Genome-Wide Association Studies of Root-Related Traits in Brassica napus L. under Low-Potassium Conditions
Abstract
:1. Introduction
2. Results
2.1. Performances of Eight Lines under K-Concentration Gradients
2.2. Phenotypic Variations of Root Traits in the Association Panel under Low-K Stress
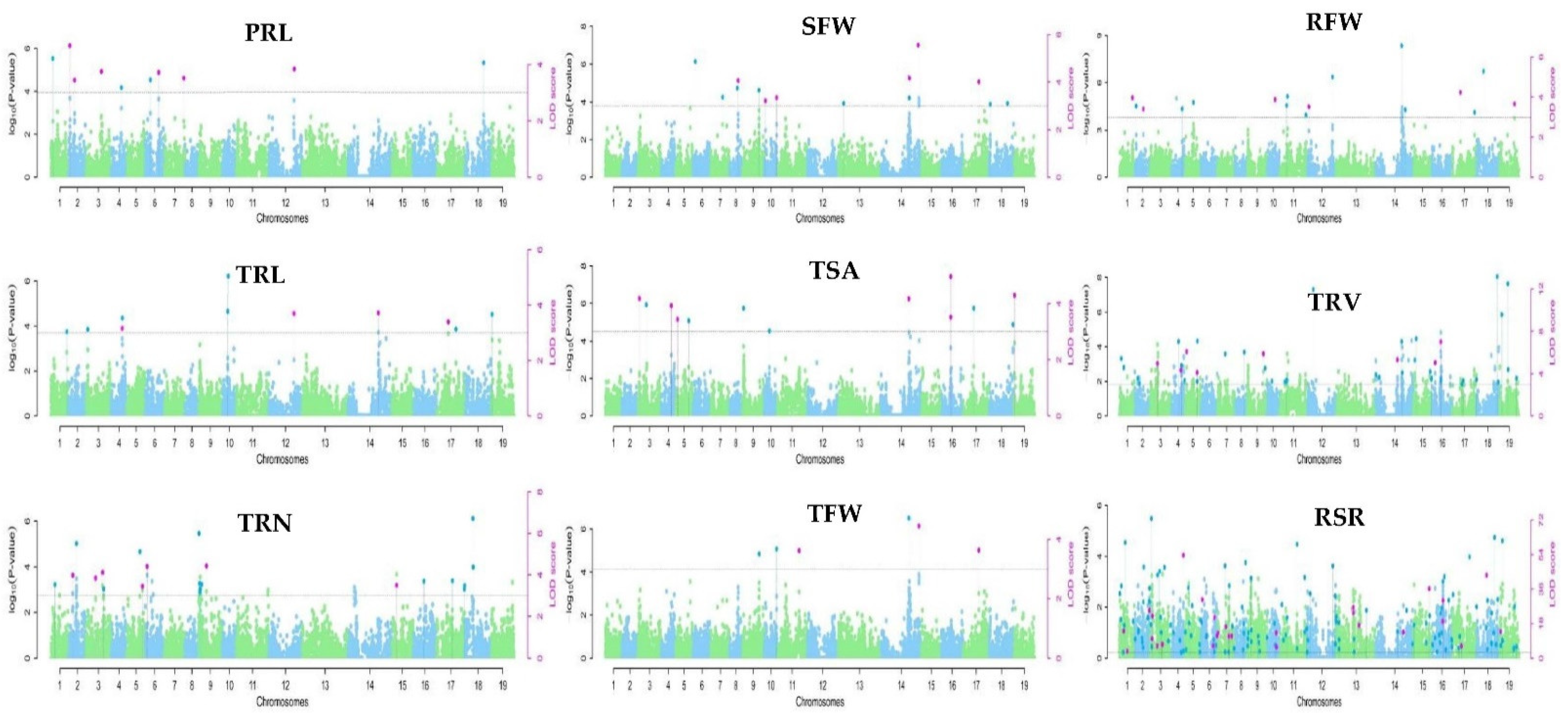
2.3. QTL Clusters Related to the Root System under Low-K Stress Were Obtained by GWAS
2.4. Candidate Genes Associated with Root-Related Traits
2.5. GO and KEGG Analysis of Potential Candidate Genes
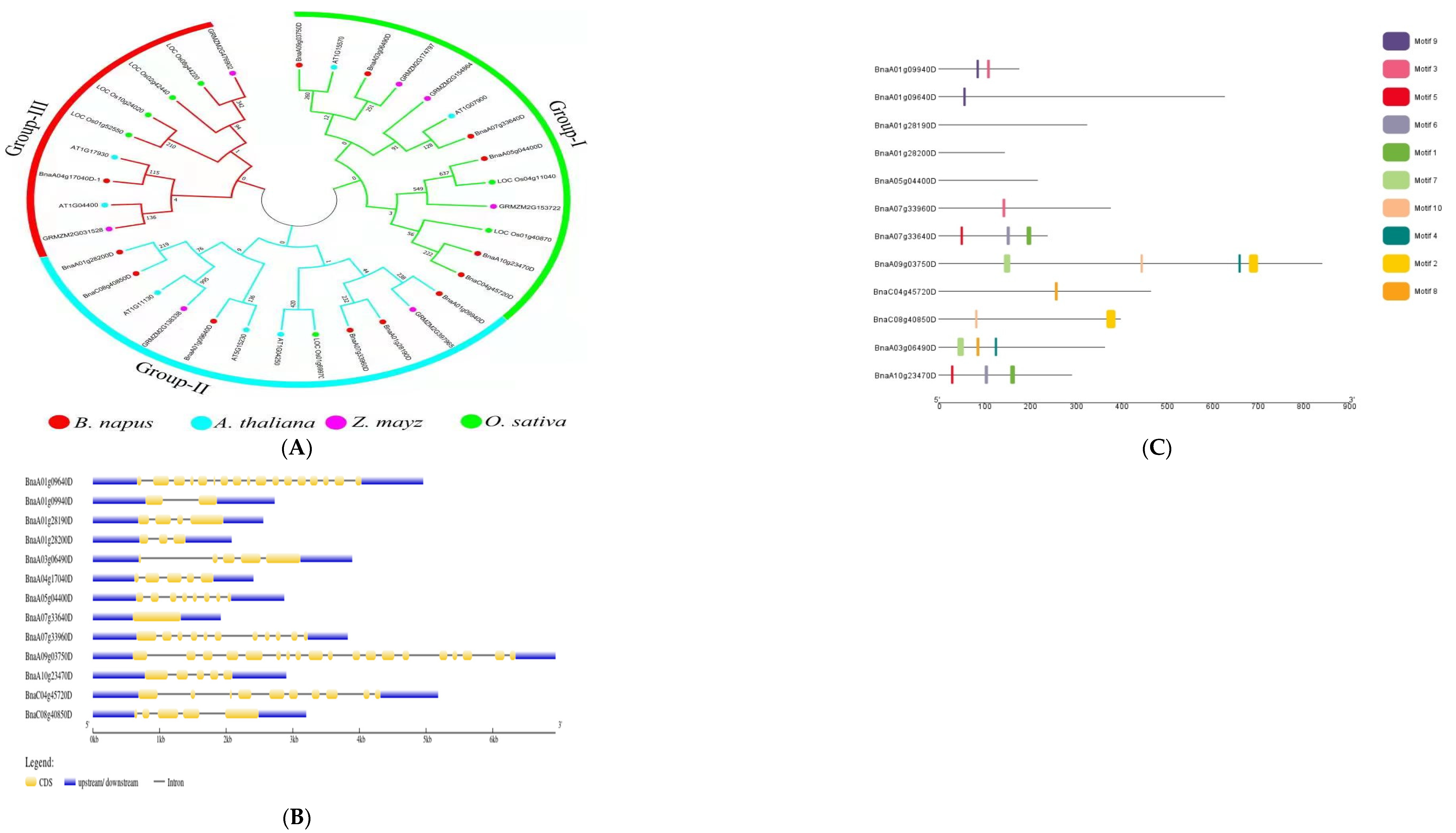
2.6. Protein Interaction Network Analysis, Phylogenetic Trees, Gene Structure Analysis, and Motif Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Experimental Design and Growth Condition
4.3. Root Phenotyping
4.4. Data Analysis
4.5. Marker–Trait Association
4.6. Exploration of Candidate Genes
4.7. GO and KEGG Analysis
4.8. Protein Interaction Network Analysis, Phylogenetic Trees, Gene-Structure Analysis, and Motif Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Wang, Y.; Wu, W.H. Genetic Approaches for Improvement of the Crop Potassium Acquisition and Utilization Efficiency. Curr. Opin. Plant Biol. 2015, 25, 46–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; He, H.; Liu, R.; Han, Q.; Shou, H.; Liu, B. Overexpression of GmAKT2 Potassium Channel Enhances Resistance to Soybean Mosaic Virus. BMC Plant Biol. 2014, 14, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, R.C.; Duveiller, E. Effect of Helminthosporium Leaf Blight on Performance of Timely and Late-Seeded Wheat under Optimal and Stressed Levels of Soil Fertility and Moisture. Field Crops Res. 2004, 89, 205–218. [Google Scholar] [CrossRef]
- Mann, R.L.; Kettlewell, P.S.; Jenkinson, P. Effect of Foliar-Applied Potassium Chloride on Septoria Leaf Blotch of Winter Wheat. Plant Pathol. 2004, 53, 653–659. [Google Scholar] [CrossRef]
- Jan, A.U.; Hadi, F.; Midrarullah; Nawaz, M.A.; Rahman, K. Potassium and Zinc Increase Tolerance to Salt Stress in Wheat (Triticum aestivum L.). Plant Physiol. Biochem. 2017, 116, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Amtmann, A.; Troufflard, S.; Armengaud, P. The Effect of Potassium Nutrition on Pest and Disease Resistance in Plants. Physiol. Plant. 2008, 133, 682–691. [Google Scholar] [CrossRef]
- Pervez, H.; Ashraf, M.; Makhdum, M. Influence of Potassium Nutrition on Gas Exchange Characteristics and Water Relations in Cotton (Gossypium hirsutum L.). Photosynthetica 2004, 42, 251–255. [Google Scholar] [CrossRef]
- Asif, M.; Yilmaz, O.; Ozturk, L. Potassium Deficiency Impedes Elevated Carbon Dioxide-Induced Biomass Enhancement in Well Watered or Drought-Stressed Bread Wheat. Zeitschrift fur Pflanzenernahrung und Bodenkunde 2017, 180, 474–481. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, H.; Chen, L.; Wang, N.; Wei, C.; Wan, X. Mesophyll Cells’ Ability to Maintain Potassium Is Correlated with Drought Tolerance in Tea (Camellia sinensis). Plant Physiol. Biochem. 2019, 136, 196–203. [Google Scholar] [CrossRef]
