Development of Ethyl Methanesulfonate Mutant Edamame Soybean (Glycine max (L.) Merr.) Populations and Forward and Reverse Genetic Screening for Early-Flowering Mutants
Abstract
:1. Introduction
2. Results and Discussion
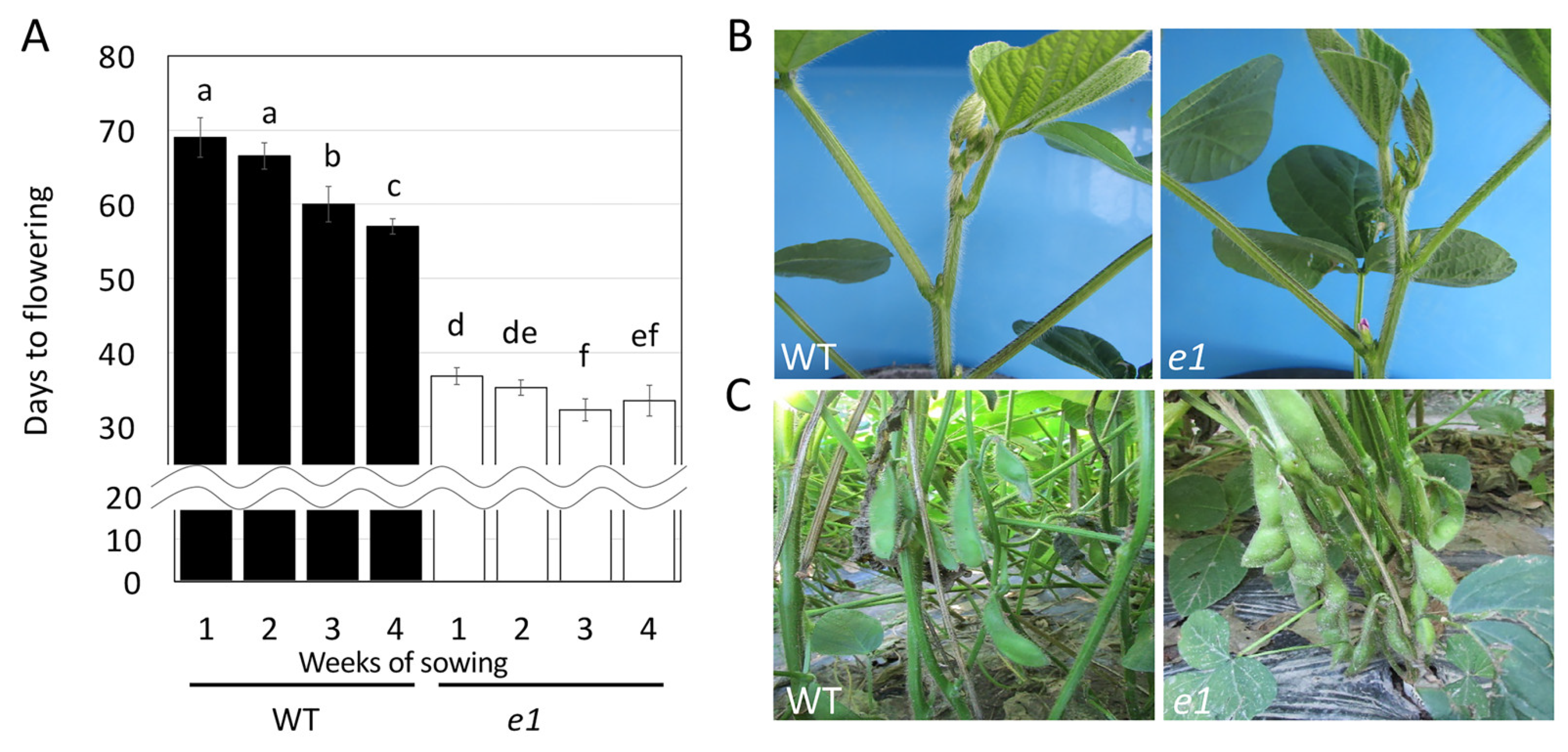
2.1. Production of EMS-Induced Edamame Hiden Mutant Populations
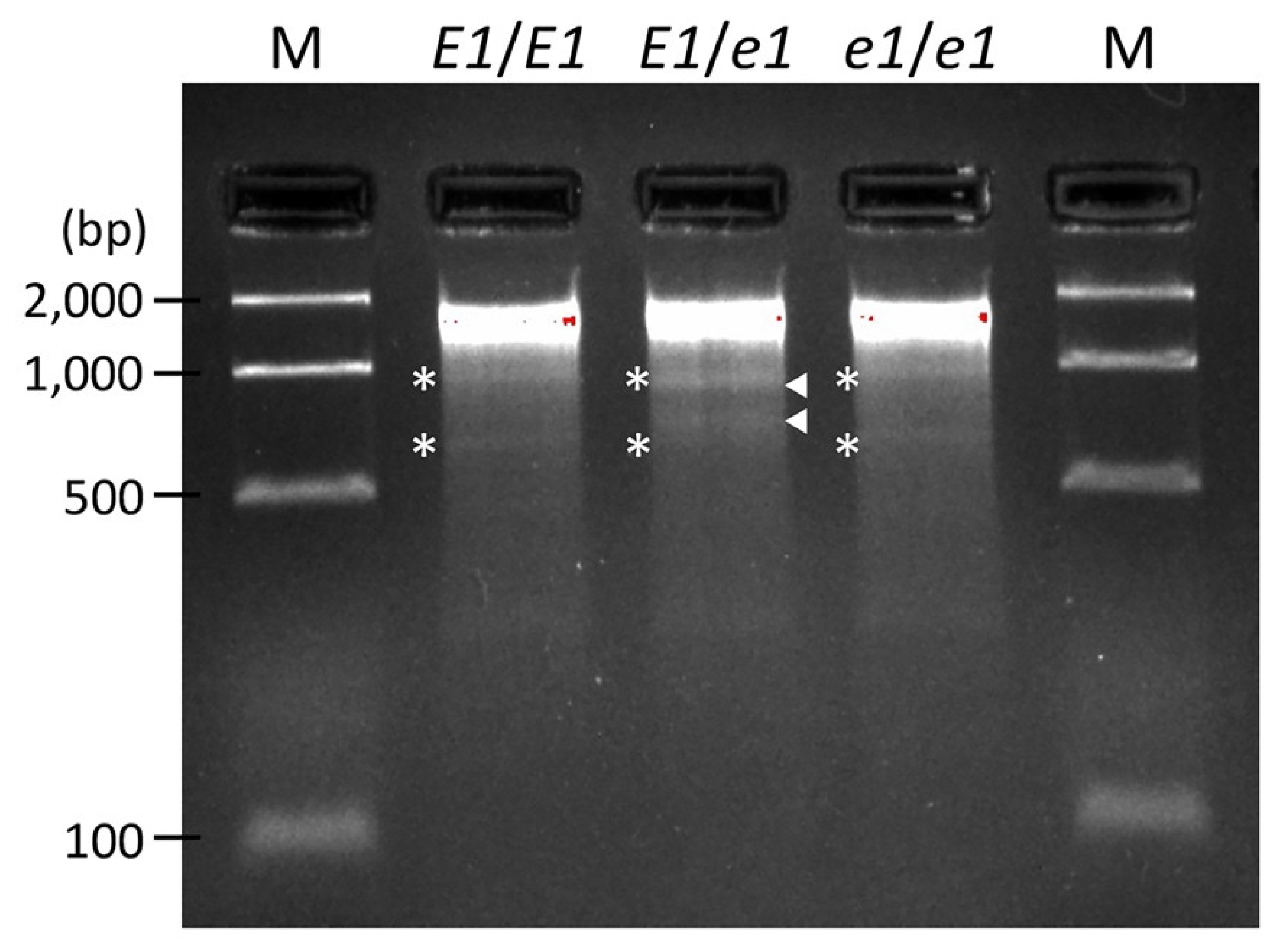
2.2. EMS-Induced DNA Mutations in Edamame Hiden Mutant Populations
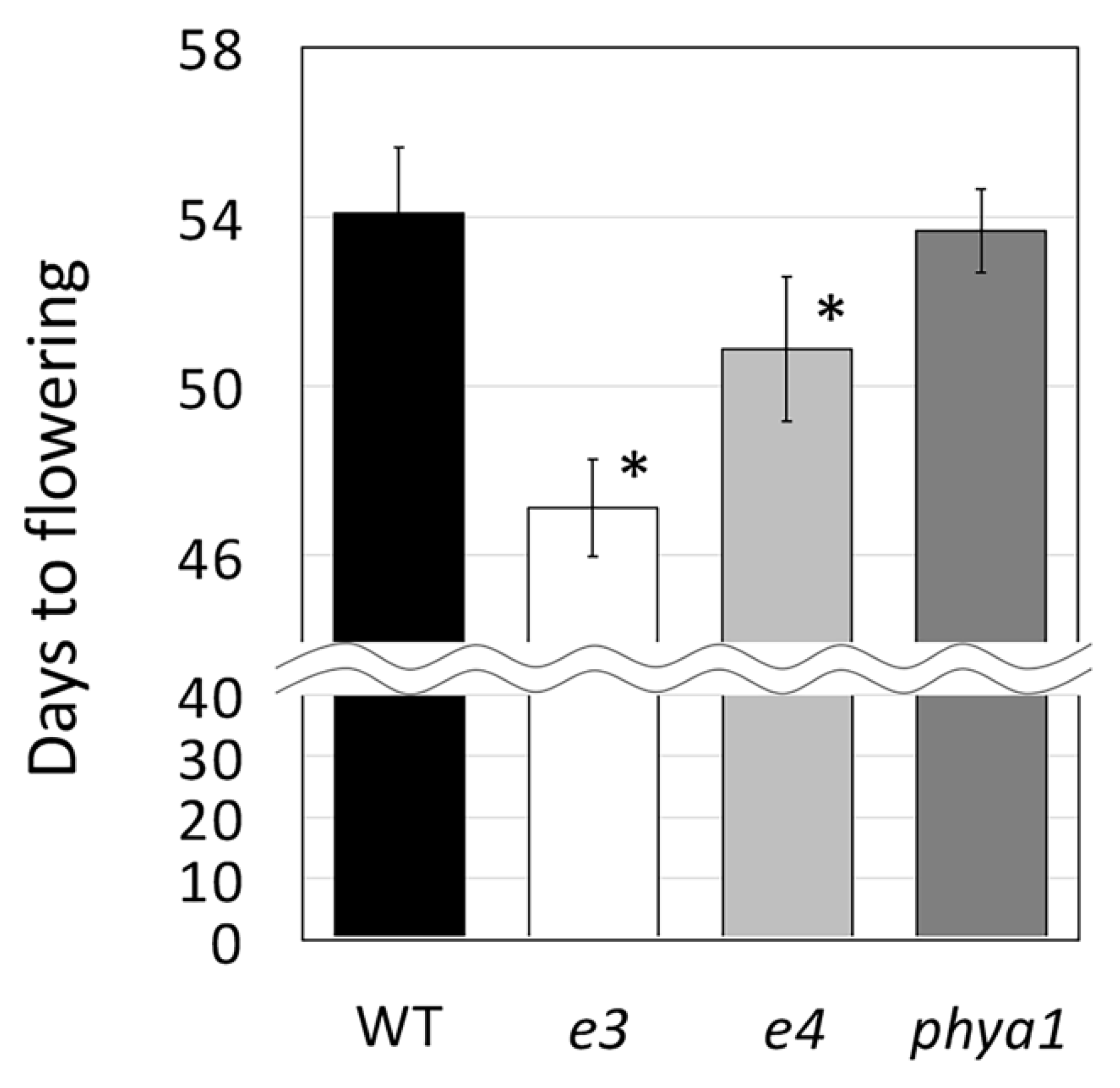
2.3. Usefulness of the Obtained Mutated Alleles of the Flowering and Maturity-Related Genes
| Target Genes | Line | Mutation | Changed Codon | Amino Acid Substitution | Position |
|---|---|---|---|---|---|
| e1 | 17-0440 | C→T | TCC→TTC | Ser→Phe | C521T |
| 17-0010 | G→A | AAG→AAA | Lys→Lys | G561A | |
| 16-0233 | G→A | GGA→GAA | Gly→Glu | G578A | |
| 17-0291 | G→A | - | - | 3′UTR | |
| 16-0694 | G→A | - | - | 3′UTR | |
| e3 | 17-0574 | G→A | AGT→AAT | Ser→Asn | G428A |
| 17-1029 | G→A | AGA→AAA | Arg→Lys | G455A | |
| 17-1220 | C→T | CCT→TCT | Pro→Ser | C841T | |
| 17-1131 | C→T | GCC→GCT | Ala→Ala | C1362T | |
| 17-1035 | G→T * | GAG→TAG | Glu→STOP | G1444T | |
| 17-0175 | G→A | AGG→AAG | Arg→Lys | G1652A | |
| 16-0927 | G→A | GGA→GAA | Gly→Glu | G2414A | |
| e4 | 16-0789 | C→T | - | - | Intron1 |
| 16-0269 | T→A * | CCT→CCA | Pro→Pro | T246A | |
| 16-0459 | G→A | CAG→CAA | Gln→Gln | G426A | |
| 16-0048 | G→A | ATG→ATA | Met→Ile | G1017A | |
| 17-0032 | C→T | - | - | Intron2 | |
| 16-0106 | C→T | CCT→TCT | Pro→Ser | C2287T | |
| 17-0251 | ΔC | - | - | C2507Δ | |
| 16-0763 | G→A | ATG→ATA | Met→Ile | G2754A | |
| 16-1031 | C→T | CTA→TTA | Leu→Leu | C2794T | |
| 17-0881 | G→A | GAT→AAT | Asp→Asn | G2866A | |
| 16-0139 | C→T | - | - | Intron3 | |
| 16-0499 | G→A | - | - | Intron4 | |
| phya1 | 17-0499 | A→G | - | - | Intron1 |
