Transcriptomic and Physiological Response of Durum Wheat Grain to Short-Term Heat Stress during Early Grain Filling
Abstract
:1. Introduction
2. Materials and Methods
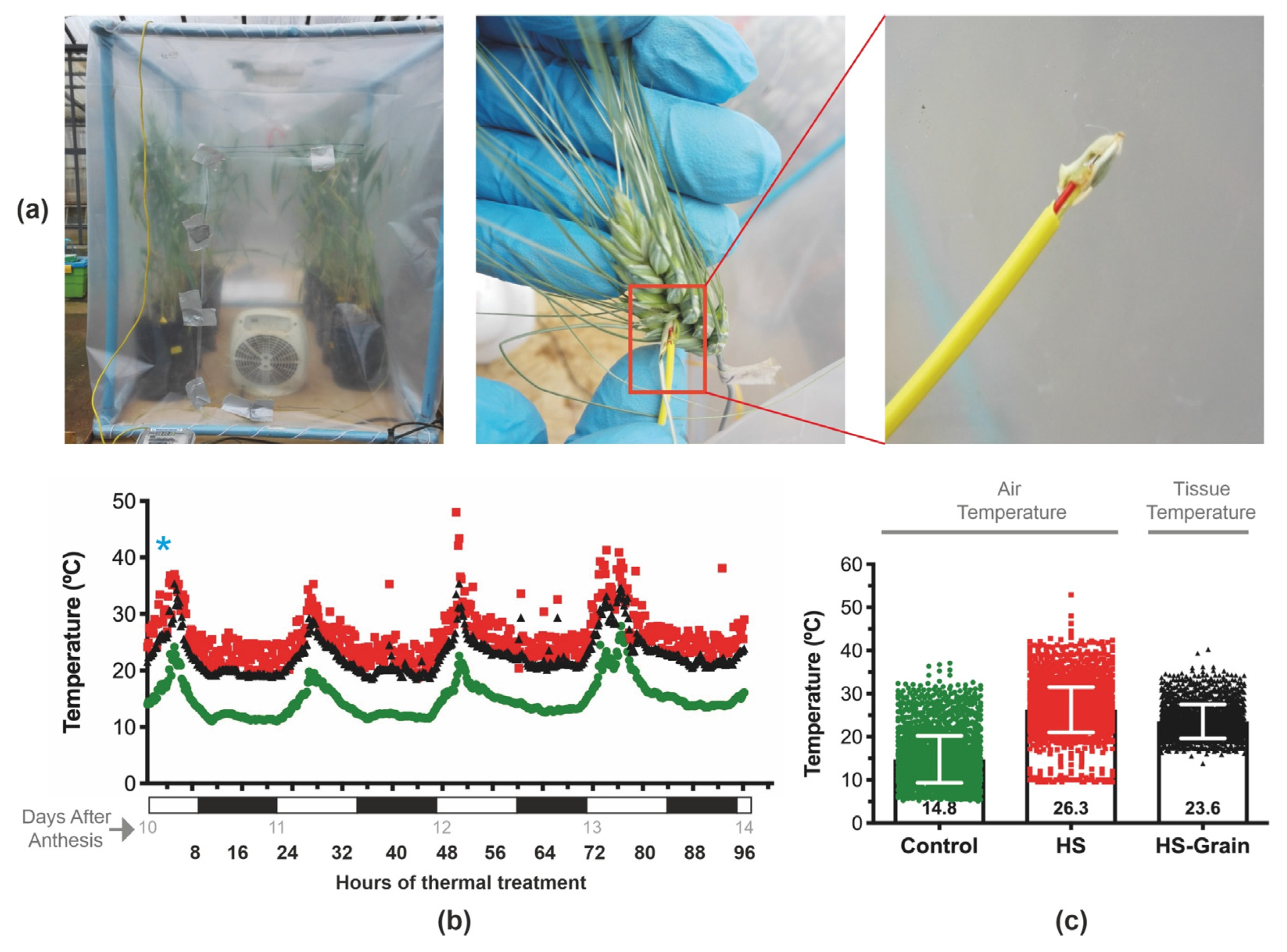
2.1. Plant Material and Thermal Treatments
2.2. Plant Sampling and Physiological Measurements
2.3. Grain Starch and Protein Concentration
2.4. RNA Isolation and Quantitative Real-Time PCR Analysis
2.5. RNA-Seq Analysis
2.6. Gene Ontology (GO) Enrichment Analysis
2.7. Transcription Factor Binding Site (TFBS) Enrichment Analysis
3. Results and Discussion
3.1. Environmental Conditions and Temperature Increment
3.2. Seed Size, Grain Weight, and Grain Quality Were Negatively Affected by Short HS at Early Grain Filling
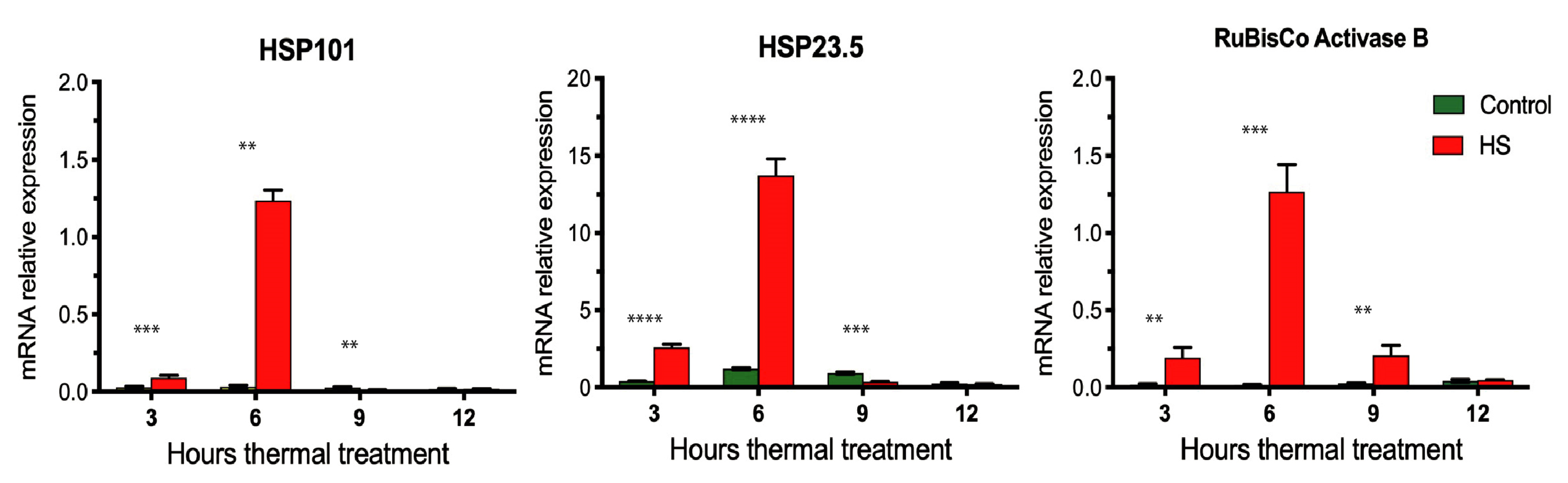
3.3. Temporal Expression Profiles of HS Marker Genes in Durum Wheat Grains
3.4. The Initial Transcriptomic Response to HS in Grain Is Characterized by the Induction of Chaperones Together with the Inhibition of Related Genes to Proteolysis and Transcriptional Regulation
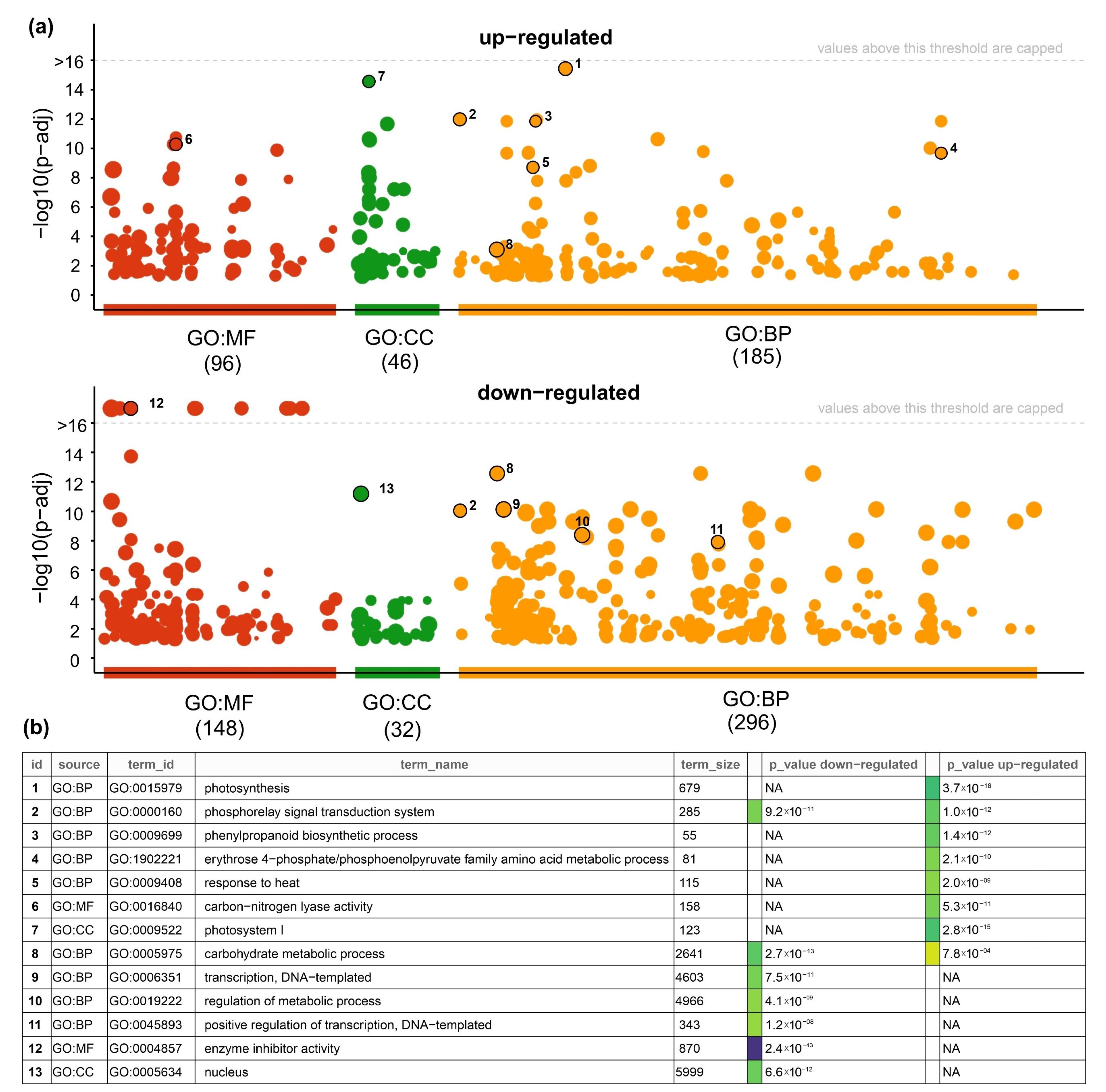
3.5. Gene Ontology (GOs) Analysis of DEGs under Short Heat Stress in Grains of Durum Wheat
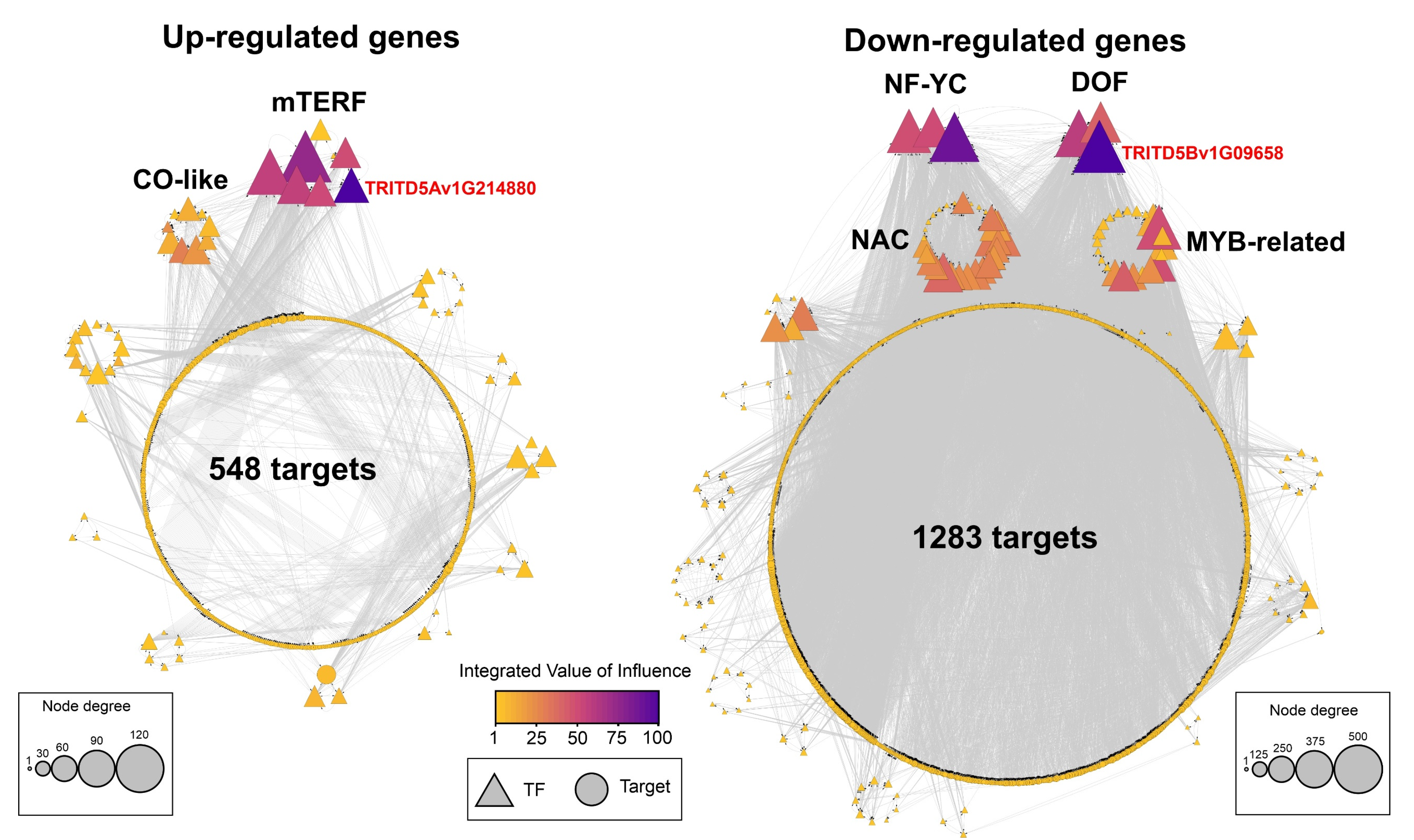
3.6. Identification of Regulatory Factors Associated with the Response to HS in Grains of Durum Wheat
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ray, D.K.; Ramankutty, N.; Mueller, N.D.; West, P.C.; Foley, J.A. Recent Patterns of Crop Yield Growth and Stagnation. Nat. Commun. 2012, 3, 1293. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; et al. Temperature Increase Reduces Global Yields of Major Crops in Four Independent Estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, R.A.; Connor, D.J. Issues for Cropping and Agricultural Science in the next 20 Years. Field Crops Res. 2018, 222, 121–142. [Google Scholar] [CrossRef]
- Masson-Delmotte, V.; Zhai, P.; Pirani, A.; Connors, S.L.; Péan, C.; Berger, S.; Caud, N.; Chen, Y.; Goldfarb, L.; Gomis, M.I.; et al. Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK, 2021. [Google Scholar]
- O’Leary, G.J.; Aggarwal, P.K.; Calderini, D.F.; Connor, D.J.; Craufurd, P.; Eigenbrode, S.D.; Han, X.; Hatfield, J.L. Challenges and Responses to Ongoing and Projected Climate Change for Dryland Cereal Production Systems throughout the World. Agronomy 2018, 8, 34. [Google Scholar] [CrossRef] [Green Version]
- Akter, N.; Rafiqul Islam, M. Heat Stress Effects and Management in Wheat: A Review. Agron. Sustain. Dev. 2017, 37, 37. [Google Scholar] [CrossRef]
- Balla, K.; Karsai, I.; Bónis, P.; Kiss, T.; Berki, Z.; Horváth, Á.; Mayer, M.; Bencze, S.; Veisz, O. Heat Stress Responses in a Large Set of Winter Wheat Cultivars (Triticum aestivum L.) Depend on the Timing and Duration of Stress. PLoS ONE 2019, 14, e0222639. [Google Scholar] [CrossRef] [PubMed]
- Lizana, X.C.; Calderini, D.F. Yield and Grain Quality of Wheat in Response to Increased Temperatures at Key Periods for Grain Number and Grain Weight Determination: Considerations for the Climatic Change Scenarios of Chile. J. Agric. Sci. 2013, 151, 209–221. [Google Scholar] [CrossRef]
