The Arabinogalactan Protein Family of Centaurium erythraea Rafn
Abstract
1. Introduction
2. Results
2.1. Motif and Amino Acid Bias (MAAB) Classification of Centaury HRGP Sequences
2.2. Identification of AGP Sequences with Low AG Glycomotif Representation
2.3. Structural Features of AGP Sequences Selected for Expression Analysis
2.4. Wounding Response of AGP Genes
2.5. Expression of AGP Genes in Centaury Organs and in Different Developmental Stages during Organogenesis and Somatic Embryogenesis in Vitro
3. Discussion
3.1. Size and Diversity of C. erythraea HRGP Superfamily
3.2. Phylogenetic Relations and Structural Features of the Selected FLA, KLA and AG Peptide Sequences
3.3. Mechanical Wounding Induces AG Peptides CeAGP6 and CeAGP7 and Downregulates Some FLAs and KLAs
3.4. Expression Profiles of AGPs in Different Plant Organs and During Somatic Embryogenesis and Organogenesis in Vitro
4. Materials and Methods
4.1. Plant Material
4.2. HRGP Sequence Identification
4.3. RNA Isolation and qPCR
4.4. Phylogenetic Analysis
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Deepak, S.; Shailasree, S.; Kini, R.K.; Muck, A.; Mithöfer, A.; Shetty, S.H. Hydroxyproline-rich Glycoproteins and Plant Defence. J. Phytopathol. 2010, 158, 585–593. [Google Scholar] [CrossRef]
- Hijazi, M.; Velasquez, S.M.; Jamet, E.; Estevez, J.M.; Albenne, C. An update on post-translational modifications of hydroxyproline-rich glycoproteins: Toward a model highlighting their contribution to plant cell wall architecture. Front. Plant Sci. 2014, 5, 395. [Google Scholar] [CrossRef]
- Kieliszewski, M.J.; Lamport, D.T.A.; Tan, L.; Cannon, M.C. Hydroxyproline-Rich Glycoproteins: Form and Function. Annu. Plant Rev. 2010, 41, 321–342. [Google Scholar] [CrossRef]
- Johnson, K.L.; Cassin, A.M.; Lonsdale, A.; Bacic, A.; Doblin, M.S.; Schultz, C.J. A motif and amino acid bias bioinformatics pipeline to identify hydroxyproline-rich glycoproteins. Plant Physiol. 2017, 174, 886–903. [Google Scholar] [CrossRef]
- Ellis, M.; Egelund, J.; Schultz, C.J.; Bacic, A. Arabinogalactan-Proteins: Key Regulators at the Cell Surface? Plant Physiol. 2010, 153, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Showalter, A.M.; Keppler, B.; Lichtenberg, J.; Gu, D.; Welch, L.R. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 2010, 153, 485–513. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Xu, J.; Yang, J.; Kieliszewski, M.J.; Showalter, A.M. The Lysine-rich Arabinogalactan-protein Subfamily in Arabidopsis: Gene Expression, Glycoprotein Purification and Biochemical Characterization. Plant Cell Physiol. 2005, 46, 975–984. [Google Scholar] [CrossRef][Green Version]
- Baldwin, T.C.; Domingo, C.; Schindler, T.; Seetharaman, G.; Stacey, N.; Roberts, K. DcAGP1, a secreted arabinogalactan protein, is related to a family of basic proline-rich proteins. Plant Mol. Biol. 2001, 45, 421–435. [Google Scholar] [CrossRef]
