Secondary Metabolites of Pseudomonas aeruginosa LV Strain Decrease Asian Soybean Rust Severity in Experimentally Infected Plants
Abstract
1. Introduction
2. Results
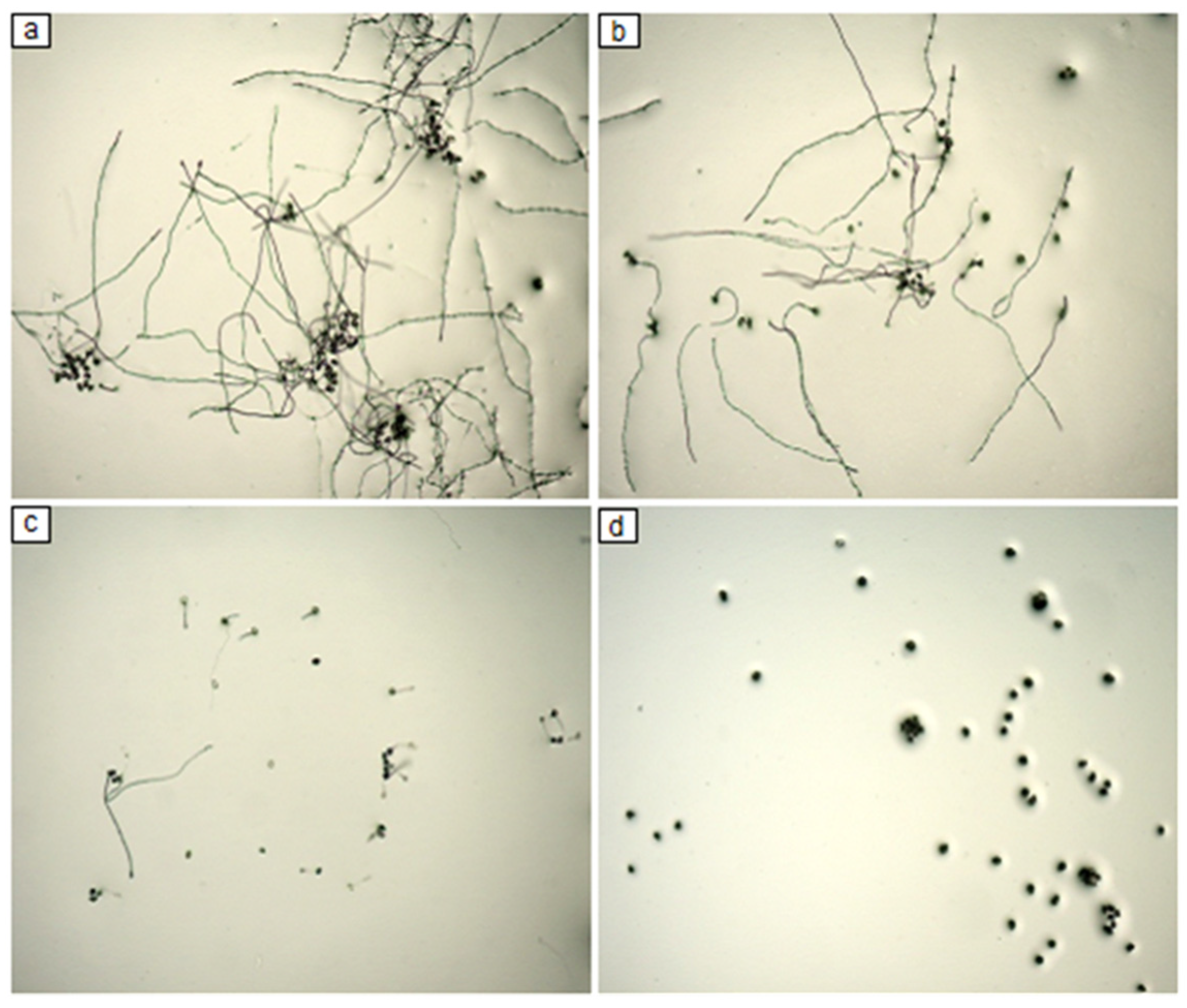
2.1. The Evaluation of F4A Fraction on P. pachyrhizi Spore Germination
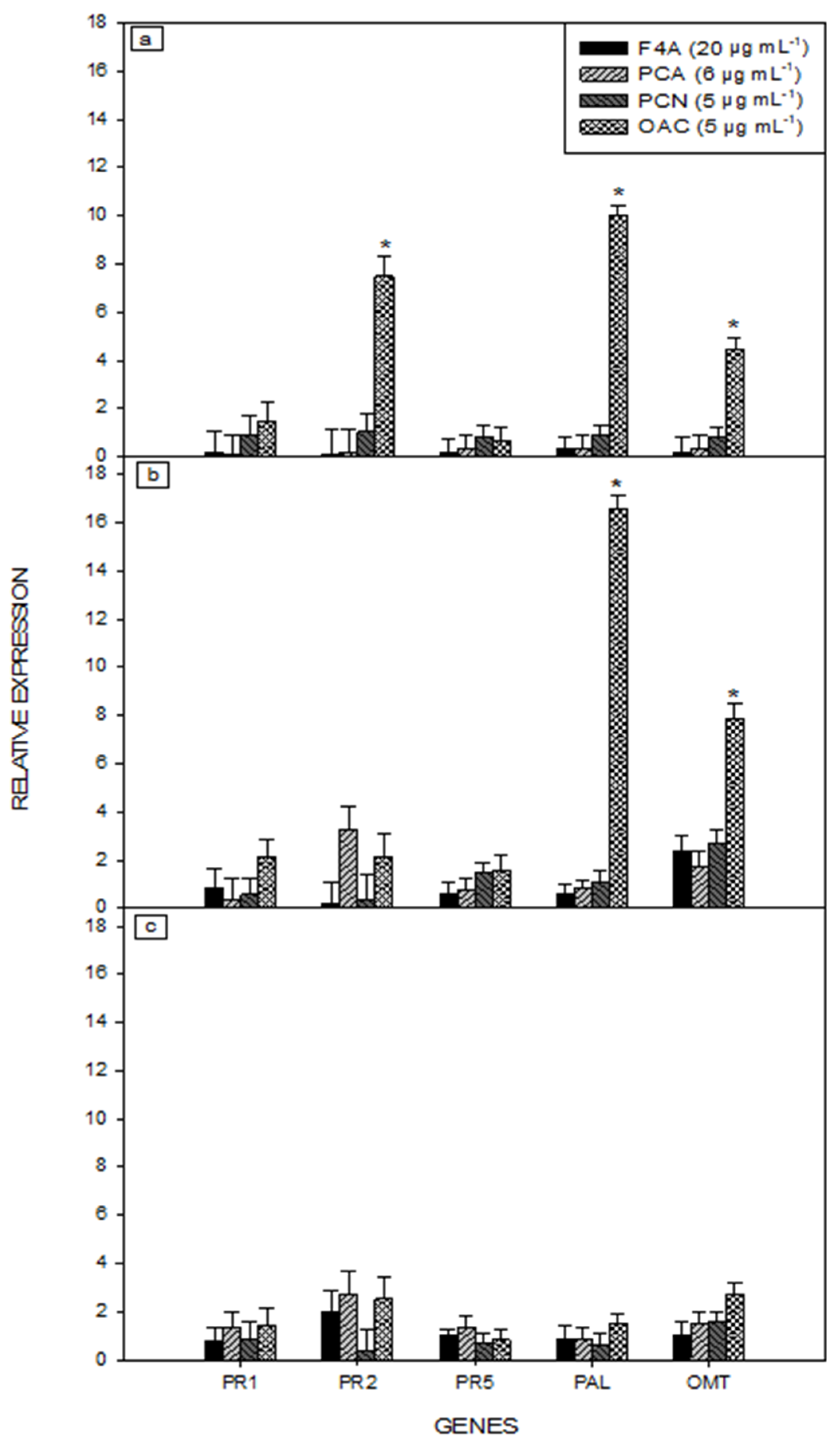
2.2. Gene Expression of Soybean Plant Treated with Secondary Metabolites of P. aeruginosa LV Strain
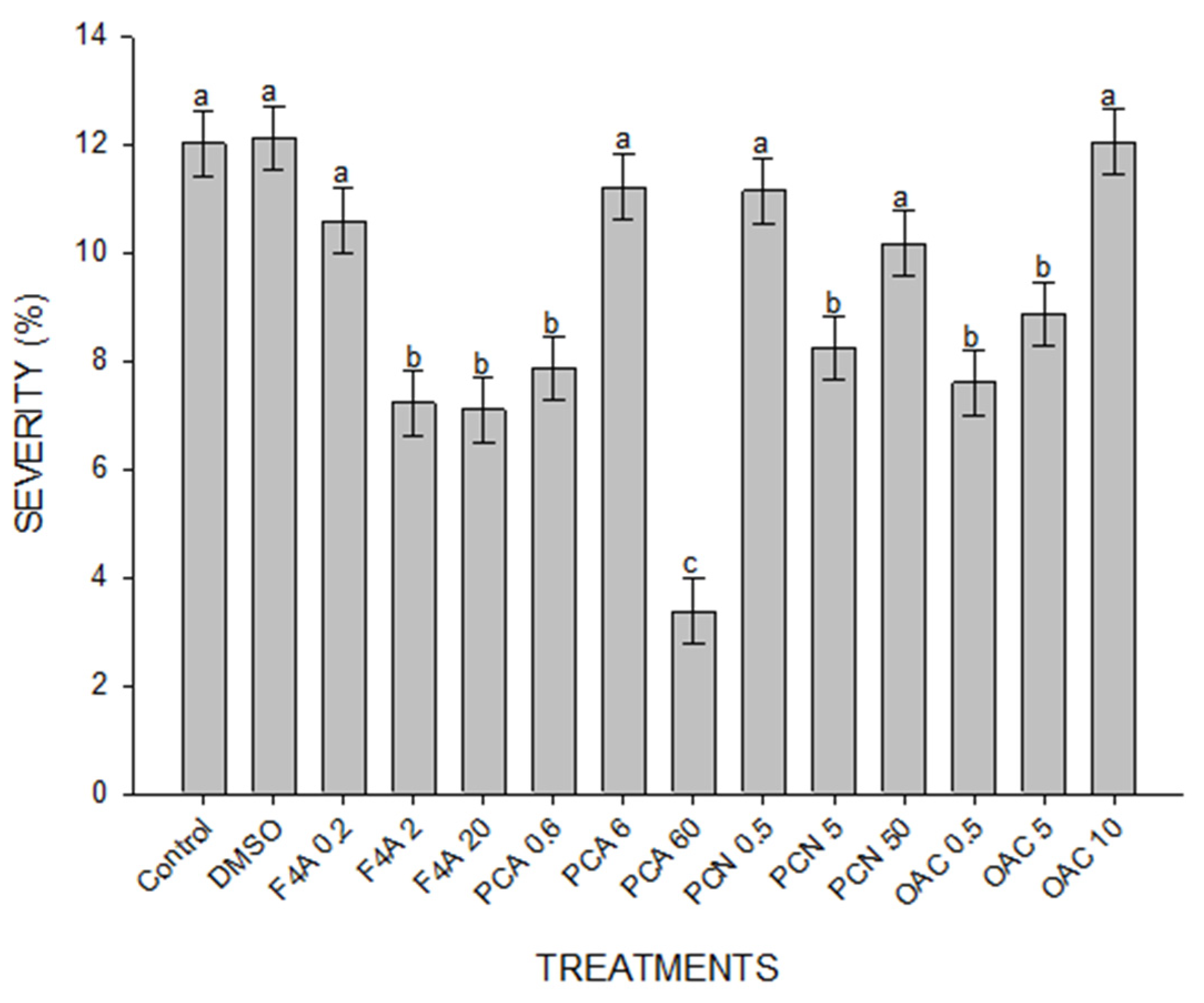
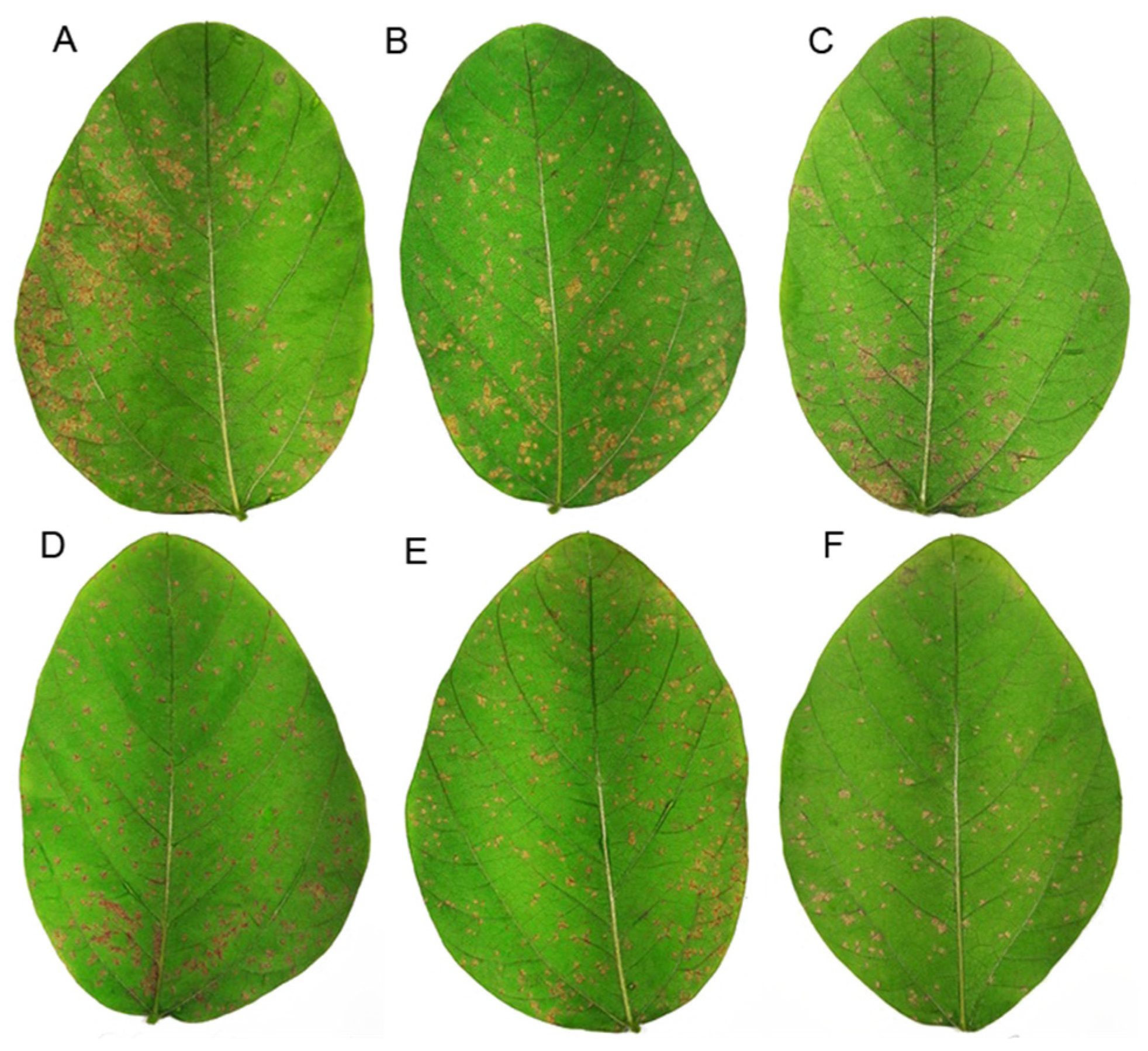
2.3. ASR Control in Soybean Plants Treated with Secondary Metabolites of P. aeruginosa LV Strain
3. Discussion
4. Materials and Methods
4.1. Metabolites Production
4.2. The Evaluation of F4A Fraction on P. pachyrhizi Spore Germination
4.3. Gene Expression of Soybean Plants Treated with Secondary Metabolites of P. aeruginosa LV Strain
4.4. ASR Control in Soybean Plants Treated with Secondary Metabolites of P. aeruginosa LV Strain
4.5. Statistical Analysis
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Langenbach, C.; Campe, R.; Beyer, S.F.; Mueller, A.N.; Conrath, U. Fighting Asian soybean rust. Front. Plant Sci. 2016, 7, 797. [Google Scholar] [CrossRef]
- Godoy, C.V.; Dinali, C.; Seixas, S.; Soares, R.M.; Marcelino-Guimarães, F.C.; Meyer, M.C.; Costamilan, L.M. Asian soybean rust in Brazil: Past, present, and future. Pesq. Agropec. Bras. 2016, 51, 407–421. [Google Scholar] [CrossRef]
- Pennisi, E. Armed and Dangerous. Science 2010, 327, 804–805. [Google Scholar] [CrossRef]
- Slaminko, T.L.; Miles, M.R.; Frederick, R.D.; Bonde, M.R.; Hartman, G.L.; Slaminko, T.L.; Miles, M.R.; Frederick, R.D.; Bonde, M.R.; Hartman, G.L. New legume hosts of Phakopsora pachyrhizi based on greenhouse evaluations. Plant Dis. 2008, 92, 767–771. [Google Scholar] [CrossRef]
