Cassava Frogskin Disease: Current Knowledge on a Re-Emerging Disease in the Americas
Abstract
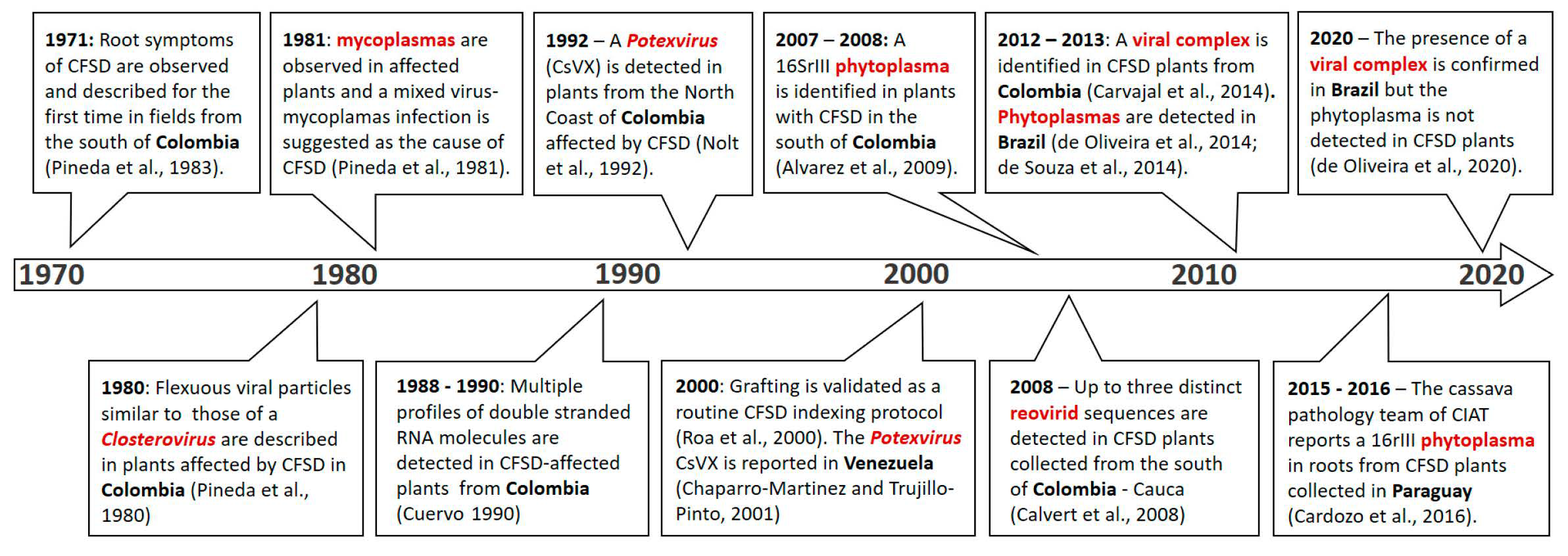
:1. Introduction
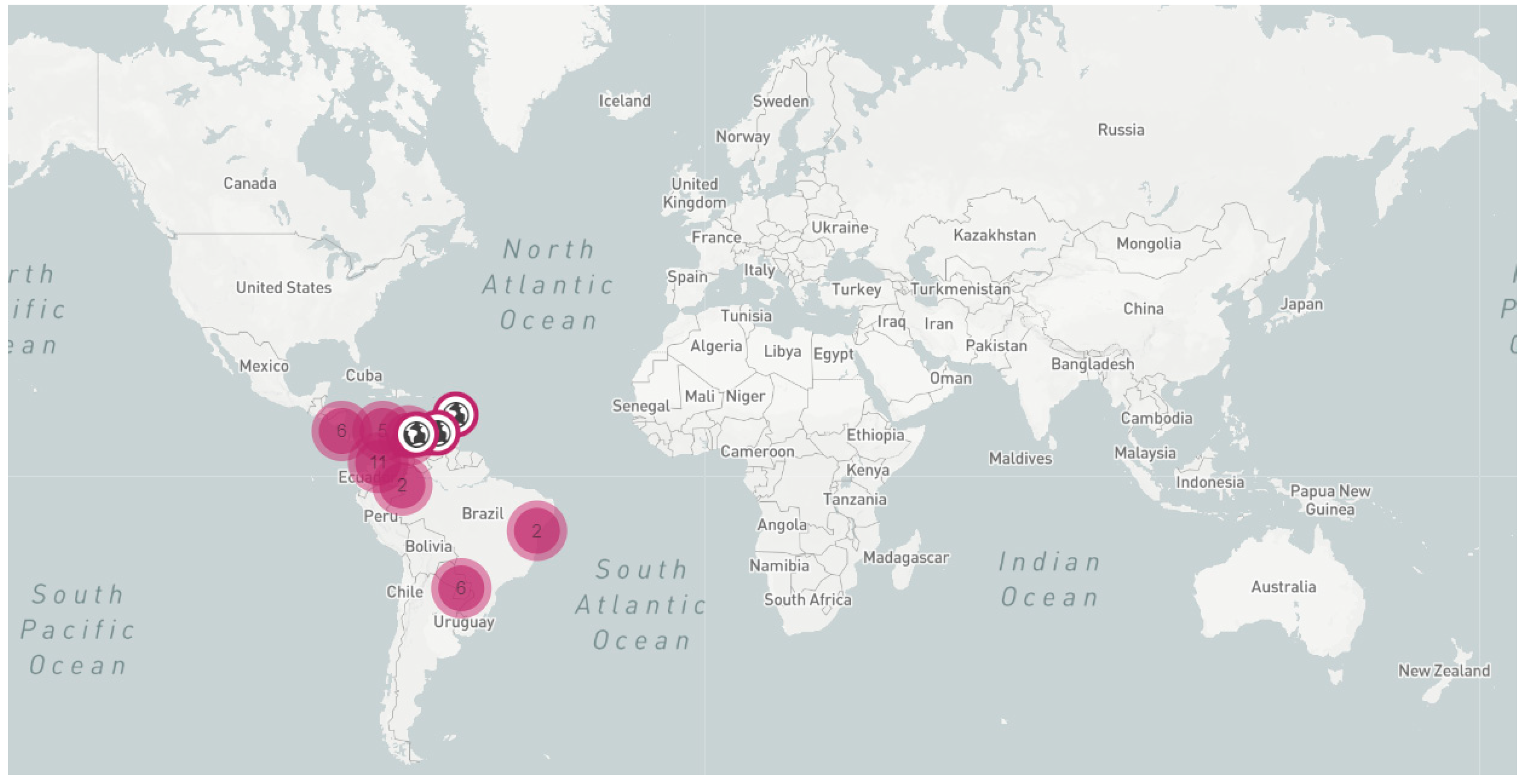
2. Symptoms, Transmission, and Geographical Distribution
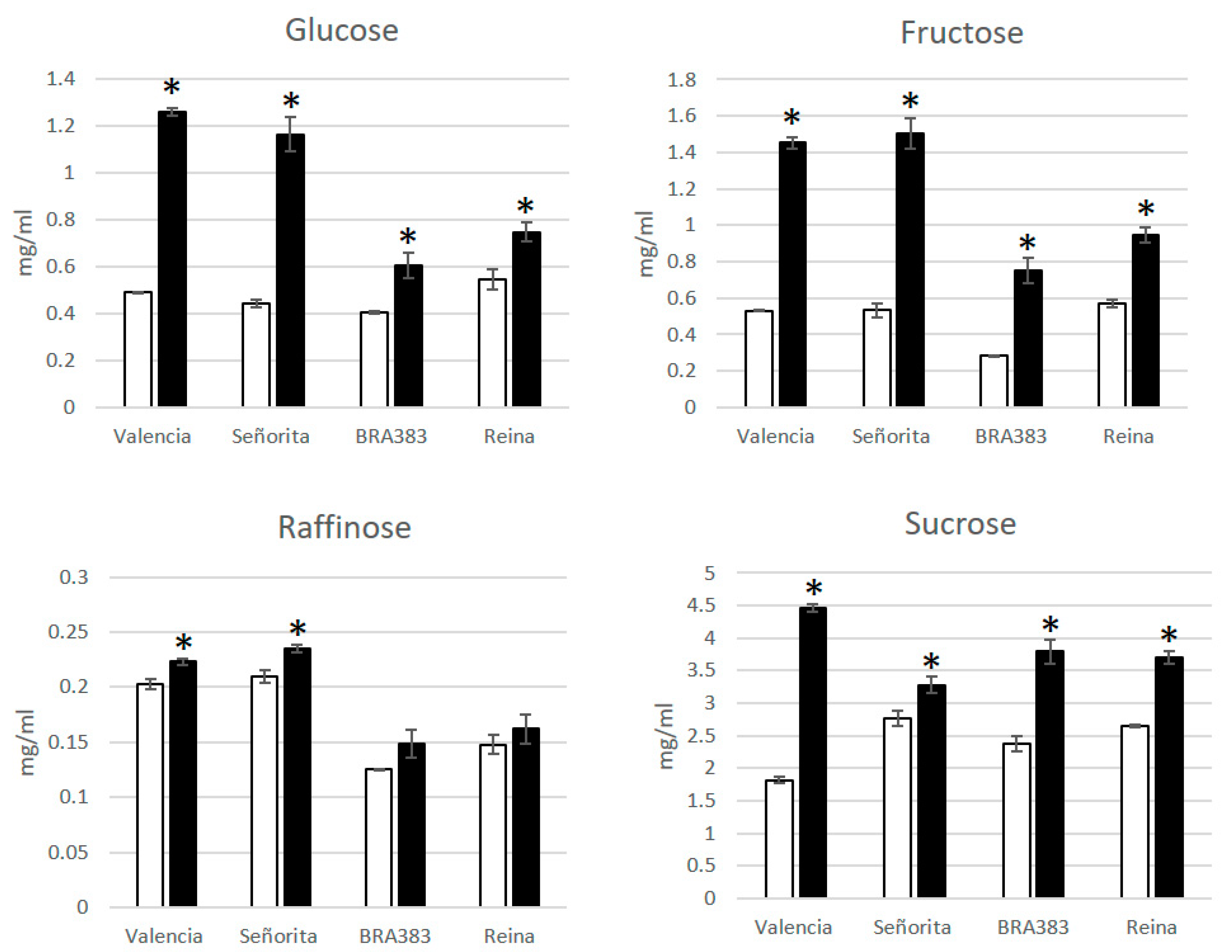
3. CFSD Affects the Yield and Sugar Content of Roots
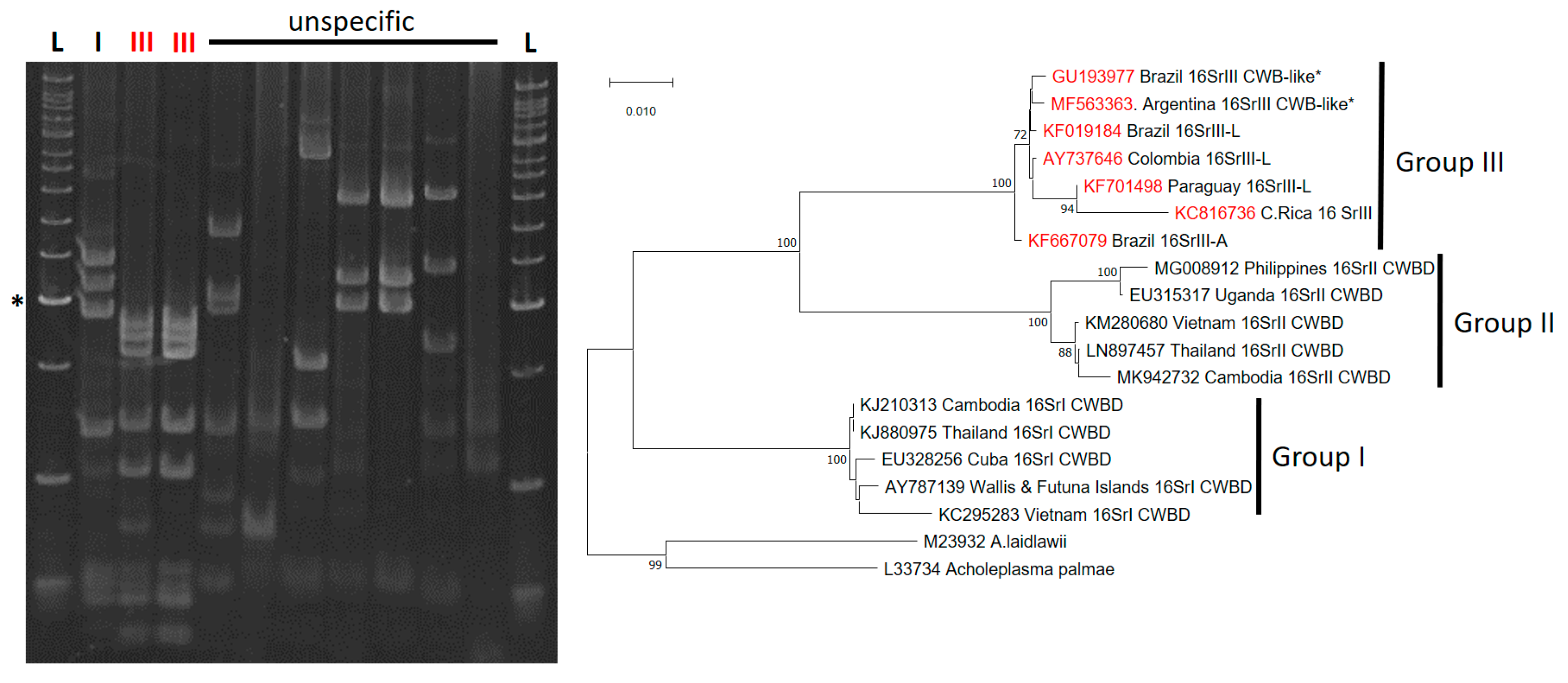
4. CFSD-Associated Pathogens and Molecular Diagnostics
5. Genetics of CFSD Resistance and Breeding
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pineda, B.; Jayasinghe, U.; Lozano, J.C. La enfermedad ‘cuero de Sapo’ en yuca (Manihot esculenta Crantz). ASIAVA 1983, 4, 10–12. [Google Scholar]
- Ceballos, H. Chapter 1: Cassava in Colombia and the World: New Prospects for a Millennial Crop. In Cassava in the Third Millenium: Modern Production, Processing, Use, and Marketing Systems; Ospina, B., Ceballos, H., Eds.; International Center for Tropical Agriculture (CIAT); Latin American and Caribbean Consortium to Support Cassava Research and Development (CLAYUCA); Technical Centre for Agricultural and Rural Cooperation (CTA): Cali, Colombia, 2012; pp. 1–11. [Google Scholar]