- Simonović, A.D.; Dragićević, M.B.; Bogdanović, M.D.; Trifunović-Momčilov, M.M.; Subotić, A.R.; Todorović, S.I. DUF1070 as a signature domain of a subclass of arabinogalactan peptides. Arch. Biol. Sci. 2016, 68, 737–746. [Google Scholar] [CrossRef]
- Simonović, A.D.; Filipović, B.K.; Trifunović, M.M.; Malkov, S.N.; Milinković, V.P.; Jevremović, S.B.; Subotić, A.R. Plant regeneration in leaf culture of Centaurium erythraea Rafn. Part 2: The role of arabinogalactan proteins. Plant Cell Tissue Organ Cult. 2015, 121, 721–739. [Google Scholar] [CrossRef]
- Showalter, A.M.; Keppler, B.D.; Liu, X.; Lichtenberg, J.; Welch, L.R. Bioinformatic Identification and Analysis of Hydroxyproline-Rich Glycoproteins in Populus trichocarpa. BMC Plant Biol. 2016, 16, 229. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yan, C.; Li, H.; Wu, W.; Liu, Y.; Wang, Y.; Chen, Q.; Ma, H. Bioinformatics Prediction and Evolution Analysis of Arabinogalactan Proteins in the Plant Kingdom. Front. Plant Sci. 2017, 8, 66. [Google Scholar] [CrossRef] [PubMed]
- Dragićević, M.B.; Paunović, D.M.; Bogdanović, M.D.; Todorović, S.I.; Simonović, A.D. ragp: Pipeline for mining of plant hydroxyproline-rich glycoproteins with implementation in R. Glycobiology 2020, 30, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Seifert, G.J.; Roberts, K. The Biology of Arabinogalactan Proteins. Annu. Rev. Plant Biol. 2007, 58, 137–161. [Google Scholar] [CrossRef] [PubMed]
- Majewska-Sawka, A.; Nothnagel, E.A. The Multiple Roles of Arabinogalactan Proteins in Plant Development. Plant Physiol. 2000, 122, 3–10. [Google Scholar] [CrossRef]
- Pereira, A.M.; Pereira, L.G.; Coimbra, S. Arabinogalactan proteins: Rising attention from plant biologists. Plant Reprod. 2015, 28, 1–15. [Google Scholar] [CrossRef]
- Costa, M.; Pereira, A.M.; Pinto, S.C.; Silva, J.; Pereira, L.G.; Coimbra, S. In silico and expression analyses of fasciclin-like arabinogalactan proteins reveal functional conservation during embryo and seed development. Plant Reprod. 2019, 32, 353–370. [Google Scholar] [CrossRef]
- Lamport, D.; Kieliszewski, M.; Showalter, A. Salt stress upregulates periplasmic arabinogalactan proteins: Using salt stress to analyse AGP function. New Phytol. 2006, 169, 479–492. [Google Scholar] [CrossRef]
- Meng, J.; Hu, B.; Yi, G.; Li, X.; Chen, H.; Wang, Y.; Yuan, W.; Xing, Y.; Sheng, Q.; Su, Z.; et al. Genome-wide analyses of banana fasciclin-like AGP genes and their differential expression under low-temperature stress in chilling sensitive and tolerant cultivars. Plant Cell Rep. 2020, 39, 693–708. [Google Scholar] [CrossRef]
- Gilson, P.; Gaspar, Y.M.; Oxley, D.; Youl, J.J.; Bacic, A. NaAGP4 is an arabinogalactan protein whose expression is suppressed by wounding and fungal infection in Nicotiana alata. Protoplasma 2001, 215, 128–139. [Google Scholar] [CrossRef]