- Yamaoka, Y. Recent outbreaks of rust diseases and the importance of basic biological research for controlling rusts. J. Gen. Plant Pathol. 2014, 80, 375–388. [Google Scholar] [CrossRef]
- Duhatschek, E.; Santos, L.A.; Faria, C.M.D.R. Sensibilidade de isolados de Phakopsora pachyrhizi provenientes da região do centro oeste do Paraná a fungicidas. Summa Phytopathol. 2018, 44, 193–194. [Google Scholar] [CrossRef][Green Version]
- Durner, J.; Shah, J.; Klessig, D.F. Salicylic acid and disease resistance in plants. Trends Plant Sci. 1997, 2, 266–274. [Google Scholar] [CrossRef]
- Sels, J.; Mathys, J.; de Coninck, B.M.A.; Cammue, B.P.A.; de Bolle, M.F.C. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar]
- Walters, D.R.; Ratsep, J.; Havis, N.D. Controlling crop diseases using induced resistance: Challenges for the future. J. Exp. Bot. 2013, 64, 1263–1280. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of Inducible Defense-related Proteins in Infected Plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed]
- Rakwal, R.; Agrawal, G.K.; Yonekura, M. Separation of proteins from stressed rice (Oryza sativa L.) leaf tissues by two-dimensional polyacrylamide gel electrophoresis: Induction of pathogenesis related and cellular protectant proteins by jasmonic acid, UV irradiation and copper chloride. Electrophoresis 1999, 20, 3472–3478. [Google Scholar] [CrossRef]
- Jwa, N.-S.; Agrawal, G.K.; Rakwal, R.; Park, C.-H.; Agrawal, V.P. Molecular Cloning and Characterization of a Novel Jasmonate Inducible Pathogenesis-Related Class 10 Protein Gene, JIOsPR10, from Rice (Oryza sativa L.) Seedling Leaves. Biochem. Biophys. Res. Commun. 2001, 286, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Senthilraja, G.; Ananda, T.; Kennedyb, J.S.; Raguchandera, T.; Samiyappan, R. Plant growth promoting rhizobacteria (PGPR) and entomopathogenic fungus bioformulation enhance the expression of defense enzymes and pathogene-sis-related proteins in groundnut plants against leaf miner insect and collar rot pathogen. Physiol. Mol. Plant Pathol. 2013, 82, 10–19. [Google Scholar] [CrossRef]