- Calvert, L.; Cuervo, M.; Lozano, I. Chapter 16: Cassava viral disease in South America. In Cassava in the Third Millenium: Modern Production, Processing, Use, and Marketing Systems; Ospina, B., Ceballos, H., Eds.; International Center for Tropical Agriculture (CIAT); Latin American and Caribbean Consortium to Support Cassava Research and Development (CLAYUCA); Technical Centre for Agricultural and Rural Cooperation (CTA): Cali, Colombia, 2012; pp. 309–321. [Google Scholar]
- Legg, J.; Kumar, L.; Makeshkumar, T.; Ferguson, M.; Kanju, E.; Ntawuruhunga, P.; Cuellar, W.J. Cassava Virus Diseases: Biology, Epidemiology and Management. Adv. Virus Res. 2015, 91, 85–142. [Google Scholar]
- Siriwan, W.; Jimenez, J.; Hemniam, N.; Saokham, K.; Lopez-Alvarez, D.; Leiva, A.M.; Cuellar, W.J. Surveillance, and diagnostics of the emergent Sri Lankan cassava mosaic virus (Fam. Geminiviridae) in Southeast Asia. Virus Res. 2020, 285, 197959. [Google Scholar] [CrossRef]
- Tuo, D.C.; Zhao, G.Y.; Yan, P.; Li, R.M.; Chen, X.; Wang, W.Q.; Zhou, P. First Report of Cassava common mosaic virus Infecting Cassava in Mainland China. Plant Dis. 2020, 104, 997. [Google Scholar] [CrossRef]
- Pineda, B.; Jayasinghe, U.; Morales, F.; Lozano, J.C. Partículas similares a virus asociadas con la enfermedad “Cuero de Sapo” en Yuca. In Proceedings of the IV National Congress of the Colombian Phytopathology Association (ASCOLFI), Medellín, Colombia, 2–5 July 1980; p. 67. (In Spanish). [Google Scholar]