- Liu, C.; Mehdy, M.C. A Nonclassical Arabinogalactan Protein Gene Highly Expressed in Vascular Tissues, AGP31, Is Transcriptionally Repressed by Methyl Jasmonic Acid in Arabidopsis. Plant Physiol. 2007, 145, 863–874. [Google Scholar] [CrossRef]
- Fragkostefanakis, S.; Dandachi, F.; Kalaitzis, P. Expression of arabinogalactan proteins during tomato fruit ripening and in response to mechanical wounding, hypoxia and anoxia. Plant Physiol. Biochem. 2012, 52, 112–118. [Google Scholar] [CrossRef]
- Nguema-Ona, E.; Vicré-Gibouin, M.; Cannesan, M.A.; Driouich, A. Arabinogalactan proteins in root-microbe interactions. Trends Plant Sci. 2013, 18, 440–449. [Google Scholar] [CrossRef]
- Šiler, B.; Živković, S.; Banjanac, T.; Cvetković, J.; Nestorović Živković, J.; Ćirić, A.; Soković, M.; Mišić, D. Centauries as underestimated food additives: Antioxidant and antimicrobial potential. Food Chem. 2014, 147, 367–376. [Google Scholar] [CrossRef]
- Filipović, B.K.; Simonović, A.D.; Trifunović, M.M.; Dmitrović, S.S.; Savić, J.M.; Jevremović, S.B.; Subotić, A.R. Plant regeneration in leaf culture of Centaurium erythraea Rafn. Part 1: The role of antioxidant enzymes. Plant Cell Tissue Organ Cult. 2015, 121, 703–719. [Google Scholar] [CrossRef]
- Simonović, A.D.; Trifunović-Momčilov, M.M.; Filipović, B.K.; Marković, M.P.; Bogdanović, M.D.; Subotić, A.R. Somatic Embryogenesis in Centaurium erythraea Rafn—Current Status and Perspectives: A Review. Plants 2021, 10, 70. [Google Scholar] [CrossRef] [PubMed]
- Trifunović, M.; Tadić, V.; Petrić, M.; Jontulović, D.; Jevremović, S.; Subotić, A. Quantification of arabinogalactan proteins during in vitro morphogenesis induced by β-d-glucosyl Yariv reagent in Centaurium erythraea root culture. Acta Physiol. Plant. 2014, 36, 1187–1195. [Google Scholar] [CrossRef]
- Trifunović, M.; Subotić, A.; Petrić, M.; Jevremović, S. The Role of Arabinogalactan Proteins in Morphogenesis of Centaurium erythraea Rafn In Vitro. In The Gentianaceae—Volume 2: Biotechnology and Applications; Rybczyński, J.J., Davey, M.R., Mikuła, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 113–138. [Google Scholar]
- Filipović, B.K.; Trifunović-Momčilov, M.M.; Simonović, A.D.; Jevremović, S.B.; Milošević, S.M.; Subotić, A.R. Immunolocalization of some arabinogalactan protein epitopes during indirect somatic embryogenesis and shoot organogenesis in leaf culture of centaury (Centaurium erythraea Rafn). In Vitro Cell. Dev. Biol. Plant 2021, 57, 470–480. [Google Scholar] [CrossRef]
- Lup, S.D.; Tian, X.; Xu, J.; Pérez-Pérez, J.M. Wound signaling of regenerative cell reprogramming. Plant Sci. 2016, 250, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Hernández, H.A.; Ledezma-Rodríguez, M.; Avilez-Montalvo, R.N.; Juárez-Gómez, Y.L.; Skeete, A.; Avilez-Montalvo, J.; De-la-Peña, C.; Loyola-Vargas, V.M. Signaling Overview of Plant Somatic Embryogenesis. Front. Plant Sci. 2019, 10, 77. [Google Scholar] [CrossRef]