- Jiang, L.; Wu, J.; Fan, S.; Li, W.; Dong, L.; Cheng, Q.; Xu, P.; Zhang, S. Isolation and characterization of a novel pathogene-sis-related protein gene (GmPRP) with induced expression in soybean (Glycine max) during infection with Phytophthora sojae. PLoS ONE 2015, 10, e0129932. [Google Scholar]
- Kitajima, S.; Sato, F. Plant Pathogenesis-Related Proteins: Molecular Mechanisms of Gene Expression and Protein Function. J. Biochem. 1999, 125, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Borsics, T.; Dodder, L.M. Dodder infection induces the expression of a pathogenesis-related gene of the family PR-10 in alfalfa. J. Exp. Bot. 2002, 53, 1831–1832. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.P.; Kaur, S.; Batish, D.R.; Kohli, R.K. Caffeic acid inhibits in vitro rooting in mung bean Vigna radiata (L.) Wilczek hypocotyls by inducing oxidative stress. Plant Growth Regul. 2008, 57, 21–30. [Google Scholar] [CrossRef]
- Dixon, R.A.; Paiva, N.L. Stress-Induced Phenylpropanoid Metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef]
- Lyne, R.L.; Mulheirn, L.I.; Leworthy, D.P. New pterocarpinoid phytoalexins of soybean. JCS Chem. Commun. 1976, 1976, 497–498. [Google Scholar] [CrossRef]
- Nwachukwu, I.D.; Luciano, F.B.; Udenigwe, C.C. The inducible soybean glyceollin phytoalexins with multifunctional health-promoting properties. Food Res. Int. 2013, 54, 1208–1216. [Google Scholar] [CrossRef]
- Kim, H.J.; Lim, J.S.; Kim, W.K.; Kim, J.S. Soybean glyceollins: Biological effects and relevance to human health. Proc. Nutr. Soc. 2012, 71, 166–174. [Google Scholar] [CrossRef]
