Silencing of a Cotton Actin-Binding Protein GhWLIM1C Decreases Resistance against Verticillium dahliae Infection
Abstract
:1. Introduction
2. Results
2.1. Expression of Cotton GhWLIM1C in Response to V. dahliae Infection
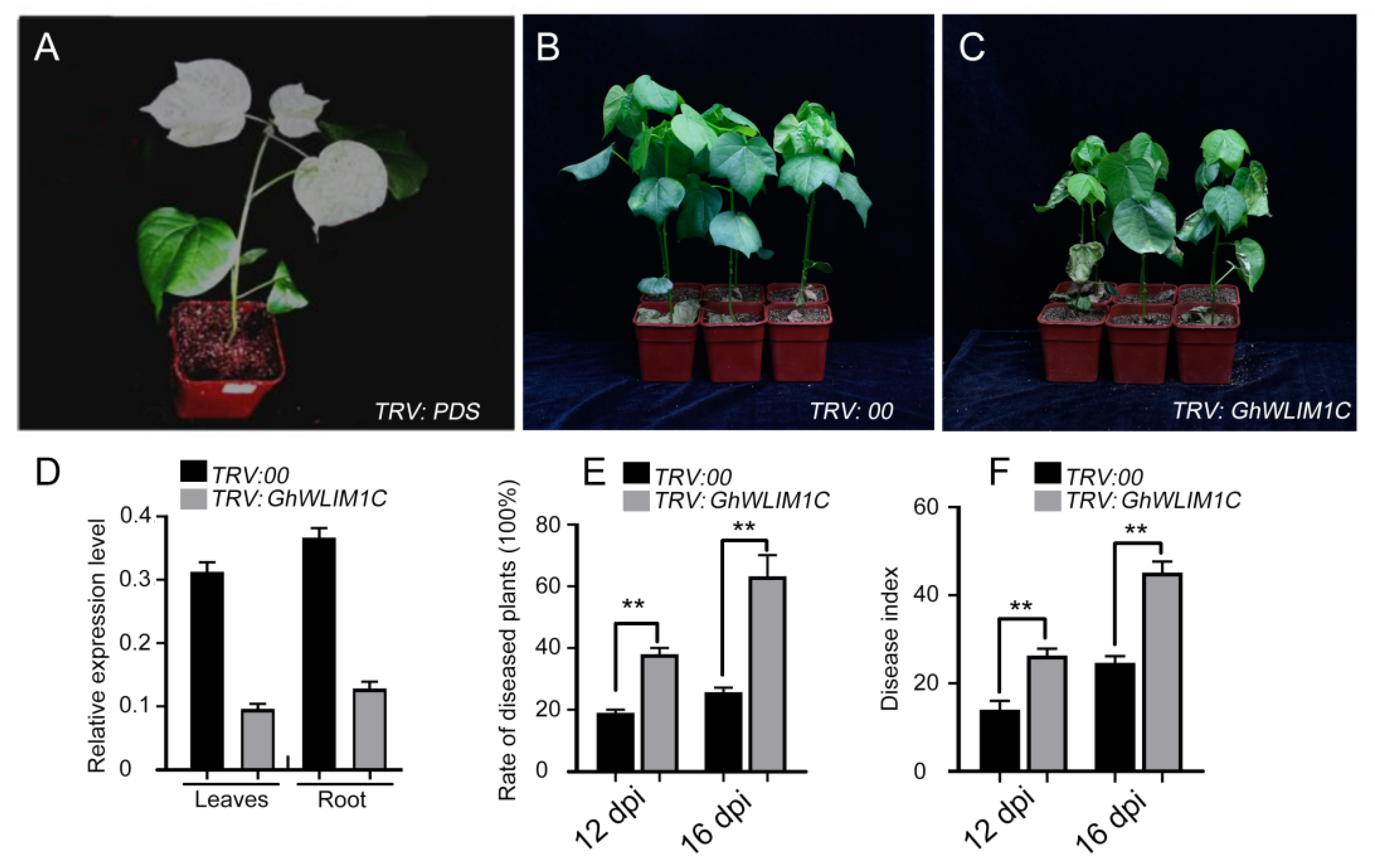
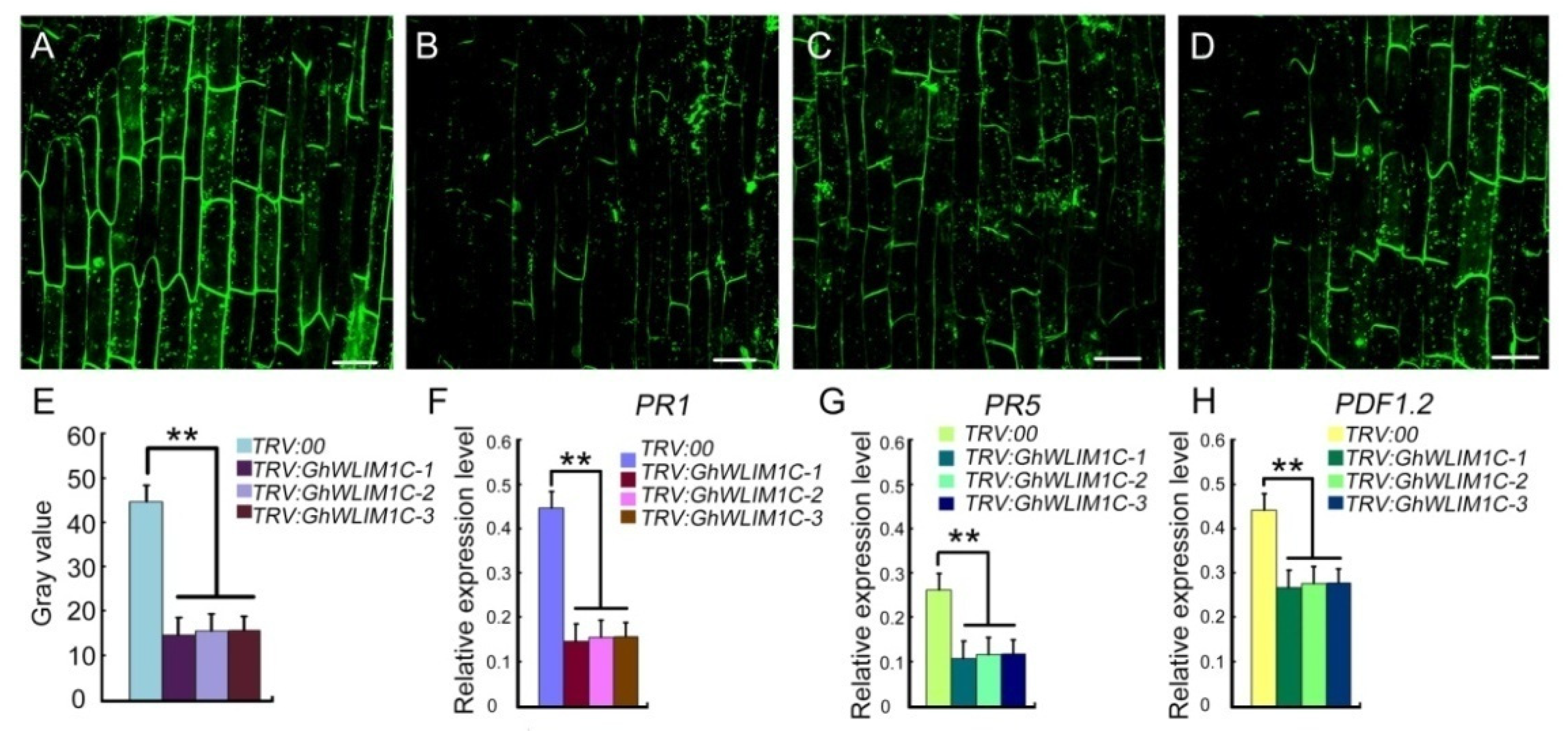
2.2. GhWLIM1C Functions in Defense against V. dahliae Infection
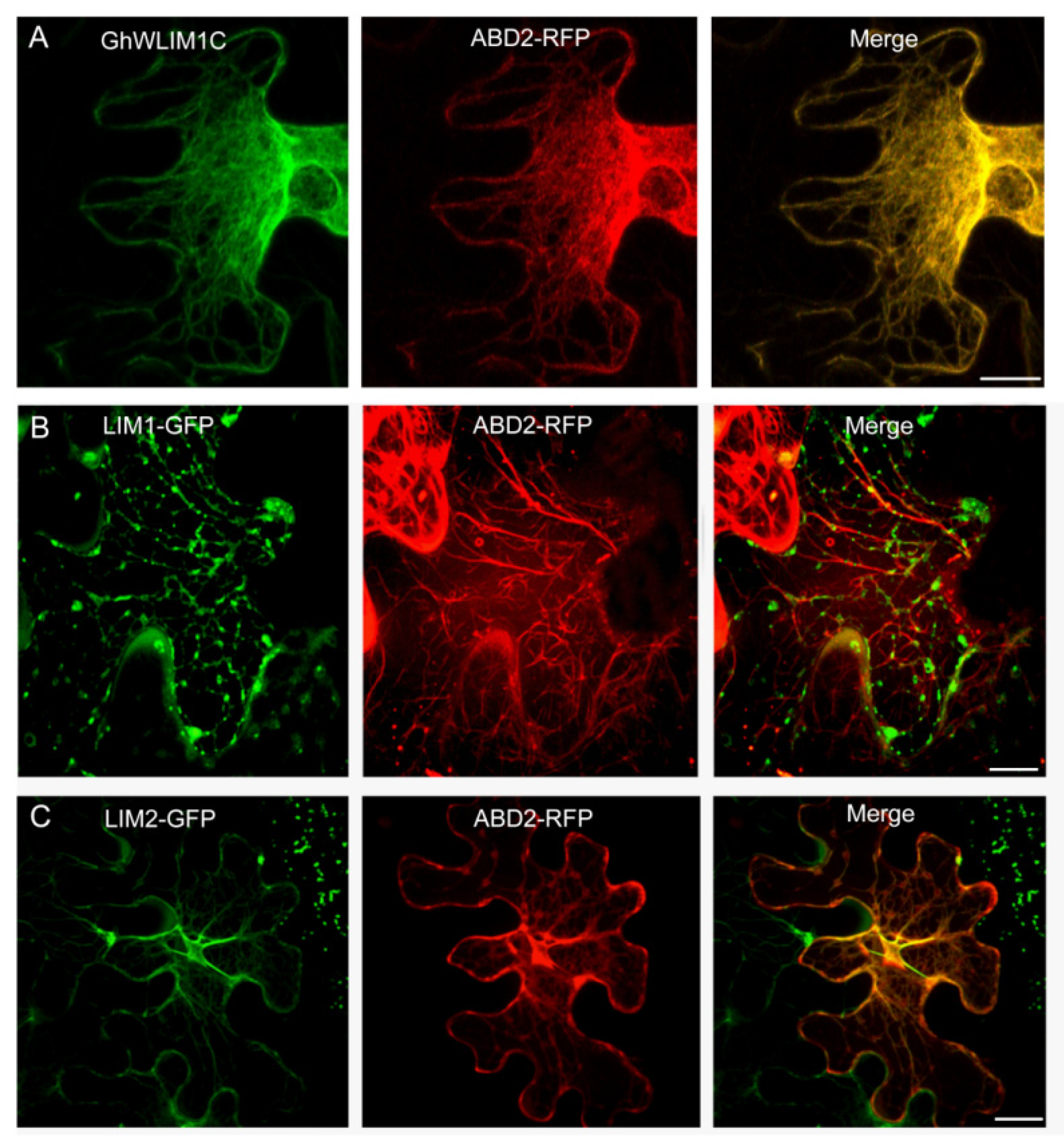
2.3. GhWLIM1C Binds to the Plant Actin Cytoskeleton In Vivo
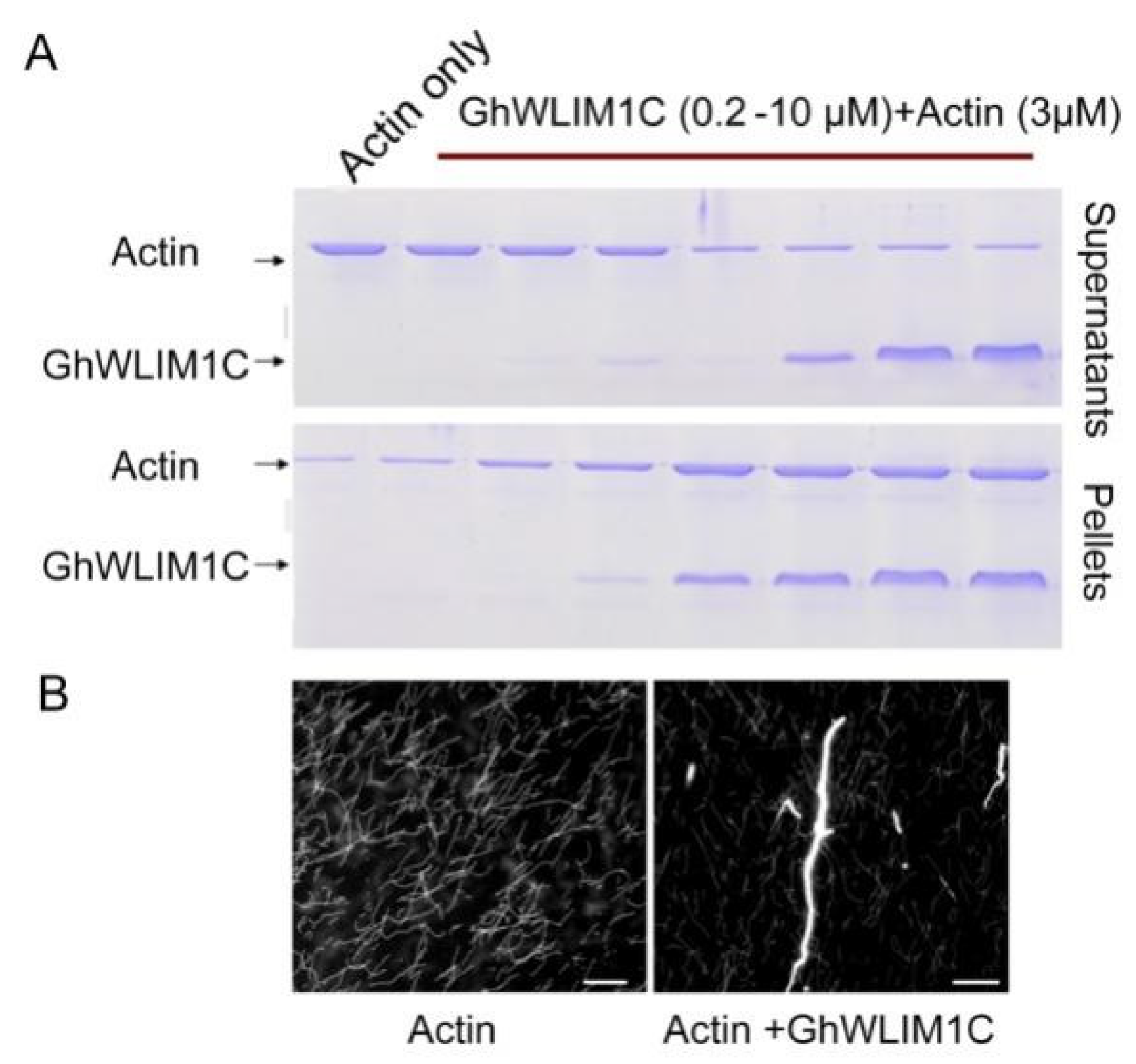
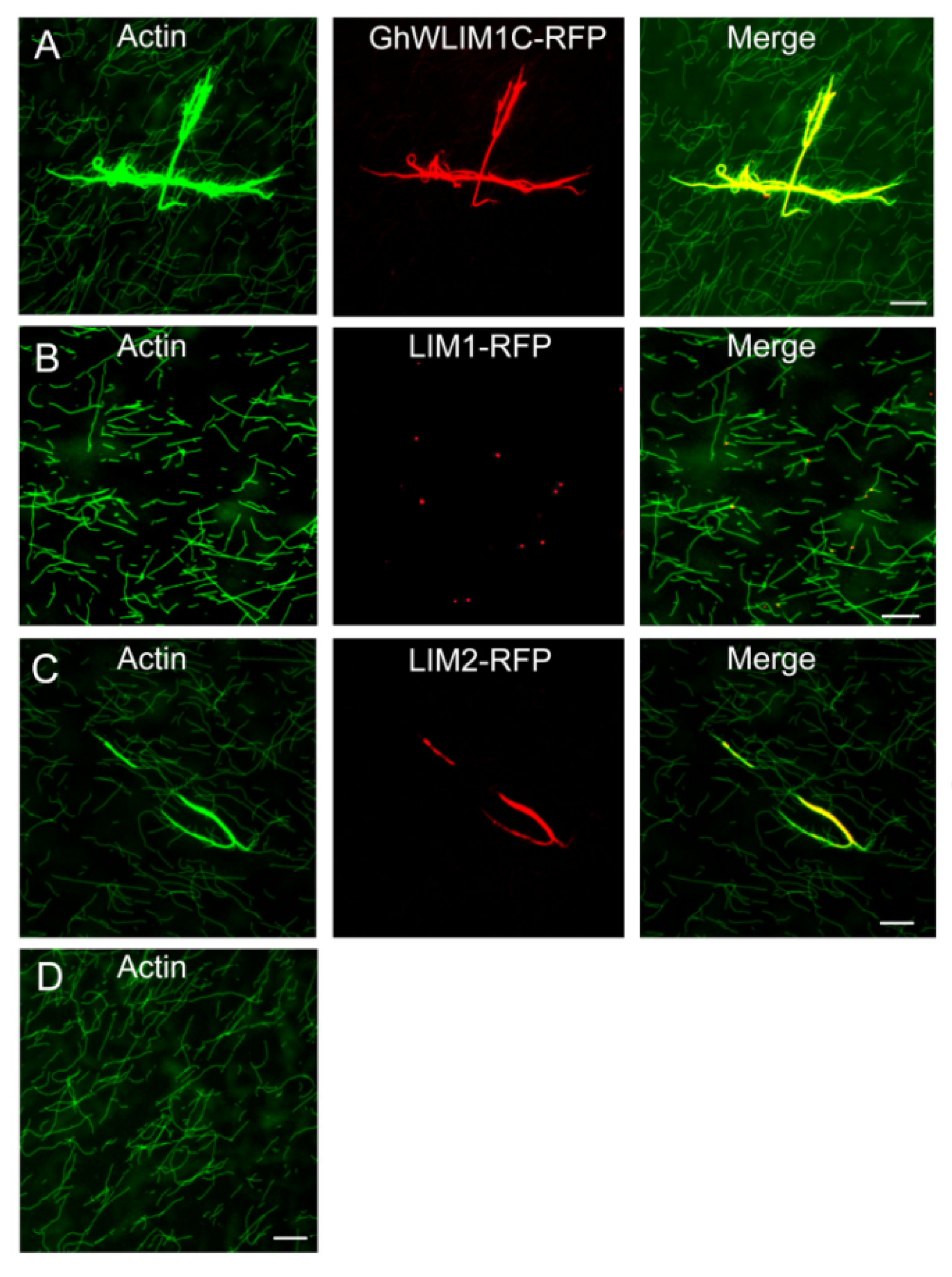
2.4. GhWLIM1C Promotes the Formation of Actin Bundles In Vitro
2.5. GhWLIM1C May Function Involving in the Activation of Plant Immune Response
3. Discussion
4. Materials and Methods
4.1. Cotton Growth and V. dahliae Culture and Infection of Cotton Seedlings
4.2. RNA Extraction and RT-qPCR Analysis
4.3. Virus-Induced Gene Silencing
4.4. Plasmid Construction and Protein Expression
4.5. Low-Speed Cosedimentation Assays
4.6. In Vitro Assembly and Observation of the Actin Cytoskeleton
4.7. Fluorescent Staining and Confocal Lasers Canning Microscopy
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bolek, Y.; El-Zik, K.M.; Pepper, A.E.; Bell, A.A.; Magill, C.W.; Thaxton, P.M.; Reddy, O.U.K. Mapping of verticillium wilt resistance genes in cotton. Plant Sci. 2005, 168, 1581–1590. [Google Scholar] [CrossRef]