- Santarem, E.R.; Pelissier, B.; Finer, J.J. Effect of explant orientation, pH, solidifying agent and wounding on initiation of soybean somatic embryos. In Vitro Cell. Dev. Biol. Plant 1997, 33, 13–19. [Google Scholar] [CrossRef]
- Bhatia, P.; Ashwath, N.; Midmore, D.J. Effects of genotype, explant orientation, and wounding on shoot regeneration in tomato. In Vitro Cell. Dev. Biol. Plant 2005, 41, 457–464. [Google Scholar] [CrossRef]
- Ćuković, K.; Dragićević, M.; Bogdanović, M.; Paunović, D.; Giurato, G.; Filipović, B.; Subotić, A.; Todorović, S.; Simonović, A. Plant regeneration in leaf culture of Centaurium erythraea Rafn. Part 3: De novo transcriptome assembly and validation of housekeeping genes for studies of in vitro morphogenesis. Plant Cell Tissue Organ Cult. 2020, 141, 417–433. [Google Scholar] [CrossRef]
- He, J.; Zhao, H.; Cheng, Z.; Ke, Y.; Liu, J.; Ma, H. Evolution Analysis of the Fasciclin-Like Arabinogalactan Proteins in Plants Shows Variable Fasciclin-AGP Domain Constitutions. Int. J. Mol. Sci. 2019, 20, 1945. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Smith, S.A. Optimizing de novo assembly of short-read RNA-seq data for phylogenomics. BMC Genom. 2013, 14, 328. [Google Scholar] [CrossRef]
- Gruenheit, N.; Deusch, O.; Esser, C.; Becker, M.; Voelckel, C.; Lockhart, P. Cutoffs and k-mers: Implications from a transcriptome study in allopolyploid plants. BMC Genom. 2012, 13, 92. [Google Scholar] [CrossRef]
- Ono, H.; Ishii, K.; Kozaki, T.; Ogiwara, I.; Kanekatsu, M.; Yamada, T. Removal of redundant contigs from de novo RNA-Seq assemblies via homology search improves accurate detection of differentially expressed genes. BMC Genom. 2015, 16, 1031. [Google Scholar] [CrossRef]
- Kerkvliet, J.; de Fouchier, A.; van Wijk, M.; Groot, A.T. The Bellerophon pipeline, improving de novo transcriptomes and removing chimeras. Ecol. Evol. 2019, 9, 10513–10521. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.L.; Cassin, A.M.; Lonsdale, A.; Wong, G.K.-S.; Soltis, D.E.; Miles, N.W.; Melkonian, M.; Melkonian, B.; Deyholos, M.K.; Leebens-Mack, J.; et al. Insights into the Evolution of Hydroxyproline-Rich Glycoproteins from 1000 Plant Transcriptomes. Plant Physiol. 2017, 174, 904–921. [Google Scholar] [CrossRef]
- Johnson, K.L.; Jones, B.J.; Bacic, A.; Schultz, C.J. The fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiol. 2003, 133, 1911–1925. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, L.; Shafee, T.; Johnson, K.L.; Bacic, A.; Classen, B. Arabinogalactan-proteins of Zostera marina L. contain unique glycan structures and provide insight into adaption processes to saline environments. Sci. Rep. 2020, 10, 8232. [Google Scholar] [CrossRef] [PubMed]
- Mashiguchi, K.; Yamaguchi, I.; Suzuki, Y. Isolation and Identification of Glycosylphosphatidylinositol-Anchored Arabinogalactan Proteins and Novel β-Glucosyl Yariv-Reactive Proteins from Seeds of Rice (Oryza sativa). Plant Cell Physiol. 2004, 45, 1817–1829. [Google Scholar] [CrossRef]