- Pineda, B.; Lozano, J.C. Investigaciones Sobre la Enfermedad del “Cuero de Sapo” en Yuca (Manihot Esculenta Crantz); Seminario interno; Centro Internacional de Agricultura Tropical (CIAT): Cali, Colombia, 1981. [Google Scholar]
- Roa, J.C.; Flor, N.C.; Ramírez, J.L.; Mafla, G.; Pineda, B.; Debouck, D.G. Evaluación Preliminar de Tres Tipos de Injertos Utilizados en la Indización de Yuca; Centro Internacional de Agricultura Tropical (CIAT): Cali, Colombia, 2000; 8p. [Google Scholar]
- Cuervo, M. Caracterizacion de los acidos nucleicos de doble cadena asociados a enfermedades similares a las ocasionadas por virus en yuca (Manihot esculenta Crantz). Fitopatol. Colomb. 1990, 14, 10–17. [Google Scholar]
- Nolt, B.L.; Pineda, L.B.; Velasco, A.C. Surveys of cassava plantations in Colombia for virus and virus-like diseases. Plant Pathol. 1992, 41, 348–354. [Google Scholar] [CrossRef]
- Alvarez, E.; Mejía, J.F.; Llano, G.A.; Loke, J.B.; Calari, A.; Duduk, B.; Bertaccini, A. Characterization of a phytoplasma associated with frogskin disease in cassava. Plant Dis. 2009, 93, 1139–1145. [Google Scholar] [CrossRef] [Green Version]
- Chaparro-Martinez, E.I.; Trujillo-Pinto, G. First report of frog skin disease in cassava (Manihot esculenta) in Venezuela. Plant Dis. 2001, 85, 1285. [Google Scholar] [CrossRef] [PubMed]