| 17-0282 | C→A * | - | - | Intron1 | |
| 17-0983 | T→C | ATT→ATC | Ile→Ile | T717C | |
| 16-0340 | ΔT | - | - | T820Δ | |
| 17-0311 | G→A | AGG→AGA | Arg→Arg | G1029A | |
| 17-0498 | G→A | - | - | Intron4 |
2.4. DNA Marker Development for Breeding Applications
3. Materials and Methods
3.1. Plant Materials
3.2. TILLING Screening
3.3. Scoring of Flowering Days
3.4. DNA Marker Analyses
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Konovsky, J.; Lumpkin, T.A.; McClary, D. Edamame: The vegetable soybean. In Understanding the Japanese Food and Agrimarket: A Multifaceted Opportunity; O’Rourke, A.D., Ed.; Haworth Press: Binghamton, NY, USA, 1994; pp. 173–181. [Google Scholar]
- Mentreddy, S.R.; Mohamed, A.I.; Joshee, N.; Yadav, A.K. Edamame: A nutritious vegetable crop. In Trends New Crop New Uses; Janick, J., Whipkey, A., Eds.; ASHS Press: Alexandria, VA, USA, 2002; pp. 432–438. [Google Scholar]
- Mahoussi, K.A.D.; Eric, E.A.; Symphorien, A.; Florent, J.Q.; Flora, J.C.; Achille, E.A.; Clement, A.; Brice, S. Vegetable soybean, edamame: Research, production, utilization and analysis of its adoption in sub-saharan Africa. J. Hortic. For. 2020, 12, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Anai, T. Potential of a mutant-based reverse genetic approach for functional genomics and molecular breeding in soybean. Breed. Sci. 2012, 61, 462–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, N.N.; Kim, D.-G.; Lyu, J.I.; Park, K.-C.; Kim, J.M.; Kim, J.-B.; Ha, B.-K.; Kwon, S.-J. Detecting genetic mobilityusing a transposon-based marker system in gamma-ray irradiated soybean mutants. Plants 2021, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Prenger, E.M.; Ostezan, A.; Mian, M.A.R.; Stupar, R.M.; Glenn, T.; Li, Z. Identification and characterization of a fast-neutron-induced mutant with elevated seed protein content in soybean. Theor. Appl. Genet. 