- Seifert, G.J. Fascinating Fasciclins: A Surprisingly Widespread Family of Proteins that Mediate Interactions between the Cell Exterior and the Cell Surface. Int. J. Mol. Sci. 2018, 19, 1628. [Google Scholar] [CrossRef] [PubMed]
- Shafee, T.; Bacic, A.; Johnson, K. Evolution of Sequence-Diverse Disordered Regions in a Protein Family: Order within the Chaos. Mol. Biol. Evol. 2020, 37, 2155–2172. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Fujita, M. Thematic Review Series: Glycosylphosphatidylinositol (GPI) Anchors: Biochemistry and Cell Biology Biosynthesis of GPI-anchored proteins: Special emphasis on GPI lipid remodeling. J. Lipid Res. 2016, 57, 6–24. [Google Scholar] [CrossRef]
- Galian, C.; Björkholm, P.; Bulleid, N.; von Heijne, G. Efficient Glycosylphosphatidylinositol (GPI) Modification of Membrane Proteins Requires a C-terminal Anchoring Signal of Marginal Hydrophobicity*. J. Biol. Chem. 2012, 287, 16399–16409. [Google Scholar] [CrossRef]
- Hervé, C.; Siméon, A.; Jam, M.; Cassin, A.; Johnson, K.L.; Salmeán, A.A.; Willats, W.G.T.; Doblin, M.S.; Bacic, A.; Kloareg, B. Arabinogalactan proteins have deep roots in eukaryotes: Identification of genes and epitopes in brown algae and their role in Fucus serratus embryo development. New Phytol. 2016, 209, 1428–1441. [Google Scholar] [CrossRef]
- Chen, Z. A superfamily of proteins with novel cysteine-rich repeats. Plant Physiol. 2001, 126, 473–476. [Google Scholar] [CrossRef]
- Zuo, C.; Liu, H.; Lv, Q.; Chen, Z.; Tian, Y.; Mao, J.; Chu, M.; Ma, Z.; An, Z.; Chen, B. Genome-Wide Analysis of the Apple (Malus domestica) Cysteine-Rich Receptor-Like Kinase (CRK) Family: Annotation, Genomic Organization, and Expression Profiles in Response to Fungal Infection. Plant Mol. Biol. Report. 2020, 38, 14–24. [Google Scholar] [CrossRef]
- Malkov, S.N.; Simonović, A.D. Shotgun assembly of Centaurium erythraea transcriptome. In Proceedings of the 19th Symposium of the Serbian Plant Physiology Society, Book of Abstracts, Banja Vrujci, Serbia, 13–15 June 2011; p. 16. [Google Scholar]
- Borassi, C.; Gloazzo Dorosz, J.; Ricardi, M.M.; Carignani Sardoy, M.; Pol Fachin, L.; Marzol, E.; Mangano, S.; Rodríguez Garcia, D.R.; Martínez Pacheco, J.; Rondón Guerrero, Y.D.C.; et al. A cell surface arabinogalactan-peptide influences root hair cell fate. New Phytol. 2020, 227, 732–743. [Google Scholar] [CrossRef] [PubMed]
- Savatin, D.V.; Gramegna, G.; Modesti, V.; Cervone, F. Wounding in the plant tissue: The defense of a dangerous passage. Front. Plant Sci. 2014, 5, 470. [Google Scholar] [CrossRef]
- Saini, S.; Sharma, I.; Pati, P.K. Versatile roles of brassinosteroid in plants in the context of its homoeostasis, signaling and crosstalks. Front. Plant Sci. 2015, 6, 950. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Nothnagel, E.A. Binding of Arabinogalactan Proteins by Yariv Phenylglycoside Triggers Wound-Like Responses in Arabidopsis Cell Cultures. Plant Physiol. 2004, 135, 1346–1366. [Google Scholar] [CrossRef] [PubMed]