- Sardans, J.; Peñuelas, J. Potassium: Trees increase their P:N ratio with size. Glob. Ecol. Biogeogr. 2015, 24, 147–156. [Google Scholar] [CrossRef] [Green Version]
- Meister, R.; Rajani, M.S.; Ruzicka, D.; Schachtman, D.P. Challenges of Modifying Root Traits in Crops for Agriculture. Trends Plant Sci. 2014, 19, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Rogers, E.D.; Benfey, P.N. Regulation of Plant Root System Architecture: Implications for Crop Advancement. Curr. Opin. Biotechnol. 2015, 32, 93–98. [Google Scholar] [CrossRef]
- Kellermeier, F.; Chardon, F.; Amtmann, A. Natural Variation of Arabidopsis Root Architecture Reveals Complementing Adaptive Strategies to Potassium Starvation. Plant Physiol. 2013, 161, 1421–1432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, R.; Schachtman, D.P. Hydrogen Peroxide Mediates Plant Root Cell Response to Nutrient Deprivation. Proc. Natl. Acad. Sci. USA 2004, 101, 8827–8832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dun, X.; Shi, J.; Liu, H.; Wang, J.; Wang, X.; Wang, H. Genetic Dissection of Root Morphological Traits as Related to Potassium Use Efficiency in Rapeseed under Two Contrasting Potassium Levels by Hydroponics. Sci. China Life Sci. 2019, 62, 746–757. [Google Scholar] [CrossRef] [PubMed]
- Hammer, G.L.; Dong, Z.; McLean, G.; Doherty, A.; Messina, C.; Schussler, J.; Zinselmeier, C.; Paszkiewicz, S.; Cooper, M. Can Changes in Canopy and/or Root System Architecture Explain Historical Maize Yield Trends in the U.S. Corn Belt? Crop Sci. 2009, 49, 299–312. [Google Scholar] [CrossRef]
- den Herder, G.; van Isterdael, G.; Beeckman, T.; de Smet, I. The Roots of a New Green Revolution. Trends Plant Sci. 2010, 15, 600–607. [Google Scholar] [CrossRef]
- Huang, X.; Han, B. Natural Variations and Genome-Wide Association Studies in Crop Plants. Annu. Rev. Plant Biol. 2014, 65, 531–551. [Google Scholar] [CrossRef]
- Patishtan, J.; Hartley, T.N.; Fonseca de Carvalho, R.; Maathuis, F.J.M. Genome-Wide Association Studies to Identify Rice Salt-Tolerance Markers. Plant Cell Environ. 2018, 41, 970–982. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, J.; He, X.; Wang, Y.; Ma, X.; Yin, D. Genome-Wide Association Study of Major Agronomic Traits Related to Domestication in Peanut. Front. Plant Sci. 2017, 8, 1611. [Google Scholar] [CrossRef] [Green Version]
- Zegeye, H.; Rasheed, A.; Makdis, F.; Badebo, A.; Ogbonnaya, F.C. Genome-Wide Association Mapping for Seedling and Adult Plant Resistance to Stripe Rust in Synthetic Hexaploid Wheat. PLoS ONE 2014, 9, e105593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newell, M.A.; Cook, D.; Tinker, N.A.; Jannink, J.L. Population Structure and Linkage Disequilibrium in Oat (Avena sativa L.): Implications for Genome-Wide Association Studies. Theor. Appl. Genet. 2011, 122, 623–632. [Google Scholar] [CrossRef]
- Li, C.; Fu, Y.; Sun, R.; Wang, Y.; Wang, Q. Single-Locus and Multi-Locus Genome-Wide Association Studies in the Genetic Dissection of Fiber Quality Traits in Upland Cotton (Gossypium hirsutum L.). Front. Plant Sci. 2018, 9, 1107. [Google Scholar] [CrossRef] [PubMed]