2019, 132, 2965–2983. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.L.; Till, B.J.; Laport, R.G.; Darlow, M.C.; Kleffner, J.M.; Jamai, A.; El-Mellouki, T.; Liu, S.; Ritchie, R.; Nielsen, N.; et al. TILLING to detect induced mutations in soybean. BMC Plant Biol. 2008, 8, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Lakhssassi, N.; Cullen, M.A.; Baz, A.E.; Vuong, T.D.; Nguyen, H.T.; Meksem, K. Assessment of phenotypic variations and correlation among seed composition traits in mutagenized soybean populations. Genes 2019, 10, 975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thapa, R.; Carrero-Colón, M.; Rainey, K.M.; Hudson, K. TILLING by sequencing: A successful approach to identify rare alleles in soybean populations. Genes 2019, 10, 1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hancock, C.N.; Zhang, F.; Floyd, K.; Richardson, A.O.; Lafayette, P.; Tucker, D.; Wessler, S.R.; Parrott, W.A. The rice miniature inverted repeat transposable element mPing is an effective insertional mutagen in soybean. Plant Physiol. 2011, 157, 552–562. [Google Scholar] [CrossRef] [Green Version]
- Mathieu, M.; Winters, E.K.; Kong, F.; Wan, J.; Wang, S.; Eckert, H.; Luth, D.; Paz, M.; Donovan, C.; Zhang, Z.; et al. Establishment of a soybean (Glycine max Merr. L) transposon-based mutagenesis repository. Planta 2009, 229, 279–289. [Google Scholar] [CrossRef]
- Lu, S.; Yin, X.; Spollen, W.; Zhang, N.; Xu, D.; Schoelz, J.; Bilyeu, K.; Zhang, Z.J. Analysis of the siRNA-mediated gene silencing process targeting three homologous genes controlling soybean seed oil quality. PLoS ONE 2015, 10, e0129010. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.N.; Kim, H.J.; Chung, Y.S.; Kim, H.U. Construction of multiple guide RNAs in CRISPR/Cas9 vector using stepwise or simultaneous golden gate cloning: Case study for targeting the FAD2 and FATB multigene in soybean. Plants 2021, 10, 2542. [Google Scholar] [CrossRef] [PubMed]