- Lygin, A.V.; Li, S.; Vittal, R.; Widholm, J.M.; Hartman, G.L.; Lozovaya, V.V. The Importance of Phenolic Metabolism to Limit the Growth of Phakopsora pachyrhizi. Phytopathology 2009, 99, 1412–1420. [Google Scholar] [CrossRef]
- Lam, K.C.; Ibrahim, R.K.; Behdad, B.; Dayanandan, S. Structure, function, and evolution of plant O-methyltransferases. Genome 2007, 50, 1001–1013. [Google Scholar] [CrossRef]
- Kim, B.; Lee, H.; Park, Y.; Lim, Y.; Ahn, J.-H. Characterization of an O-methyltransferase from soybean. Plant Physiol. Biochem. 2006, 44, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Shah, J.; Chaturvedi, R.; Chowdhury, Z.; Venables, B.; Petros, R.A. Signaling by small metabolites in systemic acquired resistance. Plant J. 2014, 79, 645–658. [Google Scholar] [CrossRef]
- Oliveira, M.; Varanda, C.; Felix, M. Induced resistance during the interaction pathogen x plant and the use of resistance inducers. Phytochem. Lett. 2016, 15, 152–158. [Google Scholar] [CrossRef]
- Dayan, F.E.; Cantrell, C.L.; Duke, S.O. Natural products in crop protection. Bioorg. Med. Chem. 2009, 17, 4022–4034. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.A.; Velazhahan, R.; Nagarithinam, S.; Vidhyasekaran, P. Induction of pathogenesis-related proteins in sugarcane leaves and cell-cultures by a glycoprotein elicitor isolated from Colletotrichum falcatum. Biol. Plant. 2008, 52, 321–328. [Google Scholar]
- Herman, M.A.B.; Davidson, J.K.; Smart, C.D. Induction of plant defense gene expression by plant activators and Pseudomonas syringae pv. tomato in greenhouse- grown tomatoes. Phytopathology 2008, 98, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Van Wees, S.C.; Van der Ent, S.; Pieterse, C.M.J. Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 2008, 11, 443–448. [Google Scholar] [CrossRef]