- García, G.A.; Dreccer, M.F.; Miralles, D.J.; Serrago, R.A. High Night Temperatures during Grain Number Determination Reduce Wheat and Barley Grain Yield: A Field Study. Glob. Change Biol. 2015, 21, 4153–4164. [Google Scholar] [CrossRef] [PubMed]
- García, G.A.; Serrago, R.A.; Dreccer, M.F.; Miralles, D.J. Post-Anthesis Warm Nights Reduce Grain Weight in Field-Grown Wheat and Barley. Field Crops Res. 2016, 195, 50–59. [Google Scholar] [CrossRef]
- Elía, M.; Slafer, G.A.; Savin, R. Yield and Grain Weight Responses to Post-Anthesis Increases in Maximum Temperature under Field Grown Wheat as Modified by Nitrogen Supply. Field Crops Res. 2018, 221, 228–237. [Google Scholar] [CrossRef] [Green Version]
- Barlow, K.; Christy, B.; O’Leary, G.; Riffkin, P.; Nuttall, J. Simulating the Impact of Extreme Heat and Frost Events on Wheat Crop Production: A Review. Field Crops Res. 2015, 171, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Fischer, R.A. Number of Kernels in Wheat Crops and the Influence of Solar Radiation and Temperature. J. Agric. Sci. 1985, 105, 447–461. [Google Scholar] [CrossRef]
- Savin, R.; Slafer, G.A. Shading Effects on the Yield of an Argentinian Wheat Cultivar. J. Agric. Sci. 1991, 116, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Calderini, D.F.; Castillo, F.M.; Arenas-M, A.; Molero, G.; Reynolds, M.P.; Craze, M.; Bowden, S.; Milner, M.J.; Wallington, E.J.; Dowle, A.; et al. Overcoming the Trade-off between Grain Weight and Number in Wheat by the Ectopic Expression of Expansin in Developing Seeds Leads to Increased Yield Potential. New Phytol. 2021, 230, 629–640. [Google Scholar] [CrossRef]
- Tashiro, T.; Wardlaw, I.F. The Effect of High Temperature at Different Stages of Ripening on Grain Set, Grain Weight and Grain Dimensions in the Semi-Dwarf Wheat ‘Banks’. Ann. Bot. 1990, 65, 51–61. [Google Scholar] [CrossRef]
- Stone, P.; Nicolas, M. Effect of Timing of Heat Stress During Grain Filling on Two Wheat Varieties Differing in Heat Tolerance. I. Grain Growth. Funct. Plant Biol. 1995, 22, 927–934. [Google Scholar] [CrossRef]
- Herrera, J.; Calderini, D.F. Pericarp Growth Dynamics Associate with Final Grain Weight in Wheat under Contrasting Plant Densities and Increased Night Temperature. Ann. Bot. 2020, 126, 1063–1076. [Google Scholar] [CrossRef] [PubMed]
- Hasan, A.K.; Herrera, J.; Lizana, C.; Calderini, D.F. Carpel Weight, Grain Length and Stabilized Grain Water Content Are Physiological Drivers of Grain Weight Determination of Wheat. Field Crops Res. 2011, 123, 241–247. [Google Scholar] [CrossRef]
- Brinton, J.; Uauy, C. A Reductionist Approach to Dissecting Grain Weight and Yield in Wheat. J. Integr. Plant Biol. 2019, 61, 337–358. [Google Scholar] [CrossRef] [Green Version]
- Stone, P.; Nicolas, M. Wheat Cultivars Vary Widely in Their Responses of Grain Yield and Quality to Short Periods of Post-Anthesis Heat Stress. Funct. Plant Biol. 1994, 21, 887–900. [Google Scholar] [CrossRef]
- Dubcovsky, J.; Dvorak, J. Genome Plasticity a Key Factor in the Success of Polyploid Wheat under Domestication. Science 2007, 316, 1862–1866. [Google Scholar] [CrossRef] [Green Version]