- Calvert, L.A.; Cuervo, M.; Lozano, I.; Villareal, N.; Arroyave, J. Identification of three strains of a virus associated with cassava plants affected by frogskin disease. J. Phytopathol. 2008, 156, 647–653. [Google Scholar] [CrossRef]
- Carvajal, M.; Olaya, C.; Lozano, I.; Cuervo, M.; Castaño, M.; Cuellar, W.J. Unraveling complex viral infections in cassava (Manihot esculenta Crantz) from Colombia. Virus Res. 2014, 186, 76–86. [Google Scholar] [CrossRef]
- de Oliveira, S.A.S.; Abreu, E.F.M.; Araújo, T.S.; Oliveira, E.J.; Andrade, E.C.; Garcia, J.M.P.; Alvarez, E. First report of a 16SrIII-L phytoplasma associated with frogskin disease in cassava (Manihot esculenta Crantz) in Brazil. Plant Dis. 2014, 98, 153. [Google Scholar] [CrossRef] [PubMed]
- de Souza, A.N.; da Silva, F.N.; Bedendo, I.P.; Carvalho, C.M. A phytoplasma belonging to a 16SrIII-A subgroup and dsRNA virus associated with Cassava frogskin disease in Brazil. Plant Dis. 2014, 98, 771–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardozo, L.; Pardo, J.M.; Zacher, M.; Torres, A.; Alvarez, E. First report of a 16SrIII phytoplasma associated with frogskin disease in cassava (Manihot esculenta) in Paraguay. Plant Dis. 2016, 100, 1492. [Google Scholar] [CrossRef]
- de Oliveira, S.A.S.; Ferreira, C.F.; Diamantino, M.S.A.S.; Santos, T.A.; Pereira, J.; de Oliveira, E. First report of cassava torrado-like virus, cassava polero-like virus and cassava new alphaflexivirus associated with cassava frogskin disease in Brazil. J. Plant Pathol. 2020, 102, 247. [Google Scholar] [CrossRef] [Green Version]