- Cai, Y.F.; He, X.H.; Mo, J.C.; Sun, Q.; Yang, J.P.; Liu, J.G. Molecular research and genetic engineering of resistance to Verticillium wilt in cotton: A review. Afr. J. Biotechnol. 2009, 8, 7363–7372. [Google Scholar]
- Vallad, G.E.; Subbarao, K.V. Colonization of resistant and susceptible lettuce cultivars by a green fluorescent protein-tagged isolate of Verticillium dahliae. Phytopathology 2008, 98, 871–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, P.; Zhao, Y.L.; Jin, Y.; Zhang, T.; Guo, H.S. Colonization process of Arabidopsis thaliana roots by a green fluorescent protein-tagged isolate of Verticillium dahliae. Protein Cell 2014, 5, 94–98. [Google Scholar] [CrossRef]
- Deng, S.; Wang, C.Y.; Zhang, X.; Wang, Q.; Lin, L. VdNUC-2, the Key Regulator of Phosphate Responsive Signaling Pathway, Is Required for Verticillium dahliae Infection. PLoS ONE 2015, 10, e0145190. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Atallah, Z.K.; Vallad, G.E.; Subbarao, K.V. Diversity, Pathogenicity; and Management of Verticillium Species. Annu. Rev. Phytopathol. 2009, 47, 39–62. [Google Scholar] [CrossRef] [Green Version]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal Effectors and Plant Susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef]