- Spindel, J.E.; Begum, H.; Akdemir, D.; Collard, B.; Redoña, E.; Jannink, J.L.; McCouch, S. Genome-Wide Prediction Models That Incorporate de Novo GWAS Are a Powerful New Tool for Tropical Rice Improvement. Heredity 2016, 116, 395–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, G.P.; Ramu, P.; Deshpande, S.P.; Hash, C.T.; Shah, T.; Upadhyaya, H.D.; Riera-Lizarazu, O.; Brown, P.J.; Acharya, C.B.; Mitchell, S.E.; et al. Population Genomic and Genome-Wide Association Studies of Agroclimatic Traits in Sorghum. Proc. Natl. Acad. Sci. USA 2013, 110, 453–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Xu, C.; Xu, S. Prediction and Association Mapping of Agronomic Traits in Maize Using Multiple Omic Data. Heredity 2017, 119, 174–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Song, Q.; Cregan, P.B.; Nelson, R.L.; Wang, X.; Wu, J.; Jiang, G.L. Genome-Wide Association Study for Flowering Time, Maturity Dates and Plant Height in Early Maturing Soybean (Glycine Max L.) Germplasm. BMC Genom. 2015, 16, 217. [Google Scholar] [CrossRef] [Green Version]
- Visioni, A.; Tondelli, A.; Francia, E.; Pswarayi, A.; Malosetti, M.; Russell, J.; Thomas, W.; Waugh, R.; Pecchioni, N.; Romagosa, I.; et al. Genome-Wide Association Mapping of Frost Tolerance in Barley (Hordeum Vulgare L.). BMC Genom. 2013, 14, 424. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for Association Mapping of Complex Traits in Diverse Samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Wang, S.B.; Feng, J.Y.; Ren, W.L.; Huang, B.; Zhou, L.; Wen, Y.J.; Zhang, J.; Dunwell, J.M.; Xu, S.; Zhang, Y.M. Improving Power and Accuracy of Genome-Wide Association Studies via a Multi-Locus Mixed Linear Model Methodology. Sci. Rep. 2016, 6, 19444. [Google Scholar] [CrossRef] [Green Version]
- Wen, Y.J.; Zhang, H.; Ni, Y.L.; Huang, B.; Zhang, J.; Feng, J.Y.; Wang, S.B.; Dunwell, J.M.; Zhang, Y.M.; Wu, R. Methodological Implementation of Mixed Linear Models in Multi-Locus Genome-Wide Association Studies. Brief. Bioinform. 2018, 19, 700–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamba, C.L.; Ni, Y.L.; Zhang, Y.M. Iterative Sure Independence Screening EM-Bayesian LASSO Algorithm for Multi-Locus Genome-Wide Association Studies. PLoS Comput. Biol. 2017, 13, e1005357. [Google Scholar] [CrossRef] [PubMed]
- Gautam, M.; Ge, X.H.; Li, Z.Y. Brassica. In Alien Gene Transfer in Crop Plants; Springer: New York, NY, USA, 2013; Volume 2, pp. 207–229. ISBN -9781461495727. [Google Scholar]
- Tao, R.; Jianwei, L.; Hui, L.; Juan, Z.; Huali, X.; Xiaowei, L.; Xiaokun, L. Potassium-fertilizer management in winter oilseed-rape production in China. J. Plant Nutr. Soil Sci. 2013, 176, 429–440. [Google Scholar]
- Arifuzzaman, M.; Mamidi, S.; Mcclean, P.; Rahman, M. QTL Mapping for Root Vigor and Days to Flowering in Brassica napus L. Can. J. Plant Sci. 2016, 97, 99–109. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Hu, D.; You, J.; Wu, D.; Cui, Y.; Dong, H.; Li, J.; Qian, W. Genome-Wide Association Study and Protein Network Analysis for Understanding Candidate Genes Involved in Root Development at the Rapeseed Seedling Stage. Plant Physiol. Biochem. 2019, 137, 42–52. [Google Scholar] [CrossRef]