- MacMillan, C.P.; Mansfield, S.D.; Stachurski, Z.H.; Evans, R.; Southerton, S.G. Fasciclin-like arabinogalactan proteins: Specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J. 2010, 62, 689–703. [Google Scholar] [CrossRef]
- Konieczny, R.; Swierczyńska, J.; Czaplicki, A.Z.; Bohdanowicz, J. Distribution of pectin and arabinogalactan protein epitopes during organogenesis from androgenic callus of wheat. Plant Cell Rep. 2007, 26, 355–363. [Google Scholar] [CrossRef]
- Wiśniewska, E.; Majewska-Sawka, A. Arabinogalactan-proteins stimulate the organogenesis of guard cell protoplasts-derived callus in sugar beet. Plant Cell Rep. 2007, 26, 1457–1467. [Google Scholar] [CrossRef]
- Orbović, V.; Göllner, E.M.; Soria, P. The effect of arabinogalactan proteins on regeneration potential of juvenile citrus explants used for genetic transformation by Agrobacterium tumefaciens. Acta Physiol. Plant. 2013, 35, 1409–1419. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction—The Phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef] [PubMed]
- Gíslason, M.H.; Nielsen, H.; Almagro Armenteros, J.J.; Johansen, A.R. Prediction of GPI-anchored proteins with pointer neural networks. Curr. Res. Biotechnol. 2021, 3, 6–13. [Google Scholar] [CrossRef]
- Walsh, I.; Martin, A.J.M.; Di Domenico, T.; Tosatto, S.C.E. ESpritz: Accurate and fast prediction of protein disorder. Bioinformatics 2011, 28, 503–509. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Gašić, K.; Hernandez, A.; Korban, S.S. RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol. Biol. Report. 2004, 22, 437–438. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Drost, H.-G.; Paszkowski, J. Biomartr: Genomic data retrieval with R. Bioinformatics 2017, 33, 1216–1217. [Google Scholar] [CrossRef]
- Wright, E.S. DECIPHER: Harnessing local sequence context to improve protein multiple sequence alignment. BMC Bioinform. 2015, 16, 322. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Whelan, S.; Goldman, N. A General Empirical Model of Protein Evolution Derived from Multiple Protein Families Using a Maximum-Likelihood Approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Le, S.Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Dayhoff, M.O.; Schwartz, R.M.; Orcutt, B.C. A model of evolutionary change in proteins. In Atlas of Protein Sequence and Structure; Dayhoff, M.O., Ed.; The National Biomedical Research Foundation: Washington, DC, USA, 1978; Volume 5, pp. 345–352. [Google Scholar]
- Henikoff, S.; Henikoff, J.G. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 1992, 89, 10915–10919. [Google Scholar] [CrossRef] [PubMed]