- Niazian, M.; Belzile, F.; Torkamaneh, D. CRISPR/Cas9 in planta hairy root transformation: A powerful platform for functional analysis of root traits in soybean. Plants 2022, 11, 1044. [Google Scholar] [CrossRef]
- Bai, M.; Yuan, J.; Kuang, H.; Gong, P.; Li, S.; Zhang, Z.; Liu, B.; Sun, J.; Yang, M.; Yang, L.; et al. Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean. Plant Biotechnol. J. 2020, 18, 721–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espina, M.J.; Ahmed, C.M.S.; Bernardini, A.; Adeleke, E.; Yadegari, Z.; Arelli, P.; Pantalone, V.; Taheri, A. Development and phenotypic screening of an ethyl methane sulfonate mutant population in soybean. Front. Plant Sci. 2018, 9, 394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshino, T.; Takagi, Y.; Anai, T. Novel GmFAD2-1b mutant alleles created by reverse genetics induce drastic elevation of oleic acid content in soybean seeds in combination with GmFAD2-1a mutant alleles. Breed. Sci. 2010, 60, 419–425. [Google Scholar] [CrossRef] [Green Version]
- Tsuda, M.; Kaga, A.; Anai, T.; Shimizu, T.; Sayama, T.; Takagi, K.; Machita, K.; Watanabe, S.; Nishimura, M.; Yamada, N.; et al. Construction of a high-density mutant library in soybean and development of a mutant retrieval method using amplicon sequencing. BMC Genom. 2015, 16, 1014. [Google Scholar] [CrossRef] [Green Version]
- Till, B.J.; Zerr, T.; Comai, L.; Henikoff, S. A protocol for TILLING and Ecotilling in plants and animals. Nat. Protoc. 2006, 1, 2465–2477. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Liu, B.; Weller, J.L.; Abe, J.; Kong, F. Molecular mechanisms for the photoperiodic regulation of flowering in soybean. J. Integr. Plant Biol. 2021, 63, 981–994. [Google Scholar] [CrossRef]