- Munhoz, L.D.; Fonteque, J.P.; Santos, I.M.O.; Navarro, M.O.P.; Simionato, A.S.; Goya, E.T.; Rezende, M.I.; Balbi-Peña, M.L.; de Oliveira, A.G.; Andrade, G. Control of bacterial stem rot on tomato by extracellular bioactive compounds produced by Pseudomonas aeruginosa LV strain. Cogent Food Agric. 2017, 3, 1282592. [Google Scholar] [CrossRef]
- Riera, N.; Wang, H.; Li, Y.; Li, J.; Pelz-Stelinski, K.; Wang, N. Induced systemic resistance against citrus canker disease by rhizobacteria. Phytopathology 2018, 108, 1038–1045. [Google Scholar] [CrossRef] [PubMed]
- Pistori, J.F.; Simionato, A.S.; Navarro, M.; Andreata, M.F.L.; Santos, I.M.O.; Meneguim, L.; Junior, R.P.L.; Oliveira, A.G.; Andrade, G. Low-molecular-weight metabolites produced by Pseudomonas aeruginosa as an alternative to control Huanglongbing in Citrus sinensis cv. Valencia. Trop. Plant Pathol. 2018, 43, 289–296. [Google Scholar] [CrossRef]
- Srivastava, P.; George, S.; Marois, J.J.; Wright, D.L.; Walker, D.R. Saccharin-induced systemic acquired resistance against rust (Phakopsora pachyrhizi) infection in soybean: Effects on growth and development. Crop. Prot. 2011, 30, 726–732. [Google Scholar] [CrossRef]
- Da Cruz, M.F.A.; Rodrigues, F.Á.; Polanco, L.R.; Curvêlo, C.R.D.S.; Nascimento, K.J.T.; Moreira, M.A.; Barros, E.G. Inducers of resistance and silicon on the activity of defense enzymes in the soybean-Phakopsora pachyrhizi interaction. Bragantia 2013, 72, 162–172. [Google Scholar] [CrossRef]
- Barros, R. Estudo sobre a aplicação foliar de acibenzolar-S-metil para indução de resistência à ferrugem asiática em soja e cercosporiose em milho. Arq. Inst. Biol. 2011, 78, 519–528. (In Spanish) [Google Scholar] [CrossRef]