- Qin, D.; Wu, H.; Peng, H.; Yao, Y.; Ni, Z.; Li, Z.; Zhou, C.; Sun, Q. Heat Stress-Responsive Transcriptome Analysis in Heat Susceptible and Tolerant Wheat (Triticum aestivum L.) by Using Wheat Genome Array. BMC Genom. 2008, 9, 432. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, H.; Khurana, N.; Tyagi, A.K.; Khurana, J.P.; Khurana, P. Identification and Characterization of High Temperature Stress Responsive Genes in Bread Wheat (Triticum aestivum L.) and Their Regulation at Various Stages of Development. Plant Mol. Biol. 2011, 75, 35–51. [Google Scholar] [CrossRef]
- Liu, Z.; Xin, M.; Qin, J.; Peng, H.; Ni, Z.; Yao, Y.; Sun, Q. Temporal Transcriptome Profiling Reveals Expression Partitioning of Homeologous Genes Contributing to Heat and Drought Acclimation in Wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, Y.; Poole, R.L.; Huttly, A.K.; Toscano-Underwood, C.; Feeney, K.; Welham, S.; Gooding, M.J.; Mills, C.; Edwards, K.J.; Shewry, P.R.; et al. Transcriptome Analysis of Grain Development in Hexaploid Wheat. BMC Genom. 2008, 9, 121. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Chen, S.; Shi, X.; Liu, D.; Zhao, P.; Lu, Y.; Cheng, Y.; Liu, Z.; Nie, X.; Song, W.; et al. Hybrid Sequencing Reveals Insight into Heat Sensing and Signaling of Bread Wheat. Plant J. 2019, 98, 1015–1032. [Google Scholar] [CrossRef] [PubMed]
- Kino, R.I.; Pellny, T.K.; Mitchell, R.A.C.; Gonzalez-Uriarte, A.; Tosi, P. High Post-Anthesis Temperature Effects on Bread Wheat (Triticum aestivum L.) Grain Transcriptome during Early Grain-Filling. BMC Plant Biol. 2020, 20, 170. [Google Scholar] [CrossRef]
- Girousse, C.; Roche, J.; Guerin, C.; Le Gouis, J.; Balzegue, S.; Mouzeyar, S.; Bouzidi, M.F. Coexpression Network and Phenotypic Analysis Identify Metabolic Pathways Associated with the Effect of Warming on Grain Yield Components in Wheat. PLoS ONE 2018, 13, e0199434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aprile, A.; Havlickova, L.; Panna, R.; Marè, C.; Borrelli, G.M.; Marone, D.; Perrotta, C.; Rampino, P.; De Bellis, L.; Curn, V.; et al. Different Stress Responsive Strategies to Drought and Heat in Two Durum Wheat Cultivars with Contrasting Water Use Efficiency. BMC Genom. 2013, 14, 821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K.; et al. Wild Emmer Genome Architecture and Diversity Elucidate Wheat Evolution and Domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum Wheat Genome Highlights Past Domestication Signatures and Future Improvement Targets. Nat. Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- IWGSC; Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the Limits in Wheat Research and Breeding Using a Fully Annotated Reference Genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Able, A.J.; Able, J.A. Integrated Analysis of Small RNA, Transcriptome, and Degradome Sequencing Reveals the Water-Deficit and Heat Stress Response Network in Durum Wheat. Int. J. Mol. Sci. 2020, 21, 6017. [Google Scholar] [CrossRef]
- Girousse, C.; Inchboard, L.; Deswarte, J.-C.; Chenu, K. How Does Post-Flowering Heat Impact Grain Growth and Its Determining Processes in Wheat? J. Exp. Bot. 2021, 72, 6596–6610. [Google Scholar] [CrossRef] [PubMed]