- Li, Y.B.; Han, L.B.; Wang, H.Y.; Zhang, J.; Sun, S.T.; Feng, D.Q.; Yang, C.L.; Sun, Y.D.; Zhong, N.Q.; Xia, G.X. The Thioredoxin GbNRX1 Plays a Crucial Role in Homeostasis of Apoplastic Reactive Oxygen Species in Response to Verticillium dahliae Infection in Cotton. Plant Physiol. 2016, 170, 2392–2406. [Google Scholar] [CrossRef] [Green Version]
- Han, L.B.; Li, Y.B.; Wang, F.X.; Wang, W.Y.; Liu, J.; Wu, J.H.; Zhong, N.Q.; Wu, S.J.; Jiao, G.L.; Wang, H.Y.; et al. The Cotton Apoplastic Protein CRR1 Stabilizes Chitinase 28 to Facilitate Defense against the Fungal Pathogen Verticillium dahliae. Plant Cell 2019, 31, 520–536. [Google Scholar] [CrossRef]
- Gu, Z.H.; Liu, T.L.; Ding, B.; Li, F.F.; Wang, Q.; Qian, S.S.; Ye, F.; Chen, T.Z.; Yang, Y.W.; Wang, J.Y.; et al. Two Lysin-Motif Receptor Kinases, Gh-LYK1 and Gh-LYK2, Contribute to Resistance against Verticillium wilt in Upland Cotton. Front. Plant Sci. 2017, 8, 2133. [Google Scholar] [CrossRef] [Green Version]
- Cheng, H.Q.; Han, L.B.; Yang, C.L.; Wu, X.M.; Zhong, N.Q.; Wu, J.H.; Wang, F.X.; Wang, H.Y.; Xia, G.X. The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection. J. Exp. Bot. 2016, 67, 1935–1950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Wang, K.; Sun, L.; Xing, H.; Wang, S.; Li, L.; Chen, S.; Guo, H.S.; Zhang, J. The plant-specific transcription factors CBP6Og and SARD1 are targeted by a Verticillium secretory protein VdSCP41 to modulate immunity. Elife 2018, 7, e34902. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.F.; Zhang, D.P.; Wang, G.X.; Wang, K.; Li, Z.; Gao, Y.H.; Li, H.C.; Bian, C.; Cheng, J.K.; Han, Y.A.; et al. Verticillium dahliae effector VDAL protects MYB6 from degradation by interacting with PUB25 and PUB26 E3 ligases to enhance Verticillium wilt resistance. Plant Cell 2021, 33, 3675–3699. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.Q.; Wheeler, T.; Li, Z.H.; Kenerley, C.M.; He, P.; Shan, L.B. Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 2011, 66, 293–305. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.Q.; Li, F.J.; Li, M.Y.; Kianinejad, A.S.; Dever, J.K.; Wheeler, T.A.; Li, Z.H.; He, P.; Shan, L.B. Cotton GhBAK1 Mediates Verticillium Wilt Resistance and Cell Death. J. Integr. Plant Biol. 2013, 55, 586–596. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.L.; Liang, S.; Wang, H.Y.; Han, L.B.; Wang, F.X.; Cheng, H.Q.; Wu, X.M.; Qu, Z.L.; Wu, J.H.; Xia, G.X. Cotton Major Latex Protein 28 Functions as a Positive Regulator of the Ethylene Responsive Factor 6 in Defense against Verticillium dahliae. Mol. Plant 2015, 8, 399–411. [Google Scholar] [CrossRef] [Green Version]
- Porter, K.; Day, B. From filaments to function: The role of the plant actin cytoskeleton in pathogen perception, signaling and immunity. J. Integr. Plant Biol. 2016, 58, 299–311. [Google Scholar] [CrossRef]