- Carvalho, B.O.; Oliveira, J.A.; Carvalho, E.R.; De Andrade, V.; Ferreira, T.F.; Reis, L.V. Action of defense activator and foliar fungicide on the control of Asiatic rust and on yield and quality of soybean seeds. J. Seed Sci. 2013, 35, 198–206. [Google Scholar] [CrossRef][Green Version]
- Lee, J.Y.; Moon, S.S.; Hwang, B.K. Isolation and in vitro and in vivo activity against Phytophthora capsici and Colletotrichum orbiculare of phenazine-1-carboxylic acid from Pseudomonas aeruginosa strain GC-B26. Pest Manag. Sci. 2003, 59, 872–882. [Google Scholar] [CrossRef]
- Schneider, K.T.; van de Mortel, M.; Bancroft, T.J.; Braun, E.; Nettleton, D.; Nelson, R.T.; Frederick, R.D.; Baum, T.J.; Graham, M.A.; Whitham, S.A. Biphasic Gene Expression Changes Elicited by Phakopsora pachyrhizi in Soybean Correlate with Fungal Penetration and Haustoria Formation. Plant Physiol. 2011, 157, 355–371. [Google Scholar] [CrossRef]
- Van De Mortel, M.; Recknor, J.C.; Graham, M.A.; Nettleton, D.; Dittman, J.D.; Nelson, R.T.; Godoy, C.; Abdelnoor, R.V.; Almeida, Á.M.R.; Baum, T.J.; et al. Distinct Biphasic mRNA Changes in Response to Asian Soybean Rust Infection. Mol. Plant Microbe Interact. 2007, 20, 887–899. [Google Scholar] [CrossRef]
- Cooper, B.; Luster, D.G.; McMahon, M.B.; Campbell, K.B. Disruption of Rpp1-mediated soybean rust immunity by virus-induced gene silencing. Plant Signal. Behav. 2013, 8, e27543. [Google Scholar] [CrossRef]
- Pandey, A.K.; Yang, C.; Zhang, C.; Graham, M.A.; Horstman, H.D.; Lee, Y.; Zabotina, O.A.; Hill, J.H.; Pedley, K.F.; Whitham, S.A. Functional Analysis of the Asian Soybean Rust Resistance Pathway Mediated by Rpp2. Mol. Plant Microbe Interact. 2011, 24, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.Z.; Ishiga, Y.; Yamanaka, N.; Ogiso-Tanaka, E.; Yamaoka, Y. Soybean leaves transcriptomic data dissects the phenylpropanoid pathway genes as a defense response against Phakopsora pachyrhizi. Plant Physiol. Biochem. 