- Gnan, S.; Priest, A.; Kover, P.X. The Genetic Basis of Natural Variation in Seed Size and Seed Number and Their Trade-Off Using Arabidopsis Thaliana MAGIC Lines. Genetics 2014, 198, 1751–1758. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, R.K.; Wilkins, O. Single Cell Gene Regulatory Networks in Plants: Opportunities for Enhancing Climate Change Stress Resilience. Plant Cell Environ. 2021, 44, 2006–2017. [Google Scholar] [CrossRef]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A Decimal Code for the Growth Stages of Cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Calderini, D.F.; Abeledo, L.G.; Savin, R.; Slafer, G.A. Effect of Temperature and Carpel Size during Pre-Anthesis on Potential Grain Weight in Wheat. J. Agric. Sci. 1999, 132, 453–459. [Google Scholar] [CrossRef] [Green Version]
- Prasad, P.V.V.; Djanaguiraman, M. Response of Floret Fertility and Individual Grain Weight of Wheat to High Temperature Stress: Sensitive Stages and Thresholds for Temperature and Duration. Funct. Plant Biol. 2014, 41, 1261–1269. [Google Scholar] [CrossRef] [Green Version]
- Savin, R.; Stone, P.; Nicolas, M. Responses of Grain Growth and Malting Quality of Barley to Short Periods of High Temperature in Field Studies Using Portable Chambers. Aust. J. Agric. Res. 1996, 47, 465–477. [Google Scholar] [CrossRef]
- Merrill, A.L.; Watt, B.K. Agriculture Handbook, 74th ed.; Agricultural Research Service, USDA: Washington, DC, USA, 1973. [Google Scholar]
- Yaffe, H.; Buxdorf, K.; Shapira, I.; Ein-Gedi, S.; Moyal-Ben Zvi, M.; Fridman, E.; Moshelion, M.; Levy, M. LogSpin: A Simple, Economical and Fast Method for RNA Isolation from Infected or Healthy Plants and Other Eukaryotic Tissues. BMC Res. Notes 2012, 5, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.; Fernald, R.D. Comprehensive Algorithm for Quantitative Real-Time Polymerase Chain Reaction. J. Comput. Biol. 2005, 12, 1047–1064. [Google Scholar] [CrossRef] [PubMed]
- Borrill, P.; Ramirez-Gonzalez, R.; Uauy, C. ExpVIP: A Customizable RNA-Seq Data Analysis and Visualization Platform. Plant Physiol. 2016, 170, 2172–2186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, H.; Bray, N.L.; Puente, S.; Melsted, P.; Pachter, L. Differential Analysis of RNA-Seq Incorporating Quantification Uncertainty. Nat. Methods 2017, 14, 687–690. [Google Scholar] [CrossRef] [PubMed]
- Harrington, S.A.; Backhaus, A.E.; Singh, A.; Hassani-Pak, K.; Uauy, C. The Wheat GENIE3 Network Provides Biologically-Relevant Information in Polyploid Wheat. G3 Genes Genomes Genet. 2020, 10, 3675–3686. [Google Scholar] [CrossRef]
- Ramírez-González, R.H.; Borrill, P.; Lang, D.; Harrington, S.A.; Brinton, J.; Venturini, L.; Davey, M.; Jacobs, J.; van Ex, F.; Pasha, A.; et al. The Transcriptional Landscape of Polyploid Wheat. Science 2018, 361, eaar6089. [Google Scholar] [CrossRef] [Green Version]