- Henty-Ridilla, J.L.; Shimono, M.; Li, J.J.; Chang, J.H.; Day, B.; Staiger, C.J. The Plant Actin Cytoskeleton Responds to Signals from Microbe-Associated Molecular Patterns. PLoS Pathog 2013, 9, e1003290. [Google Scholar] [CrossRef] [Green Version]
- Henty-Ridilla, J.L.; Li, J.J.; Day, B.; Staiger, C.J. ACTIN DEPOLYMERIZING FACTOR4 Regulates Actin Dynamics during Innate Immune Signaling in Arabidopsis. Plant Cell 2014, 26, 340–352. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Qin, L.; Liu, G.S.; Peremyslov, V.V.; Dolja, V.V.; Wei, Y.D. Myosins XI modulate host cellular responses and penetration resistance to fungal pathogens. Proc. Natl. Acad. Sci. USA 2014, 111, 13996–14001. [Google Scholar] [CrossRef] [Green Version]
- Li, J.J.; Cao, L.Y.; Staiger, C.J. Capping Protein Modulates Actin Remodeling in Response to Reactive Oxygen Species during Plant Innate Immunity. Plant Physiol. 2017, 173, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Dawid, I.B.; Toyama, R.; Taira, M. Lim Domain Proteins. Cr. Acad. Sci. III-VIE 1995, 318, 295–306. [Google Scholar]
- Han, L.B.; Li, Y.B.; Wang, H.Y.; Wu, X.M.; Li, C.L.; Luo, M.; Wu, S.J.; Kong, Z.S.; Pei, Y.; Jiao, G.L.; et al. The Dual Functions of WLIM1a in Cell Elongation and Secondary Wall Formation in Developing Cotton Fibers. Plant Cell 2013, 25, 4421–4438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moes, D.; Gatti, S.; Hoffmann, C.; Dieterle, M.; Moreau, F.; Neumann, K.; Schumacher, M.; Diederich, M.; Grill, E.; Shen, W.H.; et