- Ibrahim, S.; Li, K.; Ahmad, N.; Kuang, L.; Sadau, S.B.; Tian, Z.; Huang, L.; Wang, X.; Dun, X.; Wang, H. Genetic Dissection of Mature Root Characteristics by Genome-Wide Association Studies in Rapeseed (Brassica napus L.). Plants 2021, 10, 2569. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wang, J.; Kuang, L.; Tian, Z.; Wang, X.; Dun, X.; Tu, J.; Wang, H. Genome-Wide Association Study and Transcriptome Analysis Reveal Key Genes Affecting Root Growth Dynamics in Rapeseed. Biotechnol. Biofuels 2021, 14, 178. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Mcclean, P. Genetic Analysis on Flowering Time and Root System in Brassica napus L. Crop Sci. 2013, 53, 141–147. [Google Scholar] [CrossRef]
- Shi, L.; Shi, T.; Broadley, M.R.; White, P.J.; Long, Y.; Meng, J.; Xu, F.; Hammond, J.P. High-Throughput Root Phenotyping Screens Identify Genetic Loci Associated with Root Architectural Traits in Brassica napus under Contrasting Phosphate Availabilities. Ann. Bot. 2013, 112, 381–389. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Dun, X.; Shi, J.; Wang, X.; Liu, G.; Wang, H. Genetic Dissection of Root Morphological Traits Related to Nitrogen Use Efficiency in Brassica napus L. under Two Contrasting Nitrogen Conditions. Front. Plant Sci. 2017, 8, 1709. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Ding, G.; Shi, L.; Feng, J.; Xu, F.; Meng, J. Quantitative Trait Loci for Root Morphology in Response to Low Phosphorus Stress in Brassica napus. Theor. Appl. Genet. 2010, 121, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Thomas, C.L.; Xiang, J.; Long, Y.; Wang, X.; Zou, J.; Luo, Z.; Ding, G.; Cai, H.; Graham, N.S.; et al. QTL Meta-Analysis of Root Traits in Brassica napus under Contrasting Phosphorus Supply in Two Growth Systems. Sci. Rep. 2016, 6, 33113. [Google Scholar] [CrossRef] [PubMed]
- Trovato, M.; Mattioli, R.; Costantino, P. Multiple Roles of Proline in Plant Stress Tolerance and Development. Rend. Lincei 2008, 19, 325–346. [Google Scholar] [CrossRef]
- Biancucci, M.; Mattioli, R.; Moubayidin, L.; Sabatini, S.; Costantino, P.; Trovato, M. Proline Affects the Size of the Root Meristematic Zone in Arabidopsis. BMC Plant Biol. 2015, 15, 263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosquete, M.R.; Waidmann, S.; Kleine-Vehn, J. PIN7 Auxin Carrier Has a Preferential Role in Terminating Radial Root Expansion in Arabidopsis thaliana. Int. J. Mol. Sci. 2018, 19, 1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porco, S.; Larrieu, A.; Du, Y.; Gaudinier, A.; Goh, T.; Swarup, K.; Swarup, R.; Kuempers, B.; Bishopp, A.; Lavenus, J.; et al. Lateral Root Emergence in Arabidopsis Is Dependent on Transcription Factor LBD29 Regulation of Auxin Influx Carrier LAX3. Development 2016, 143, 3340–3349. [Google Scholar] [CrossRef] [Green Version]