- Xia, Z.; Watanabe, S.; Yamada, T.; Tsubokura, Y.; Nakashima, H.; Zhai, H.; Anai, T.; Sato, S.; Yamazaki, T.; Lü, S.; et al. Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. USA 2012, 109, E2155–E2164. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, S.; Xia, Z.; Hideshima, R.; Tsubokura, Y.; Sato, S.; Yamanaka, N.; Takahashi, R.; Anai, T.; Tabata, S.; Kitamura, K.; et al. A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 2011, 188, 395–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, S.; Hideshima, R.; Xia, Z.; Tsubokura, Y.; Sato, S.; Nakamoto, Y.; Yamanaka, N.; Takahashi, R.; Ishimoto, M.; Anai, T.; et al. Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 2009, 182, 1251–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Kanazawa, A.; Matsumura, H.; Takahashi, R.; Harada, K.; Abe, J. Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 2008, 180, 995–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greene, E.A.; Codomo, C.A.; Taylor, N.E.; Henikoff, J.G.; Till, B.J.; Reynolds, S.H.; Enns, L.C.; Burtner, C.; Johnson, J.E.; Odden, A.R.; et al. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 2003, 164, 731–740. [Google Scholar] [CrossRef]
- Satoh, H.; Matsusaka, H.; Kumamaru, T. Use of N-methyl-N-nitrosourea treatment of fertilized egg cells for saturation mutagenesis of rice. Breed. Sci. 2010, 60, 475–485. [Google Scholar] [CrossRef] [Green Version]
- Kawakami, T.; Goto, H.; Abe, Y.; Chuba, M.; Watanabe, M.; Hoshino, T. High frequency of transversion mutations in the rice (Oryza sativa L.) mutant population produced by diepoxybutane mutagenesis. Genet. Resour. Crop Evol. 2020, 67, 1355–1365. [Google Scholar] [CrossRef]