al. A LIM Domain Protein from Tobacco Involved in Actin-Bundling and Histone Gene Transcription. Mol. Plant 2013, 6, 483–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Q.; Zhao, Y. The diverse bilofunctions of LIM domain proteins: Determined by subcellular localization and protein-protein interaction. Biol. Cell 2007, 99, 489–502. [Google Scholar] [CrossRef]
- Kawaoka, A.; Kaothien, P.; Yoshida, K.; Endo, S.; Yamada, K.; Ebinuma, H. Functional analysis of tobacco LIM protein Ntlim1 involved in lignin biosynthesis. Plant J. 2000, 22, 289–301. [Google Scholar] [CrossRef]
- Thomas, C.; Hoffmann, C.; Dieterle, M.; Van Troys, M.; Ampe, C.; Steinmetza, A. Tobacco WLIM1 is a novel F-actin binding protein involved in actin cytoskeleton remodeling. Plant Cell 2006, 18, 2194–2206. [Google Scholar] [CrossRef] [Green Version]
- Pang, J.H.; Zhu, Y.; Li, Q.; Liu, J.Z.; Tian, Y.C.; Liu, Y.L.; Wu, J.H. Development of Agrobacterium-Mediated Virus-Induced Gene Silencing and Performance Evaluation of Four Marker Genes in Gossypium barbadense. PLoS ONE 2013, 8, e73211. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; He, X.; Luo, X.Y.; Xu, L.; Liu, L.L.; Min, L.; Jin, L.; Zhu, L.F.; Zhang, X.L. Cotton WRKY1 Mediates the Plant Defense-to-Development Transition during Infection of Cotton by Verticillium dahliae by Activating JASMONATE ZIM-DOMAIN1 Expression. Plant Physiol. 2014, 166, 2179–2194. [Google Scholar] [CrossRef] [Green Version]