- Fei, Q.; Wei, S.; Zhou, Z.; Gao, H.; Li, X. Adaptation of Root Growth to Increased Ambient Temperature Requires Auxin and Ethylene Coordination in Arabidopsis. Plant Cell Rep. 2017, 36, 1507–1518. [Google Scholar] [CrossRef]
- Seifert, G.J.; Barber, C.; Wells, B.; Dolan, L.; Roberts, K. Galactose Biosynthesis in Arabidopsis: Genetic Evidence for Substrate Channeling from UDP-D-Galactose into Cell Wall Polymers. Curr. Biol. 2002, 12, 1840–1845. [Google Scholar] [CrossRef] [Green Version]
- Xie, D.W.; Wang, X.N.; Fu, L.S.; Sun, J.; Zheng, W.; Li, Z.F. Identification of the Trehalose-6-Phosphate Synthase Gene Family in Winter Wheat and Expression Analysis under Conditions of Freezing Stress. J. Genet. 2015, 94, 55–65. [Google Scholar] [CrossRef]
- Li, W.; Huai, X.; Li, P.; Raza, A.; Mubarik, M.S.; Habib, M.; Faiz, S.; Zhang, B.; Pan, J.; Khan, R.S.A. Genome-Wide Characterization of Glutathione Peroxidase (GPX) Gene Family in Rapeseed (Brassica napus L.) Revealed Their Role in Multiple Abiotic Stress Response and Hormone Signaling. Antioxidants 2021, 10, 1481. [Google Scholar] [CrossRef]
- Wang, T.; Wei, L.; Wang, J.; Xie, L.; Li, Y.Y.; Ran, S.; Ren, L.; Lu, K.; Li, J.; Timko, M.P.; et al. Integrating GWAS, Linkage Mapping and Gene Expression Analyses Reveals the Genetic Control of Growth Period Traits in Rapeseed (Brassica napus L.). Biotechnol. Biofuels 2020, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Liu, Y.; Gruber, B.D.; Neumann, K.; Kilian, B.; Graner, A.; von Wirén, N. Genetic Dissection of Root System Architectural Traits in Spring Barley. Front. Plant Sci. 2019, 10, 400. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Thuillet, A.C.; Yu, J.; Pressoir, G.; Romero, S.M.; Mitchell, S.E.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Maize Association Population: A High-Resolution Platform for Quantitative Trait Locus Dissection. Plant J. 2005, 44, 1054–1064. [Google Scholar] [CrossRef] [PubMed]
- Stich, B.; Möhring, J.; Piepho, H.P.; Heckenberger, M.; Buckler, E.S.; Melchinger, A.E. Comparison of Mixed-Model Approaches for Association Mapping. Genetics 2008, 178, 1745–1754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naz, A.A.; Arifuzzaman, M.; Muzammil, S.; Pillen, K.; Léon, J. Wild Barley Introgression Lines Revealed Novel QTL Alleles for Root and Related Shoot Traits in the Cultivated Barley (Hordeum vulgare L.). BMC Genet. 2014, 15, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, W.; Wang, B.; Hauck, A.L.; Dong, X.; Li, J.; Lai, J. Genetic Dissection of Maize Seedling Root System Architecture Traits Using an Ultra-High Density Bin-Map and a Recombinant Inbred Line Population. J. Integr. Plant Biol. 2016, 58, 266–279. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kuang, L.; Wang, X.; Liu, G.; Dun, X.; Wang, H. Temporal Genetic Patterns of Root Growth in Brassica napus L. Revealed by a Low-Cost, High-Efficiency Hydroponic System. Theor. Appl. Genet. 2019, 132, 2309–2323. [Google Scholar] [CrossRef]