- Morita, R.; Kusaba, M.; Iida, S.; Yamaguchi, H.; Nishio, T.; Nishimura, M. Molecular characterization of mutations induced by gamma irradiation in rice. Genes Genet. Syst. 2009, 84, 361–370. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, T.; Watanabe, S.; Takagi, Y.; Anai, T. A novel GmFAD3-2a mutant allele developed through TILLING reduces α-linolenic acid content in soybean seed oil. Breed. Sci. 2014, 64, 371–377. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, T.; Iijima, N.; Hata, M.; Watanabe, A.; Kawakami, T.; Anai, T. Molecular characterization of high stearic acid soybean mutants and post-transcriptional control of GmSACPD genes in the mutant with a single nucleotide deletion. Plant Gene 2020, 21, 100207. [Google Scholar] [CrossRef]
- Yamagata, Y.; Yoshimura, A.; Anai, T.; Watanabe, S. Selection criteria for SNP loci to maximize robustness of high-resolution melting analysis for plant breeding. Breed. Sci. 2018, 68, 488–498. [Google Scholar] [CrossRef] [Green Version]
- Simko, I. High-resolution DNA melting analysis in plant research. Trends Plant Sci. 2016, 21, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Yu, J.; Wang, H.; Luth, D.; Bai, G.; Wang, K.; Chen, R. Increasing seed size and quality by manipulating BIG SEEDS1 in legume species. Proc. Natl. Acad. Sci. USA 2016, 113, 12414–12419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, C.X.; Paddock, K.J.; Zhang, Z.; Stacey, M.G. GmKIX8-1 regulates organ size in soybean and is the causative gene for the major seed weight QTL qSw17-1. New Phytol. 2021, 229, 920–934. [Google Scholar] [CrossRef] [PubMed]



| Gene | Primer | Sequences | PCR Length (bp) | Total Length for TILLING (bp) | Number of Mutant |
|---|---|---|---|---|---|
| E1 | F2 | CAGAGAGTTTCAACACTAATTCGTC | 1,632 | 1,623 | 5 |
| R2 | AGGTCTCTATGTATGTTTCATGCAG | ||||
| E3 | 1F2 | ATTCTTGTTTACCGTACTCTGGATG | 2,398 | 4,516 | 7 |
| 1R1 | TCCTTCAATCTTCAAATCACCTAGC | ||||