- Salavaty, A.; Ramialison, M.; Currie, P.D. Integrated Value of Influence: An Integrative Method for the Identification of the Most Influential Nodes within Networks. Patterns 2020, 1, 100052. [Google Scholar] [CrossRef] [PubMed]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Vilo, J.; Peterson, H. gprofiler2—An R Package for Gene List Functional Enrichment Analysis and Namespace Conversion Toolset g:Profiler. F1000Research 2020, 9, ELIXIR-709. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fornes, O.; Castro-Mondragon, J.A.; Khan, A.; van der Lee, R.; Zhang, X.; Richmond, P.A.; Modi, B.P.; Correard, S.; Gheorghe, M.; Baranašić, D.; et al. JASPAR 2020: Update of the Open-Access Database of Transcription Factor Binding Profiles. Nucleic Acids Res. 2020, 48, D87–D92. [Google Scholar] [CrossRef] [PubMed]
- Gearing, L.J.; Cumming, H.E.; Chapman, R.; Finkel, A.M.; Woodhouse, I.B.; Luu, K.; Gould, J.A.; Forster, S.C.; Hertzog, P.J. CiiiDER: A Tool for Predicting and Analysing Transcription Factor Binding Sites. PLoS ONE 2019, 14, e0215495. [Google Scholar] [CrossRef] [Green Version]
- Panozzo, J.F.; Eagles, H.A.; Cawood, R.J.; Wootton, M. Wheat Spike Temperatures in Relation to Varying Environmental Conditions. Aust. J. Agric. Res. 1999, 50, 997–1006. [Google Scholar] [CrossRef]
- Graybosch, R.A.; Peterson, C.J.; Baenziger, P.S.; Shelton, D.R. Environmental Modification of Hard Red Winter Wheat Flour Protein Composition. J. Cereal Sci. 1995, 22, 45–51. [Google Scholar] [CrossRef] [Green Version]
- Jagadish, S.V.K.; Way, D.A.; Sharkey, T.D. Plant Heat Stress: Concepts Directing Future Research. Plant Cell Environ. 2021, 44, 1992–2005. [Google Scholar] [CrossRef]
- Guilioni, L.; Lhomme, J.P. Modelling the Daily Course of Capitulum Temperature in a Sunflower Canopy. Agric. For. Meteorol. 2006, 138, 258–272. [Google Scholar] [CrossRef]
- Keeling, P.L.; Bacon, P.J.; Holt, D.C. Elevated Temperature Reduces Starch Deposition in Wheat Endosperm by Reducing the Activity of Soluble Starch Synthase. Planta 1993, 191, 342–348. [Google Scholar] [CrossRef]
- Kumar, R.R.; Goswami, S.; Shamim, M.; Mishra, U.; Jain, M.; Singh, K.; Singh, J.P.; Dubey, K.; Singh, S.; Rai, G.K.; et al. Biochemical Defense Response: Characterizing the Plasticity of Source and Sink in Spring Wheat under Terminal Heat Stress. Front. Plant Sci. 2017, 8, 1603. [Google Scholar] [CrossRef] [Green Version]
- Jenner, C. Starch Synthesis in the Kernel of Wheat under High Temperature Conditions. Funct. Plant Biol. 1994, 21, 791–806. [Google Scholar] [CrossRef]
- Stone, P.; Nicolas, M. Effect of Timing of Heat Stress during Grain Filling on Two Wheat Varieties Differing in Heat Tolerance. II. Fractional Protein Accumulation. Funct. Plant Biol. 1996, 23, 739–749. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Khanna-Chopra, R. Effect of Heat Stress on Grain Starch Content in Diploid, Tetraploid and Hexaploid Wheat Species. J. Agron. Crop Sci. 2003, 189, 242–249. [Google Scholar] [CrossRef]
- Dupont, F.M.; Hurkman, W.J.; Vensel, W.H.; Tanaka, C.; Kothari, K.M.; Chung, O.K.; Altenbach, S.B. Protein Accumulation and Composition in Wheat Grains: Effects of Mineral Nutrients and High Temperature. Eur. J. Agron. 2006, 25, 96–107. [Google Scholar] [CrossRef]
