C.; Schlueter, J.A.; Jackson, S.A.; Blair, M.W. Integration of physical and genetic maps of common bean through BAC-derived microsatellite markers. BMC Genom. 2010, 11, 436. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Park, S.J.; Poysa, V.; Gepts, P. Integration of simple sequence repeat (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris L.). J. Hered. 2000, 91, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- International Board for Plant Genetic Resources (IBPGR). Phaseolus Vulgaris Descriptors; International Board for Plant Genetic Resources: Rome, Italy, 1982. [Google Scholar]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Raymond, M.; Rousset, F. GENEPOP (Version 1.2): Population Genetics Software for Exact Tests and Ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Bowcock, A.M.; Ruiz-Linares, A.; Tomfohrde, J.; Minch, E.; Kidd, J.R.; Cavalli-Sforza, L.L. High resolution of human evolutionary trees with polymorphic microsatellites. Nature 1994, 368, 455–457. [Google Scholar] [CrossRef]
- Minch, E.; Ruiz-Linares, A.; Goldstein, D.; Feldman, M.; Cavalli-Sforza, L.L. MICROSAT: A Computer Program for Calculating Various Statistics on Microsatellite Allele Data, Version 1.5b; Department of Genetics, Stanford University: Stanford, CA, USA, 1997. [Google Scholar]
- Felsenstein, J. PHYLIP (Phylogeny Inference Package), Version 3.6. 2004. Available online: http://evolution.genetics.washington.edu/phylip.html (accessed on 15 September 2020).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; Von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Kalinowski, S.T. HP-RARE 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 2005, 5, 187–189. [Google Scholar] [CrossRef]
- SAS Institute Inc. Base SAS® 9.3 Procedures Guide: Statistical Procedures; SAS Institute Inc.: Cary, NC, USA, 2011. [Google Scholar]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of Molecular Variance Inferred From Metric Distances Among DNA Haplotypes: Application to Human Mitochondrial DNA Restriction Data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin (version 3.5): A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2005, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.C.F. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000; Université de Montpellier: Montpellier, France, 2004; Available online: http://www.genetix.univ-montp2.fr/genetix/genetix.Htm (accessed on 10 October 2020).

| Chromosome | Chromosome Length (kbp) a | No. of SNPs | Mean no. of SNPs per Mbp |
|---|---|---|---|
| 1 | 52,183.5 | 610 | 11.69 |
| 2 | 49,033.7 | 817 | 16.66 |
| 3 | 52,218.6 | 756 | 14.48 |
| 4 | 45,793.2 | 382 | 8.34 |
| 5 | 40,237.5 | 486 | 12.08 |
| 6 | 31,973.2 | 532 | 16.64 |
| 7 | 51,698.4 | 605 | 11.70 |
| 8 | 59,634.6 | 640 | 10.73 |
| 9 | 37,399.6 | 596 | 15.94 |
| 10 | 43,213.2 | 417 | 9.65 |
| 11 | 50,203.6 | 470 | 9.36 |
| Total | 51,3589.1 | 6311 | 12.29 |
| No. | Landrace | Picture | Habit a | No. of Accessions | Centre of Origin b | Genetic Group c | Morphogenetic Group |
|---|---|---|---|---|---|---|---|
| 1 | “Kukuruzar” |  | I | 19 | A | H1A | M01 |
| 2 | “Tetovac” |  | I | 15 | A | H1A | M02 |
| 3 | “Biser” |  | D | 5 | A | H1A | M03 |
| 4 | “Sivi” |  | I | 3 | B | H2B1 | M04 |
| 5 | ”Sivi prošarani” |  | I | 1 | B | H2B1 | M05 |
| 6 7 8 | “Trešnjevac” “Trešnjevac” “Trešnjevac” |  | I I D | 4 11 25 | A B B | H1A H2B1 H3B2 | M06_1 M06_2 M06_3 |
| 9 10 | “Puter” “Puter” |  | I D | 2 7 | B B | H2B1 H3B2 | M07_1 M07_2 |
| 11 12 | “Dan noć” “Dan noć” |  | I D | 2 3 | B B | H2B1 H3B2 | M08_1 M08_2 |
| 13 | “Zelenčec” |  | D | 22 | B | H3B2 | M09 |
| 14 | “Bijeli” |  | D | 3 | B | H3B2 | M10 |
| Group | n | Na | Nar | Npr | Npar | HO | HE |
|---|---|---|---|---|---|---|---|
| True-types (Origin) | |||||||
| A: Mesoamerican | 43 | 2.923 | 2.923 | 38 | 1.521 | 0.000 | 0.259 |