- Jimenez, J. Caracterización del Virus Cassava Torrado-Like Virus en Yuca (Manihot esculenta Crantz) e Identificación de Proteínas Virales Involucradas en la Supresión del Silenciamiento de ARN. Master’s Thesis, Universidad Nacional de Colombia, Palmira, Colombia, 2017. [Google Scholar]
- Leiva, A.M. Análisis de Diversidad de Aislados Latinoamericanos del Virus del Mosaico Común de la Yuca (CsCMV. Género Potexvirus) Caracterización Biológica, Diagnóstico Molecular y Obtención de Genoma Completa. Master’s Thesis, Universidad Nacional de Colombia, Palmira, Colombia, 2015. [Google Scholar]
- Jimenez, J.; Leiva, A.M.; Olaya, C.; Acosta-Trujillo, D.; Cuellar, W.J. An optimized nucleic acid isolation protocol for virus detection in cassava (Manihot esculenta Crantz.). MethodsX 2021, 8, 101496. [Google Scholar] [CrossRef]
- Lozano, I.; Leiva, A.M.; Jimenez, J.; Fernandez, E.; Carvajal, M.; Cuervo, M.; Cuellar, W.J. Resolution of cassava-infecting alphaflexiviruses: Molecular and biological characterization of a novel group of potexviruses lacking the TGB3 gene. Virus Res. 2017, 241, 53–61. [Google Scholar] [CrossRef]