- Spiertz, J.H.J.; Hamer, R.J.; Xu, H.; Primo Martin, C.; Don, C.; Putten, P.E.L. van der Heat Stress in Wheat (Triticum aestivum L.): Effects on Grain Growth and Quality Traits. Eur. J. Agron. 2006, 25, 89–95. [Google Scholar] [CrossRef]
- Wang, X.; Hou, L.; Lu, Y.; Wu, B.; Gong, X.; Liu, M.; Wang, J.; Sun, Q.; Vierling, E.; Xu, S. Metabolic Adaptation of Wheat Grain Contributes to a Stable Filling Rate under Heat Stress. J. Exp. Bot. 2018, 69, 5531–5545. [Google Scholar] [CrossRef] [Green Version]
- Wardlaw, I.F. Interaction Between Drought and Chronic High Temperature During Kernel Filling in Wheat in a Controlled Environment. Ann. Bot. 2002, 90, 469–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slafer, G.A.; Savin, R.; Pinochet, D.; Calderini, D.F. Chapter 3—Wheat. In Crop Physiology Case Histories for Major Crops; Sadras, V.O., Calderini, D.F., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 98–163. ISBN 978-0-12-819194-1. [Google Scholar]
- Vierling, E. The Roles of Heat Shock Proteins in Plants. Annu. Rev. Plant Biol. 1991, 42, 579–620. [Google Scholar] [CrossRef]
- Marmiroli, N.; Lorenzoni, C.; Cattivelli, L.; Stanca, A.M.; Terzi, V. Induction of Heat Shock Proteins and Acquisition of Thermotolerance in Barley (Hordeum vulgare L.) Variations Associated with Growth Habit and Plant Development. J. Plant Physiol. 1989, 135, 267–273. [Google Scholar] [CrossRef]
- Sun, L.; Liu, Y.; Kong, X.; Zhang, D.; Pan, J.; Zhou, Y.; Wang, L.; Li, D.; Yang, X. ZmHSP16.9, a Cytosolic Class I Small Heat Shock Protein in Maize (Zea Mays), Confers Heat Tolerance in Transgenic Tobacco. Plant Cell Rep. 2012, 31, 1473–1484. [Google Scholar] [CrossRef] [PubMed]
- Queitsch, C.; Hong, S.W.; Vierling, E.; Lindquist, S. Heat Shock Protein 101 Plays a Crucial Role in Thermotolerance in Arabidopsis. Plant Cell 2000, 12, 479–492. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.-M.; Li, J.; He, J.; Liu, H.; Zhang, H.-M. A Class I Cytosolic HSP20 of Rice Enhances Heat and Salt Tolerance in Different Organisms. Sci. Rep. 2020, 10, 1383. [Google Scholar] [CrossRef]
- Basha, E.; Jones, C.; Blackwell, A.E.; Cheng, G.; Waters, E.R.; Samsel, K.A.; Siddique, M.; Pett, V.; Wysocki, V.; Vierling, E. An Unusual Dimeric Small Heat Shock Protein Provides Insight into the Mechanism of This Class of Chaperones. J. Mol. Biol. 2013, 425, 1683–1696. [Google Scholar] [CrossRef] [Green Version]
- Law, R.D.; Crafts-Brandner, S.J. High Temperature Stress Increases the Expression of Wheat Leaf Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase Activase Protein. Arch. Biochem. Biophys. 2001, 386, 261–267. [Google Scholar] [CrossRef]
- Feldman, D.E.; Frydman, J. Protein Folding in Vivo: The Importance of Molecular Chaperones. Curr. Opin. Struct. Biol. 2000, 10, 26–33. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of Plant Heat-Shock Proteins and Molecular Chaperones in the Abiotic Stress Response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Kaur, R.; Sinha, K.; Bhunia, R.K. Can Wheat Survive in Heat? Assembling Tools towards Successful Development of Heat Stress Tolerance in Triticum aestivum L. Mol. Biol. Rep. 2019, 46, 2577–2593. [Google Scholar] [CrossRef]