| 2F1 | ACTCAAACACTCTTGTGTGATATGC | 2,706 | |||
| 2R2 | TTCATGTCACGTTTATTTTGCAGGC | ||||
| E4 | F2 | TCCTTACATGTACTTAACATCCTATCC | 2,808 | 5,905 | 12 |
| R1 | GTTTAAATCCATACTCTCGGTATCTTTG | ||||
| F4 | TACTGAGAAATGCATTCAAAGATAC | 3,141 | |||
| R3 | GAATCAGCGGTTTCTAATTTCTACG | ||||
| PhyA1 | F1 | TTAGACTCGCACAACAATAATCTTTC | 2,984 | 6,104 | 6 |
| SR2 | GCATTTCTCAGTATTATCTGCAAGG | ||||
| F3 | AGGGAAGAGGATATAAGGTGAGTTG | 2,698 | |||
| R4 | GCAGGTTAGAAGAATACAAGCCCAC | ||||
| F5 | GAGTTAGTATGTGCCTGTGTCTATC | 2,694 | |||
| R6 | GCTCTAGTGAGAATGAGAACAGGTC | ||||
| Total | 18,148 | 30 | |||
| Size of mutant library | 2,455 | ||||
| Mutant density | 1.5 | Mbp | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koshika, N.; Shioya, N.; Fujimura, T.; Oguchi, R.; Ota, C.; Kato, E.; Takahashi, R.; Kimura, S.; Furuno, S.; Saito, K.; et al. Development of Ethyl Methanesulfonate Mutant Edamame Soybean (Glycine max (L.) Merr.) Populations and Forward and Reverse Genetic Screening for Early-Flowering Mutants. Plants 2022, 11, 1839. https://doi.org/10.3390/plants11141839
Koshika N, Shioya N, Fujimura T, Oguchi R, Ota C, Kato E, Takahashi R, Kimura S, Furuno S, Saito K, et al. Development of Ethyl Methanesulfonate Mutant Edamame Soybean (Glycine max (L.) Merr.) Populations and Forward and Reverse Genetic Screening for Early-Flowering Mutants. Plants. 2022; 11(14):1839. https://doi.org/10.3390/plants11141839
Chicago/Turabian StyleKoshika, Natsume, Naohiro Shioya, Takashi Fujimura, Rina Oguchi, Chie Ota, Emi Kato, Reiko Takahashi, Shuichi Kimura, Shinsuke Furuno, Koichi Saito, and et al. 2022. "Development of Ethyl Methanesulfonate Mutant Edamame Soybean (Glycine max (L.) Merr.) Populations and Forward and Reverse Genetic Screening for Early-Flowering Mutants" Plants 11, no. 14: 1839. https://doi.org/10.3390/plants11141839