- Pardo, J.M. Desarrollo de una Metodología de PCR en Tiempo Real para Detección y Cuantificación de Fitoplasma 16SrIII-L y Reovirus CFSV Asociados con la Enfermedad del Cuero de Sapo en Yuca. Master’s Thesis, Universidad Nacional de Colombia, Palmira, Colombia, 2013. [Google Scholar]
- Ceballos, H.; Hershey, C. Cassava (Manihot esculenta Crantz). In Genetic Improvement of Tropical Crops; Campos, H., Caligari, P., Eds.; Springer: Cham, Switzerland, 2017; pp. 129–180. [Google Scholar]
- Leiva, A.M.; Jimenez, J.; Sandoval, H.; Perez, S.; Cuellar, W.J. Complete genome sequence of a novel secovirid infecting cassava in the Americas. Archives of Virology. Arch. Virol. 2021, 167, 665–668. [Google Scholar] [CrossRef]
- Pardo, J.M.; Truke, M.J.; Alvarez, E.; Cardozo, L.; Varela, I. A real-time PCR assay to detect and quantify 16SRIII-L and phytoplasmas associated with cassava frogskin disease in Costa Rica and Paraguay. In Proceedings of the 50th Annual meeting Caribbean Food Crops Society—Enhancing Family Farms through Sustainable Energy, Research and Technology, St. Thomas, VI, USA, 7–11 July 2014. [Google Scholar]
- Cuellar, W.J.; Mwanzia, L.; Lourido, D.; Martinez, A.F.; Rodriguez, R.; Garcia, C. PestDisPlace: Monitoring the Distribution of Pests and Diseases; Version 3.0; International Center for Tropical Agriculture (CIAT): Palmira, Colombia, 2018; Available online: https://pestdisplace.org (accessed on 20 April 2022).