- Bernardino, K.C.; Pastina, M.M.; Menezes, C.B.; de Sousa, S.M.; MacIel, L.S.; Geraldo Carvalho, G.C.; Guimarães, C.T.; Barros, B.A.; da Costa E Silva, L.; Carneiro, P.C.S.; et al. The Genetic Architecture of Phosphorus Efficiency in Sorghum Involves Pleiotropic QTL for Root Morphology and Grain Yield under Low Phosphorus Availability in the Soil. BMC Plant Biol. 2019, 19, 87. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, W.; Zhang, N.; Chen, M.; Zheng, S.; Zhao, C.; Han, J.; Liu, J.; Zhang, X.; Song, L.; et al. Identification of QTL Regions for Seedling Root Traits and Their Effect on Nitrogen Use Efficiency in Wheat (Triticum aestivum L.). Theor. Appl. Genet. 2018, 131, 2677–2698. [Google Scholar] [CrossRef]
- Gong, X.; McDonald, G. QTL Mapping of Root Traits in Phosphorus-Deficient Soils Reveals Important Genomic Regions for Improving NDVI and Grain Yield in Barley. Theor. Appl. Genet. 2017, 130, 1885–1902. [Google Scholar] [CrossRef]
- Islam, A.; Zhang, Y.; Anis, G.; Rani, M.H.; Anley, W.; Yang, Q.; Liu, L.; Shen, X.; Cao, L.; Cheng, S.; et al. Fine Mapping and Candidate Gene Analysis of QRN5a, a Novel QTL Promoting Root Number in Rice under Low Potassium. Theor. Appl. Genet. 2021, 134, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Zhang, F.; Chong, X.; Song, A.; Guan, Z.; Fang, W.; Chen, F. Genome-Wide Association Study Identifies Favorable SNP Alleles and Candidate Genes for Waterlogging Tolerance in Chrysanthemums. Hortic. Res. 2019, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wang, B.; Chen, X.; Wu, J.; King, G.J.; Xiao, Y.; Liu, K. Evaluation of Linkage Disequilibrium Pattern and Association Study on Seed Oil Content in Brassica napus Using DdRAD Sequencing. PLoS ONE 2016, 11, e0189308. [Google Scholar] [CrossRef] [Green Version]
- Hoagland, D.R.; Arnon, D.I. Preparing the Nutrient Solution. Water-Cult. Method Grow. Plants Without Soil 1950, 347, 29–31. [Google Scholar]
- Pace, J.; Gardner, C.; Romay, C.; Ganapathysubramanian, B.; Lübberstedt, T. Genome-Wide Association Analysis of Seedling Root Development in Maize (Zea mays L.). BMC Genom. 2015, 16, 47. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.; Jian, H.; Lu, K.; Filardo, F.; Yin, N.; Liu, L.; Qu, C.; Li, W.; Du, H.; Li, J. Genome-Wide Association Analysis and Differential Expression Analysis of Resistance to Sclerotinia Stem Rot in Brassica napus. Plant Biotechnol. J. 2016, 14, 1368–1380. [Google Scholar] [CrossRef] [Green Version]
- Jeanmougin, F.; Thompson, J.D.; Gouy, M.; Higgins, D.G.; Gibson, T.J. Multiple Sequence Alignment with Clustal X. Trends Biochem. Sci. 1998, 23, 403–405. [Google Scholar] [CrossRef]

| Classification | Trait Description | Abbreviations | Units |
|---|---|---|---|
| Root-related traits | Primary root length | PRL | cm |
| Total root volume | TRV | cm3 | |
| Total root surface area | TSA | cm2 | |
| Total root length | TRL | cm | |
| Total root number | TRN | N | |
| Biomass-related traits | Root fresh weight | RFW | g |
| Shoot fresh weight | SFW | g | |
| Total fresh weights | TFW | g | |
| Root–shoot fresh weight ratio | RSR |
| Trait | Mean | SD | Min | Max | Skewness | Kurtosis | CV (%) | H2 (%) |