2018, 132, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Fraser, C.M.; Chapple, C. The Phenylpropanoid Pathway in Arabidopsis. Arab. Book 2011, 9, e0152. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.J.; Alkharouf, N.W.; Schneider, K.T.; Matthews, B.F.; Frederick, R.D. Expression patterns in soybean resistant to Phakopsora pachyrhizi reveal the importance of peroxidases and lipoxygenases. Funct. Integr. Genom. 2008, 8, 341–359. [Google Scholar] [CrossRef]
- Zelezniak, A.; Sheridan, S.; Patil, K.R. Contribution of Network Connectivity in Determining the Relationship between Gene Expression and Metabolite Concentration Changes. PLoS Comput. Biol. 2014, 10, e1003572. [Google Scholar] [CrossRef]
- Rampazo, L.G.L. Evaluation the Effect of Biological Agents and its Products on the Incidence of Citrus Canker Lesions. Ph.D. Thesis, State University of Londrina, Londrina, Brazil, 2004. [Google Scholar]
- Vale, F.X.R.; Fernandes, E.R.F.; Liberato, J.R. QUANT—A Software for plant disease severity assessment. In Proceedings of the 8th International Congress of Plant Pathology, Christchurch, New Zealand, 2–7 February 2003; p. 105. [Google Scholar]


| Oligonucleotide | Sequence 5 ‘–3 ‘ |
|---|---|
| β-actin F | GAGCTATGAATTGCCTGATGG |
| β-actin R | CGTTTCATGAATTCCAGTAGC |
| PR-1 F | AGAGGCAGAGGTGGGTTCT |
| PR-1 R | TCACCAACAAAGTTGCCAG |
| PR-2 F | TGAAATAAGGGCCACGAGTCCAAATG |
| PR-2 R | ATGGTACATGCAGACTTCGAATGCAGAT |
| PR-5 F | CTCATGCACCAGTATTCCC |
| PR-5 R | AAGCTTTGTAGTTGGTCC |
| PAL F | CAAACATCGGCAGATTACTCC |
| PAL R | CTGGAATGTCTTGGAGATTGG |
| OMT F | TGGCTAGTCACTCCATGCTATC |
| OMT R | AACGAGACACCATCAGCATC |
| Treatment | Product | Concentration (µg mL−1) |
|---|---|---|
| 1 | H2O | |
| 2 | H2O + DMSO + mineral oil | |
| 3 | F4A | 0.2 |
| 4 | F4A | 2 |
| 5 | F4A | 20 |
| 6 | PCA | 0.6 |