| B: Andean | 79 | 3.462 | 3.306 | 52 | 1.904 | 0.000 | 0.336 |
| p | ns | ns | |||||
| True-types (Genetic groups) | |||||||
| H1A | 43 | 2.923 | 2.700 | 38 | 1.413 | 0.000 | 0.259 |
| H2B1 | 19 | 2.423 | 2.423 | 16 | 0.684 | 0.000 | 0.301 |
| H3B2 | 60 | 2.462 | 2.232 | 20 | 0.683 | 0.000 | 0.247 |
| p | ns | ns | |||||
| Off-types | |||||||
| X1 | 35 | 3.538 | - | - | - | 0.000 | 0.412 |
| X2 | 9 | 2.846 | - | - | - | 0.000 | 0.489 |
| X3 | 8 | 2.962 | - | - | - | 0.000 | 0.648 |
| Group | n | Na | Nar a | Npr | Npar | HO | HE | |||
|---|---|---|---|---|---|---|---|---|---|---|
| True-types (Origin) | ||||||||||
| A: Mesoamerican | 43 | 1.618 | 1.618 | 1962 | 0.361 | 0.010 | 0.164 | |||
| B: Andean | 79 | 1.689 | 1.639 | 2411 | 0.382 | 0.003 | 0.116 | |||
| p | * | *** | *** | |||||||
| True-types (Genetic groups) | ||||||||||
| H1A | 43 | 1.618 | 1.556 | a | 1962 | 0.382 | 0.010 | a | 0.164 | a |
| H2B1 | 19 | 1.418 | 1.418 | b | 76 | 0.015 | 0.003 | b | 0.141 | b |
| H3B2 | 60 | 1.557 | 1.426 | b | 132 | 0.023 | 0.003 | b | 0.079 | c |
| p | *** | *** | *** | |||||||
| Off-types b | ||||||||||
| X1 | 35 | 1.786 | - | - | - | 0.004 | 0.161 | |||
| X2 | 9 | 1.934 | - | - | - | 0.003 | 0.360 | |||
| X3 | 8 | 1.945 | - | - | - | 0.003 | 0.480 | |||
| Group | n | T1 Days to Flowering a | T2 Duration of Flowering | T3 Seed Length (mm) | T4 Seed Width (mm) | T5 Seed Height (mm) | T6 100 seed Weight (g) | T7 Elongation | T8 Flatness | T9 Flatness Index | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| True-types (Origin) | |||||||||||||||||||
| A | 43 | 62.51 | 26.67 | 15.16 | 5.25 | 8.19 | 45.71 | 1.85 | 1.57 | 2.24 | |||||||||
| B | 79 | 55.11 | 21.02 | 14.68 | 6.43 | 8.14 | 50.14 | 1.81 | 1.27 | 1.78 | |||||||||
| p | *** | *** | ns | *** | ns | ** | ns | *** | *** | ||||||||||
| True-types (Genetic groups) | |||||||||||||||||||
| H1A | 43 | 62.51 | a | 26.67 | a | 15.16 | a | 5.25 | c | 8.19 | b | 45.71 | b | 1.85 | a | 1.57 | a | 2.24 | a |
| H2B1 | 19 | 62.24 | a | 27.06 | a | 15.68 | a | 6.97 | a | 9.14 | a | 61.32 | a | 1.71 | b | 1.32 | b | 1.79 | b |
| H3B2 | 60 | 52.85 | b | 19.11 | b | 14.36 | b | 6.26 | b | 7.82 | c | 46.59 | b | 1.85 | a | 1.25 | b | 1.78 | b |
| p | *** | *** | ** | *** | *** | *** | * | *** | *** | ||||||||||
| Off-types b | |||||||||||||||||||
| X1 | 35 | 57.28 | 23.07 | 14.37 | 6.37 | 8.28 | 49.66 | 1.75 | 1.30 | 1.79 | |||||||||
| X2 | 9 | 55.80 | 24.22 | 14.24 | 6.31 | 7.78 | 46.17 | 1.84 | 1.24 | 1.76 | |||||||||
| X3 | 8 | 55.98 | 23.48 | 14.98 | 5.91 | 8.03 | 48.05 | 1.88 | 1.38 | 1.98 | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vidak, M.; Šatović, Z.; Liber, Z.; Grdiša, M.; Gunjača, J.; Kilian, A.; Carović-Stanko, K. Assessment of the Origin and Diversity of Croatian Common Bean Germplasm Using Phaseolin Type, SSR and SNP Markers and Morphological Traits. Plants 2021, 10, 665. https://doi.org/10.3390/plants10040665
Vidak M, Šatović Z, Liber Z, Grdiša M, Gunjača J, Kilian A, Carović-Stanko K. Assessment of the Origin and Diversity of Croatian Common Bean Germplasm Using Phaseolin Type, SSR and SNP Markers and Morphological Traits. Plants. 2021; 10(4):665. https://doi.org/10.3390/plants10040665
Chicago/Turabian StyleVidak, Monika, Zlatko Šatović, Zlatko Liber, Martina Grdiša, Jerko Gunjača, Andrzej Kilian, and Klaudija Carović-Stanko. 2021. "Assessment of the Origin and Diversity of Croatian Common Bean Germplasm Using Phaseolin Type, SSR and SNP Markers and Morphological Traits" Plants 10, no. 4: 665. https://doi.org/10.3390/plants10040665
APA StyleVidak, M., Šatović, Z., Liber, Z., Grdiša, M., Gunjača, J., Kilian, A., & Carović-Stanko, K. (2021). Assessment of the Origin and Diversity of Croatian Common Bean Germplasm Using Phaseolin Type, SSR and SNP Markers and Morphological Traits. Plants, 10(4), 665. https://doi.org/10.3390/plants10040665