- Tomlinson, K.R.; Bailey, A.M.; Alicai, T.; Seal, S.; Foster, G.D. Cassava brown streak disease: Historical timeline, current knowledge and future prospects. Mol. Plant Pathol. 2018, 19, 1282–1294. [Google Scholar] [CrossRef] [Green Version]
- CIAT. Annual Report: Improved Cassava for the Developing World; International Center for Tropical Agriculture (CIAT): Cali, Colombia, 2007; 299p. [Google Scholar]
- Olaya, C.A. Caracterización Genómica Parcial y Biológica de Reovirus que Infecta Yuca (Manihot esculenta Crantz). Master’s Thesis, Universidad Nacional de Colombia, Palmira, Colombia, 2013. [Google Scholar]
- Alarcón, F.; Dufour, D. Chapter 26: Sour cassava starch in Colombia. In Cassava in the Third Millenium: Modern Production, Processing, Use, and Marketing Systems; Ospina, B., Ceballos, H., Eds.; International Center for Tropical Agriculture (CIAT); Latin American and Caribbean Consortium to Support Cassava Research and Development (CLAYUCA); Technical Centre for Agricultural and Rural Cooperation (CTA): Cali, Colombia, 2012; pp. 496–525. [Google Scholar]
- Alvarez, E.; Pardo, J.M.; Dufour, D.; Moreno, J.L.; Alvarez, E. The metabolism of carbohydrates in roots of cassava (Manihot esculenta Crantz) infected with frogskin disease. Phytopathology 2014, 104, 7. [Google Scholar]
- Kogovšek, P.; Pompe-Novak, M.; Petek, M.; Fragner, L.; Weckwerth, W.; Gruden, K. Primary metabolism, phenylpropanoids and antioxidant pathways are regulated in potato as a response to Potato virus Y infection. PLoS ONE 2016, 11, e0146135. [Google Scholar] [CrossRef]
- Xue, C.; Liu, Z.; Dai, L.; Bu, J.; Liu, M.; Zhao, Z.; Jiang, Z.; Gao, W.; Zhao, J. Changing host photosynthetic, carbohydrate, and energy metabolisms play important roles in Phytoplasma infection. Phytopathology 2018, 108, 1067–1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shalitin, D.; and Wolf, S. Cucumber mosaic virus infection affects sugar transport in melon plants. Plant Physiol. 2000, 123, 597–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munyikwa, T.R.I.; Langeveld, S.; Salehuzzaman, S.N.I.M.; Jacobsen, E.; Visser, R.G.F. Cassava starch biosynthesis: New avenues for modifying starch quantity and quality. Euphytica 1997, 96, 65–75. [Google Scholar] [CrossRef]
- Geigenberger, P. Regulation of starch biosynthesis in response to a fluctuating environment. Plant Physiol. 2011, 155, 1566–1577. [Google Scholar] [CrossRef] [Green Version]
- Venturini, M.T.; Araújo, T.D.S.; Abreu, E.F.M.; Andrade, E.C.D.; Santos, V.D.S.; Silva, M.R.D.; Oliveira, E.J.D. Crop losses in Brazilian cassava varieties induced by the Cassava common mosaic virus. Sci. Agric. 2016, 73, 520–524. [Google Scholar] [CrossRef]
- Collavino, A.; Zanini, A.A.; Medina, R.; Schaller, S.; Di Feo, L. Cassava common mosaic virus infection affects growth and yield components of cassava plants (Manihot esculenta) in Argentina. Plant Pathol 2021, 00, 1–10. [Google Scholar] [CrossRef]
- Stapleton, G. Global starch market outlook and competing starch raw materials for by product segment and region. In Pricing Outlook and Cassava Growth Potential. Cassava Starch World 2010; Centre for Management Technology (CMT): Phnom Penh, Cambodia, 2012. [Google Scholar]
- Christensen, N.M.; Axelsen, B.; Nicolaisen, N.; Schulz, A. Phytoplasmas and their interactions with hosts. Trends Plant Sci. 2005, 10, 526–535. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Oshima, K.; Ammar, E.D.; Kakizawa, S.; Kingdom, H.N.; Namba, S. Phytoplasmas: Bacteria that manipulate plants and insects. Mol. Plant Pathol. 2008, 9, 403–423. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, W.; Lee, I.M.; Shao, J.; Suo, X.; Davis, R.E. Construction of an interactive online phytoplasma classification tool, iPhyClassifier, and its application in analysis of the peach X-disease phytoplasma group (16SrIII). Int. J. Syst. Evol. Microbiol. 2009, 59, 2582–2593. [Google Scholar] [CrossRef]
- Alvarez, E.; Pardo, J.M.; Mejía, J.F.; Bertaccini, A.; Thanh, N.D.; Trinh, X.H. Detection and identification of ‘Candidatus Phytoplasma asteris’-related phytoplasmas associated with a witches’ broom disease of cassava in Vietnam. Phytopathog. Mollicutes 2013, 3, 77–81. [Google Scholar] [CrossRef]