- Schliep, K.P. phangorn: Phylogenetic analysis in R. Bioinformatics 2010, 27, 592–593. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, S. kmer: An R Package for Fast Alignment-Free Clustering of Biological Sequences; R Package Version 1.1.1. Available online: https://CRAN.R-project.org/package=kmer (accessed on 12 April 2021).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 23 March 2021).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. gplots: Various R Programming Tools for Plotting Data. R Package Version 3.1.1. 2020. Available online: https://CRAN.R-project.org/package=gplots (accessed on 23 March 2021).
- Pardy, C. mpmi: Mixed-Pair Mutual Information Estimators. R Package Version 0.43.1. 2020. Available online: https://CRAN.R-project.org/package=mpmi (accessed on 23 March 2021).






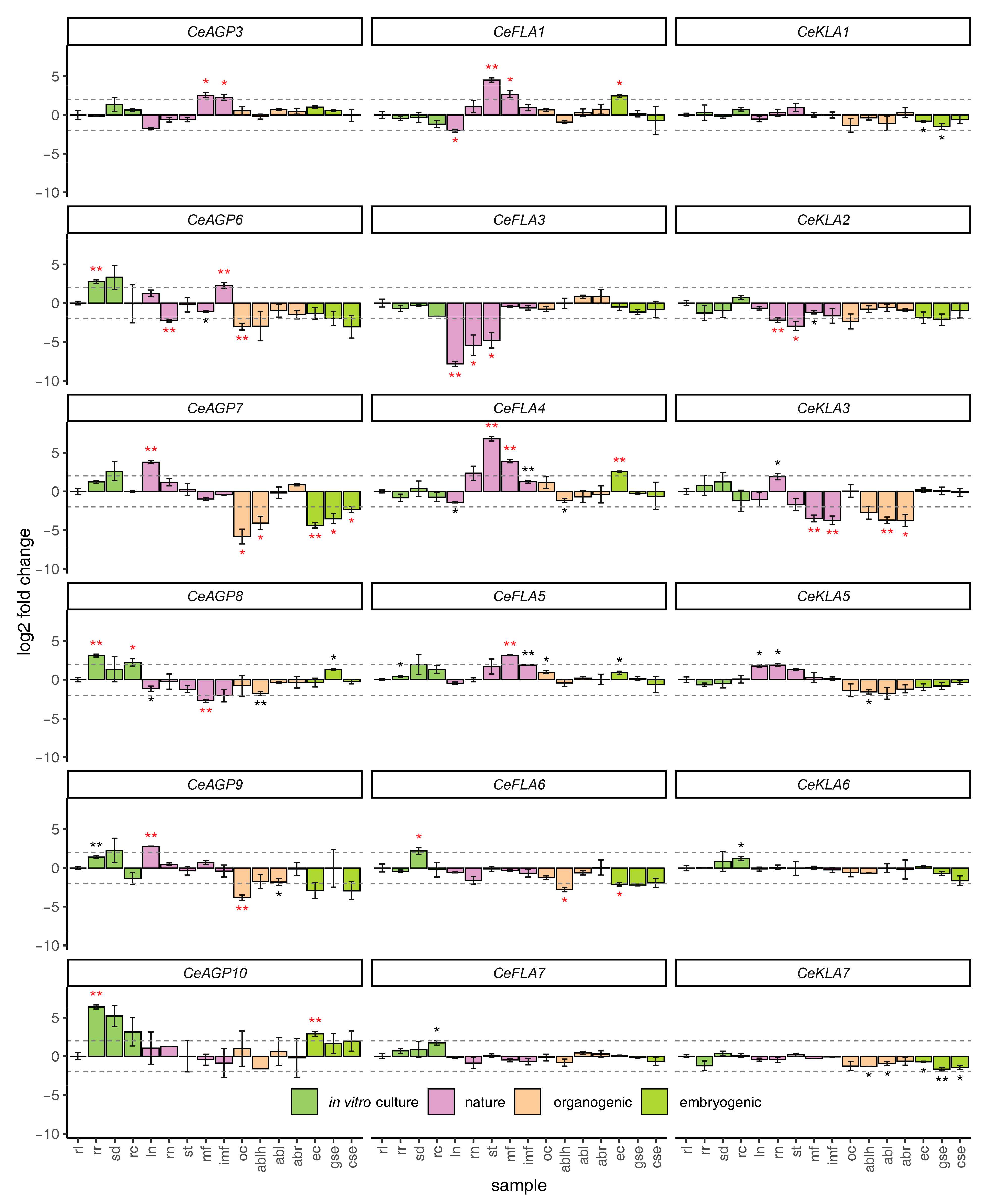

| Sample | Treatment | Light Conditions | Abbreviation | |
|---|---|---|---|---|
| Plants and roots grown in vitro | rosette leaves | Solid hormone-free medium | 16 h light 8 h darkness | rl |
| rosette roots | rr | |||
| seedlings | sd | |||
| root cultures | rc | |||
| Flowering plants from nature | leaves | none | natural | ln |
| roots | rn | |||
| stems | st | |||
| mature flowers | mf | |||
| immature flowers | imf | |||
| Organogenesis | organogenic callus | 0.2 mgl−1 2,4-D 0.5 mgl−1 CPPU | 16 h light 8 h darkness | oc |
| adventitious buds formed on leaf explants | ablh | |||
| adventitious buds formed on leaf explants | hormone-free medium | abl | ||
| adventitious buds formed on root explants | abr | |||
| Somatic embryogenesis | embryogenic callus | 0.2 mgl−1 2,4-D 0.5 mgl−1 CPPU | darkness | ec |
| globular embryos | gse | |||
| cotyledonary embryos | cse |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paunović, D.M.; Ćuković, K.B.; Bogdanović, M.D.; Todorović, S.I.; Trifunović-Momčilov, M.M.; Subotić, A.R.; Simonović, A.D.; Dragićević, M.B. The Arabinogalactan Protein Family of Centaurium erythraea Rafn. Plants 2021, 10, 1870. https://doi.org/10.3390/plants10091870
Paunović DM, Ćuković KB, Bogdanović MD, Todorović SI, Trifunović-Momčilov MM, Subotić AR, Simonović AD, Dragićević MB. The Arabinogalactan Protein Family of Centaurium erythraea Rafn. Plants. 2021; 10(9):1870. https://doi.org/10.3390/plants10091870
Chicago/Turabian StylePaunović, Danijela M., Katarina B. Ćuković, Milica D. Bogdanović, Slađana I. Todorović, Milana M. Trifunović-Momčilov, Angelina R. Subotić, Ana D. Simonović, and Milan B. Dragićević. 2021. "The Arabinogalactan Protein Family of Centaurium erythraea Rafn" Plants 10, no. 9: 1870. https://doi.org/10.3390/plants10091870
APA StylePaunović, D. M., Ćuković, K. B., Bogdanović, M. D., Todorović, S. I., Trifunović-Momčilov, M. M., Subotić, A. R., Simonović, A. D., & Dragićević, M. B. (2021). The Arabinogalactan Protein Family of Centaurium erythraea Rafn. Plants, 10(9), 1870. https://doi.org/10.3390/plants10091870