| 7 | PCA | 6 |
| 8 | PCA | 60 |
| 9 | PCN | 0.5 |
| 10 | PCN | 5 |
| 11 | PCN | 50 |
| 12 | OAC | 0.5 |
| 13 | OAC | 5 |
| 14 | OAC | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
dos Santos, I.M.O.; Abe, V.Y.; de Carvalho, K.; Barazetti, A.R.; Simionato, A.S.; de Almeida Pega, G.E.; Matis, S.H.; Cano, B.G.; Cely, M.V.T.; Marcelino-Guimarães, F.C.; et al. Secondary Metabolites of Pseudomonas aeruginosa LV Strain Decrease Asian Soybean Rust Severity in Experimentally Infected Plants. Plants 2021, 10, 1495. https://doi.org/10.3390/plants10081495
dos Santos IMO, Abe VY, de Carvalho K, Barazetti AR, Simionato AS, de Almeida Pega GE, Matis SH, Cano BG, Cely MVT, Marcelino-Guimarães FC, et al. Secondary Metabolites of Pseudomonas aeruginosa LV Strain Decrease Asian Soybean Rust Severity in Experimentally Infected Plants. Plants. 2021; 10(8):1495. https://doi.org/10.3390/plants10081495
Chicago/Turabian Styledos Santos, Igor Matheus Oliveira, Valéria Yukari Abe, Kenia de Carvalho, André Riedi Barazetti, Ane Stéfano Simionato, Guilherme E. de Almeida Pega, Sergio Henrique Matis, Barbara Gionco Cano, Martha Viviana Torres Cely, Francismar Correa Marcelino-Guimarães, and et al. 2021. "Secondary Metabolites of Pseudomonas aeruginosa LV Strain Decrease Asian Soybean Rust Severity in Experimentally Infected Plants" Plants 10, no. 8: 1495. https://doi.org/10.3390/plants10081495
APA Styledos Santos, I. M. O., Abe, V. Y., de Carvalho, K., Barazetti, A. R., Simionato, A. S., de Almeida Pega, G. E., Matis, S. H., Cano, B. G., Cely, M. V. T., Marcelino-Guimarães, F. C., Chryssafidis, A. L., & Andrade, G. (2021). Secondary Metabolites of Pseudomonas aeruginosa LV Strain Decrease Asian Soybean Rust Severity in Experimentally Infected Plants. Plants, 10(8), 1495. https://doi.org/10.3390/plants10081495