- Agarwal, P.; Baranwal, V.K.; Khurana, P. Genome-Wide Analysis of BZIP Transcription Factors in Wheat and Functional Characterization of a TabZIP under Abiotic Stress. Sci. Rep. 2019, 9, 4608. [Google Scholar] [CrossRef] [Green Version]
- Kumar, D.; Chattopadhyay, S. Glutathione Modulates the Expression of Heat Shock Proteins via the Transcription Factors BZIP10 and MYB21 in Arabidopsis. J. Exp. Bot. 2018, 69, 3729–3743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Sharma, S.; Chunduri, V.; Kaur, A.; Kaur, S.; Malhotra, N.; Kumar, A.; Kapoor, P.; Kumari, A.; Kaur, J.; et al. Genome-Wide Identification and Characterization of Heat Shock Protein Family Reveals Role in Development and Stress Conditions in Triticum aestivum L. Sci. Rep. 2020, 10, 7858. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Fukao, Y.; Hayashi, M.; Fukazawa, M.; Suzuki, I.; Nishimura, M. Cytosolic HSP90 Regulates the Heat Shock Response That Is Responsible for Heat Acclimation in Arabidopsis Thaliana. J. Biol. Chem. 2007, 282, 37794–37804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Shi, Y.; Yang, S. Molecular Regulation of Plant Responses to Environmental Temperatures. Mol. Plant 2020, 13, 544–564. [Google Scholar] [CrossRef] [PubMed]
- Rangan, P.; Furtado, A.; Henry, R. Transcriptome Profiling of Wheat Genotypes under Heat Stress during Grain-Filling. J. Cereal Sci. 2020, 91, 102895. [Google Scholar] [CrossRef]
- Wang, L.; Ma, K.-B.; Lu, Z.-G.; Ren, S.-X.; Jiang, H.-R.; Cui, J.-W.; Chen, G.; Teng, N.-J.; Lam, H.-M.; Jin, B. Differential Physiological, Transcriptomic and Metabolomic Responses of Arabidopsis Leaves under Prolonged Warming and Heat Shock. BMC Plant Biol. 2020, 20, 86. [Google Scholar] [CrossRef]
- Zhang, Z.; Dong, J.; Ji, C.; Wu, Y.; Messing, J. NAC-Type Transcription Factors Regulate Accumulation of Starch and Protein in Maize Seeds. Proc. Natl. Acad. Sci. USA 2019, 116, 11223. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Q.; Wang, Y.; Li, H.; Zhang, C.; Wei, B.; Wang, Y.; Huang, H.; Li, Y.; Yu, G.; Liu, H.; et al. Transcription Factor ZmNAC126 Plays an Important Role in Transcriptional Regulation of Maize Starch Synthesis-Related Genes. Crop J. 2020, 9, 192–203. [Google Scholar] [CrossRef]
- Gao, Y.; An, K.; Guo, W.; Chen, Y.; Zhang, R.; Zhang, X.; Chang, S.; Rossi, V.; Jin, F.; Cao, X.; et al. The Endosperm-Specific Transcription Factor TaNAC019 Regulates Glutenin and Starch Accumulation and Its Elite Allele Improves Wheat Grain Quality. Plant Cell 2021, 33, 603–622. [Google Scholar] [CrossRef] [PubMed]
- Ohama, N.; Kusakabe, K.; Mizoi, J.; Zhao, H.; Kidokoro, S.; Koizumi, S.; Takahashi, F.; Ishida, T.; Yanagisawa, S.; Shinozaki, K.; et al. The Transcriptional Cascade in the Heat Stress Response of Arabidopsis Is Strictly Regulated at the Level of Transcription Factor Expression. Plant Cell 2016, 28, 181–201. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, A.M.; Arsovski, A.A.; Lempe, J.; Bubb, K.L.; Weirauch, M.T.; Sabo, P.J.; Sandstrom, R.; Thurman, R.E.; Neph, S.; Reynolds, A.P.; et al. Mapping and Dynamics of Regulatory DNA and Transcription Factor Networks in A. Thaliana. Cell Rep. 2014, 8, 2015–2030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkins, O.; Hafemeister, C.; Plessis, A.; Holloway-Phillips, M.-M.; Pham, G.M.; Nicotra, A.B.; Gregorio, G.B.; Jagadish, S.V.K.; Septiningsih, E.M.; Bonneau, R.; et al. EGRINs (Environmental Gene Regulatory Influence Networks) in Rice That Function in the Response to Water Deficit, High Temperature, and Agricultural Environments. Plant Cell 2016, 28, 2365–2384. [Google Scholar] [CrossRef] [Green Version]