- Jun, Z.; Zhang, Z.Y.; Gao, Y.L.; Zhou, L.; Fang, L.; Chen, X.D.; Ning, Z.Y.; Chen, T.Z.; Guo, W.Z.; Zhang, T.Z. Overexpression of GbRLK, a putative receptor-like kinase gene, improved cotton tolerance to Verticillium wilt. Sci. Rep. 2015, 5, 15048. [Google Scholar] [CrossRef] [Green Version]
- Winder, S.J.; Ayscough, K.R. Actin-binding proteins. J. Cell Sci. 2005, 118, 651–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bach, I. The LIM domain: Regulation by association. Mech. Develop. 2000, 91, 5–17. [Google Scholar] [CrossRef]
- Jurata, L.W.; Gill, G.N. Structure and function of LIM domains. Curr. Top Microbiol. 1998, 228, 75–113. [Google Scholar]
- Tran, T.C.; Singleton, C.A.; Fraley, T.S.; Greenwood, J.A. Cysteine-rich protein I (CRPI) regulates actin filament bundling. BMC Cell Biol. 2005, 6, 45. [Google Scholar] [CrossRef] [Green Version]
- Papuga, J.; Hoffmann, C.; Dieterle, M.; Moes, D.; Moreau, F.; Tholl, S.; Steinmetz, A.; Thomas, C. Arabidopsis LIM Proteins: A Family of Actin Bundlers with Distinct Expression Patterns and Modes of Regulation. Plant Cell 2010, 22, 3034–3052. [Google Scholar] [CrossRef] [Green Version]
- Han, L.B.; Li, Y.B.; Sun, Y.D.; Wang, H.Y.; Kong, Z.S.; Xia, G.X. The two domains of cotton WLIM1a protein are functionally divergent. Sci. China Life Sci. 2016, 59, 206–212. [Google Scholar] [CrossRef]