|---|---|---|---|---|---|---|---|---|
| PRL | 23.57 | 2.77 | 13.72 | 31.62 | −0.05 | 0.35 | 11.73 | 56.9 |
| SFW | 2.47 | 0.44 | 1.20 | 3.72 | 0.01 | −0.07 | 17.83 | 60.4 |
| RFW | 0.43 | 0.08 | 0.23 | 0.71 | 0.28 | 0.65 | 18.38 | 60.3 |
| TRL | 711.5 | 115.1 | 409.8 | 1156.3 | 0.47 | 1.16 | 16.17 | 53.1 |
| TSA | 51.7 | 8.54 | 30.2 | 87.4 | 0.42 | 0.94 | 16.51 | 52.6 |
| TRV | 0.31 | 0.06 | 0.14 | 0.53 | 0.29 | 0.17 | 19.15 | 49.4 |
| TRN | 1084.1 | 310.5 | 540.3 | 3417.2 | 2.22 | 11.39 | 28.64 | 52.1 |
| TFW | 2.90 | 0.49 | 1.43 | 4.35 | −0.01 | −0.12 | 17.03 | 60.2 |
| RSR | 0.18 | 0.03 | 0.09 | 0.31 | 0.89 | 1.54 | 18.27 | 62.1 |
| Cluster | Trait | Lead SNP | Position (bp) | Gene Model | Distance to Lead SNP (Kb) | Gene Symbol | At Homolog Genes | Annotation |
|---|---|---|---|---|---|---|---|---|
| qRT.A02-4 | RFW, RSR | Bn-A02-p13803435 | 10,432,009 | BnaA02g17300D | −29.99 | SAUR | AT1G75580 | SAUR-like auxin-responsive protein family |
| qRT.A02-11 | TRN, RSR | Bn-A02-p8199891 | 5,191,339 | BnaA02g10340D | −94.04 | CRF3 | AT5G53290 | Cytokinin response factor 3 |
| qRT.A03-12 | TSA, TRV, RSR | Bn-A03-p10023639 | 9,214,456 | BnaA03g19500D | −22.93 | CKX1 | AT2G41510 | Cytokinin oxidase/dehydrogenase 1 |
| qRT.A03-17 | TRL, TSA | seq-new-rs32620 | 2,981,029 | BnaA03g06500D | 76.26 | UGP2 | AT5G17310 | UDP-glucose pyrophosphorylase 2 |
| BnaA03g06730D | −35.11 | MYB56 | AT5G17800 | MYB domain protein 56 | ||||
| BnaA03g06800D | −61.10 | SAUR | AT5G18010 | SAUR-like auxin-responsive protein family | ||||
| qRT.A04-4 | RSR, TRL, TSA, TRV | Bn-A04-p12314909 | 13,306,243 | BnaA04g16140D | 97.43 | GH3 | AT1G48670 | Auxin-responsive GH3 family protein |
| qRT.A05-11 | TSA, TRV | Bn-A05-p2142102 | 2,273,504 | BnaA05g04380D | −85.25 | ERF13 | AT2G44840 | Ethylene-responsive element binding factor 13 |
| qRT.A06-10 | RSR, TRN | seq-new-rs33430 | 2,265,805 | BnaA06g03580D | 71.58 | GH3 | AT1G23160 | Auxin-responsive GH3 family protein |
| BnaA06g03620D | 33.41 | RACK1B_AT | AT1G48630 | Receptor for activated C kinase 1B | ||||
| qRT.A07-1 | PRL, RSR | Bn-A07-p21573107 | 23,110,821 | BnaA07g33740D | 25.48 | ARF17 | AT1G77850 | Auxin response factor 17 |
| qRT.A07-6 | TRV, RSR | seq-new-rs40608 | 9,704,794 | BnaA07g10150D | 25.95 | PIN7 | AT1G23080 | PIN-FORMED 7 |
| qRT.A08-1 | RSR, SFW | seq-new-rs39127 | 12,905,937 | BnaA08g15590D | −35.17 | CP1 | AT4G36880 | Cysteine proteinase1 |
| BnaA08g15600D | −56.19 | GASA1 | AT1G75750 | GAST1 protein homolog 1 | ||||
| qRT.A09-4 | TFW, SFW | seq-new-rs26492 | 25,679,360 | BnaA09g35190D | 21.94 | SDIR1 | AT3G55530 | SALT- AND DROUGHT-INDUCED RING FINGER1 |
| BnaA09g35230D | −3.72 | P5CS2 | AT3G55610 | Delta 1-pyrroline-5-carboxylate synthase 2 | ||||
| qRT.A10-1 | SFW, TFW | Bn-A10-p15967013 | 15,593,735 | BnaA10g23640D | 16.74 | GA20OX3 | AT5G07200 | Gibberellin 20-oxidase 3 |
| BnaA10g23650D | 9.42 | RR21 | AT5G07210 | Response regulator 21 | ||||
| BnaA10g23740D | −42.10 | ML4 | AT5G07290 | MEI2-like 4 | ||||
| qRT.A10-3 | RSR, RFW | Bn-A10-p10120142 | 11,533,850 | BnaA10g14470D | 28.88 | EIL2 | AT5G21120 | ETHYLENE-INSENSITIVE3-like 2 |