- Huynh-Thu, V.A.; Irrthum, A.; Wehenkel, L.; Geurts, P. Inferring Regulatory Networks from Expression Data Using Tree-Based Methods. PLoS ONE 2010, 5, e12776. [Google Scholar] [CrossRef]
- Robles, P.; Quesada, V. Research Progress in the Molecular Functions of Plant MTERF Proteins. Cells 2021, 10, 205. [Google Scholar] [CrossRef]
- Wobbe, L. The Molecular Function of Plant MTERFs as Key Regulators of Organellar Gene Expression. Plant Cell Physiol. 2020, 61, 2004–2017. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Schulz, V.; Brings, L.; Schoeller, T.; Kühn, K.; Vierling, E. MTERF18 and ATAD3 Are Required for Mitochondrial Nucleoid Structure and Their Disruption Confers Heat Tolerance in Arabidopsis Thaliana. New Phytol. 2021, 232, 2026–2042. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, N.; Deng, X.; Liu, D.; Li, M.; Cui, D.; Hu, Y.; Yan, Y. Genome-Wide Analysis of Wheat DNA-Binding with One Finger (Dof) Transcription Factor Genes: Evolutionary Characteristics and Diverse Abiotic Stress Responses. BMC Genom. 2020, 21, 276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, M.P.; Onodera, Y.; Touno, S.M.; Takaiwa, F. Synergism between RPBF Dof and RISBZ1 BZIP Activators in the Regulation of Rice Seed Expression Genes. Plant Physiol. 2006, 141, 1694–1707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrero, D.M.L.; Piattoni, C.V.; Asencion Diez, M.D.; Rojas, B.E.; Hartman, M.D.; Ballicora, M.A.; Iglesias, A.A. Phosphorylation of ADP-Glucose Pyrophosphorylase During Wheat Seeds Development. Front. Plant Sci. 2020, 11, 1058. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arenas-M, A.; Castillo, F.M.; Godoy, D.; Canales, J.; Calderini, D.F. Transcriptomic and Physiological Response of Durum Wheat Grain to Short-Term Heat Stress during Early Grain Filling. Plants 2022, 11, 59. https://doi.org/10.3390/plants11010059
Arenas-M A, Castillo FM, Godoy D, Canales J, Calderini DF. Transcriptomic and Physiological Response of Durum Wheat Grain to Short-Term Heat Stress during Early Grain Filling. Plants. 2022; 11(1):59. https://doi.org/10.3390/plants11010059
Chicago/Turabian StyleArenas-M, Anita, Francisca M. Castillo, Diego Godoy, Javier Canales, and Daniel F. Calderini. 2022. "Transcriptomic and Physiological Response of Durum Wheat Grain to Short-Term Heat Stress during Early Grain Filling" Plants 11, no. 1: 59. https://doi.org/10.3390/plants11010059
APA StyleArenas-M, A., Castillo, F. M., Godoy, D., Canales, J., & Calderini, D. F. (2022). Transcriptomic and Physiological Response of Durum Wheat Grain to Short-Term Heat Stress during Early Grain Filling. Plants, 11(1), 59. https://doi.org/10.3390/plants11010059