- Li, P.; Day, B. Battlefield Cytoskeleton: Turning the Tide on Plant Immunity. Mol. Plant Microbe Interact. 2019, 32, 25–34. [Google Scholar] [CrossRef] [Green Version]
- Qu, Z.L.; Wang, H.Y.; Xia, G.X. GhHb1: A nonsymbiotic hemoglobin gene of cotton responsive to infection by Verticillium dahliae. Biochim. Biophys. Acta 2005, 1730, 103–113. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, T.; Qin, M.; Zhu, S.; Li, Y. Silencing of a Cotton Actin-Binding Protein GhWLIM1C Decreases Resistance against Verticillium dahliae Infection. Plants 2022, 11, 1828. https://doi.org/10.3390/plants11141828
Cao T, Qin M, Zhu S, Li Y. Silencing of a Cotton Actin-Binding Protein GhWLIM1C Decreases Resistance against Verticillium dahliae Infection. Plants. 2022; 11(14):1828. https://doi.org/10.3390/plants11141828
Chicago/Turabian StyleCao, Tingyan, Minghui Qin, Shuai Zhu, and Yuanbao Li. 2022. "Silencing of a Cotton Actin-Binding Protein GhWLIM1C Decreases Resistance against Verticillium dahliae Infection" Plants 11, no. 14: 1828. https://doi.org/10.3390/plants11141828
APA StyleCao, T., Qin, M., Zhu, S., & Li, Y. (2022). Silencing of a Cotton Actin-Binding Protein GhWLIM1C Decreases Resistance against Verticillium dahliae Infection. Plants, 11(14), 1828. https://doi.org/10.3390/plants11141828