| qRT.C01-7 | RSR, TRV, RFW | seq-new-rs38417 | 12,806,484 | BnaC01g18450D | −24.27 | LTI65 | AT5G52300 | LOW-TEMPERATURE-INDUCED 65 |
| qRT.C04-2 | SFW, TFW | Bn-scaff_16888_1-p1169101 | 45,355,403 | BnaC04g45700D | 33.58 | AUX1 | AT2G38120 | AUXIN RESISTANT 1 |
| BnaC04g45720D | 23.19 | CAX1 | AT2G38170 | Cation exchanger 1 | ||||
| BnaC04g45770D | −11.70 | AT2G38240 | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | |||||
| qRT.C04-8 | RSR, TRV | seq-new-rs30573 | 25,264,080 | BnaC04g24310D | −42.70 | EIF3E | AT3G57290 | Eukaryotic translation initiation factor 3E |
| qRT.C06-10 | TRV, RSR | seq-new-rs48045 | 5,209,723 | BnaC06g04590D | −14.14 | AT1G51460 | ABC-2 type transporter family protein | |
| BnaC06g04620D | −63.33 | AT1G51538 | Aminotransferase-like, plant mobile domain family protein | |||||
| qRT.C07-1 | TRV, TRN | seq-new-rs28637 | 44,197,394 | BnaC07g46640D | 28.74 | LCR59 | AT4G30070 | Low-molecular-weight cysteine-rich 59 |
| qRT.C07-2 | RSR, TRL | seq-new-rs46639 | 35,175,730 | BnaC07g30830D | 55.17 | LR | AT5G23400 | Leucine-rich repeat (LRR) family protein |
| BnaC07g30930D | −19.94 | SAR1 | AT1G33410 | SUPPRESSOR OF AUXIN RESISTANCE1 | ||||
| qRT.C07-6 | TRL, TSA, TRV | Bn-scaff_18202_1-p1536412 | 22,287,865 | BnaC07g16350D | −20.37 | ABA1 | AT5G67030 | ABA DEFICIENT 1 |
| qRT.C08-5 | RSR, SFW | seq-new-rs34390 | 29,522,206 | BnaC08g29060D | 64.89 | AFB3 | AT1G12820 | Auxin signaling F-box 3 |
| BnaC08g29120D | 41.24 | LBD29 | AT3G58190 | Lateral organ boundaries-domain 29 | ||||
| qRT.C09-1 | TRV, RSR | seq-new-rs41567 | 42,567,030 | BnaC09g40100D | −55.68 | WOX12 | AT5G17810 | WUSCHEL related homeobox 12 |
| qRT.C09-2 | RFW, RSR | seq-new-rs25004 | 38,586,454 | BnaC09g35190D | −14.05 | AT5G59845 | Gibberellin-regulated family protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibrahim, S.; Ahmad, N.; Kuang, L.; Tian, Z.; Sadau, S.B.; Iqbal, M.S.; Wang, X.; Wang, H.; Dun, X. Genome-Wide Association Studies of Root-Related Traits in Brassica napus L. under Low-Potassium Conditions. Plants 2022, 11, 1826. https://doi.org/10.3390/plants11141826
Ibrahim S, Ahmad N, Kuang L, Tian Z, Sadau SB, Iqbal MS, Wang X, Wang H, Dun X. Genome-Wide Association Studies of Root-Related Traits in Brassica napus L. under Low-Potassium Conditions. Plants. 2022; 11(14):1826. https://doi.org/10.3390/plants11141826
Chicago/Turabian StyleIbrahim, Sani, Nazir Ahmad, Lieqiong Kuang, Ze Tian, Salisu Bello Sadau, Muhammad Shahid Iqbal, Xinfa Wang, Hanzhong Wang, and Xiaoling Dun. 2022. "Genome-Wide Association Studies of Root-Related Traits in Brassica napus L. under Low-Potassium Conditions" Plants 11, no. 14: 1826. https://doi.org/10.3390/plants11141826
APA StyleIbrahim, S., Ahmad, N., Kuang, L., Tian, Z., Sadau, S. B., Iqbal, M. S., Wang, X., Wang, H., & Dun, X. (2022). Genome-Wide Association Studies of Root-Related Traits in Brassica napus L. under Low-Potassium Conditions. Plants, 11(14), 1826. https://doi.org/10.3390/plants11141826