- Lim, P.O.; Sears, B.B. 16S rRNA sequence indicates that plant-pathogenic mycoplasma-like organisms are evolutionarily distinct from animal mycoplasmas. J. Bacteriol. 1989, 171, 5901–5906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abou-Jawdah, Y.; Aknadibossian, V.; Jawhari, M.; Tawidian, P.; Abrahamian, P. Real-Time PCR Protocol for Phytoplasma Detection and Quantification. In Phytoplasmas. Methods in Molecular Biology; Musetti, R., Pagliari, L., Eds.; Humana Press: New York, NY, USA, 2019; Volume 1875. [Google Scholar]
- Cuervo, M. Caracterización Molecular de Algunos Aislamientos del Virus del Cuero de Sapo de la Yuca Recolectados en Diferentes Zonas de Colombia. Master’s Thesis, Universidad Nacional de Colombia, Palmira, Colombia, 2006. [Google Scholar]
- King, A.M.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier: Amsterdam, The Netherlands, 2012; Volume 9. [Google Scholar]
- Quito-Avila, D.F.; Jelkmann, W.; Tzanetakis, I.E.; Keller, K.; Martin, R.R. Complete sequence and genetic characterization of Raspberry latent virus, a novel member of the family Reoviridae. Virus Res. 2011, 155, 397–405. [Google Scholar] [CrossRef]
- Spear, A.; Sisterson, M.S.; Stenger, D.C. Reovirus genomes from plant-feeding insects represent a newly discovered lineage within the family Reoviridae. Virus Res. 2012, 163, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Kreuze, J.F.; Perez, A.; Untiveros, M.; Quispe, D.; Fuentes, S.; Barker, I.; Simon, R. Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: A generic method for diagnosis, discovery and sequencing of viruses. Virology 2009, 388, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuellar, W.J.; Cruzado, K.R.; Untiveros, M.; Fuentes, S.; Soto, M.; Kreuze, J.F. Genome characterization of a Peruvian isolate of sweet potato chlorotic stunt virus (SPCSV): Further variability and a model for p22 acquisition. Virus Res. 2011, 157, 111–115. [Google Scholar] [CrossRef] [Green Version]
- Villamil-Garzón, A.; Cuellar, W.J.; Guzman-Barney, M. Natural co-infection of Solanum tuberosum crops by the Potato yellow vein virus and potyvirus in Colombia. Agron. Colomb. 2014, 32, 213–223. [Google Scholar] [CrossRef]
- Tuo, D.; Zhou, P.; Yan, P.; Cui, H.; Liu, Y.; Wang, H.; Shen, W. A cassava common mosaic virus vector for virus-induced gene silencing in cassava. Plant Methods 2021, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, M.J.S.; Oliveira, E.J.; Souza, A.S.; Pereira, J.S.; Diamantino, M.S.A.S.; Oliveira, S.A.S. Cleaning cassava genotypes infected with cassava frogskin disease via in vitro shoot tip culture. Genet. Mol. Res. 2017, 16, gmr16029556. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.-R.; Cui, Z.-H.; Li, J.-W.; Hao, X.-Y.; Zhao, L.; Wang, Q.-C. In vitro thermotherapy-based methods for plant virus eradication. Plant Methods 2018, 14, 87. [Google Scholar] [CrossRef] [Green Version]
- Nestel, B.; Cock, J. Cassava; the Development of an International Research Network; International Development Research Center: Ottawa, ON, Canada, 1976; Available online: http://hdl.handle.net/10625/1169 (accessed on 20 April 2022).
- Ceballos, H.; Rojanaridpiched, C.; Phumichai, C.; Becerra, L.A.; Kittipadakul, P.; Iglesias, C.; Vernon, E.G. Excellence in Cassava Breeding: Perspectives for the Future. Crop Breed. Genet. Genom. 2020, 2, e200008. [Google Scholar]
- Alvarez, E.; Gómez, Y.; Zacher, M.B.; Campos, R. Innovaciones Tecnológicas en el Manejo Integrado del Cuero de Sapo de la Yuca (Manihot esculenta Crantz): Estrategias para Reducir el Impacto de la Enfermedad por Efectos del Cambio Climático en Colombia, Costa Rica y Paraguay; International Center for Tropical Agriculture (CIAT): Cali, Colombia, 2015; p. 44. [Google Scholar]
- Sanchez, T.; Dufour, D.; Moreno, J.L.; Pizarro, M.; Aragón, I.J.; Dominguez, M.; Ceballos, H. Changes in extended shelf life of cassava roots during storage in ambient conditions. Postharvest Biol. Technol. 2013, 86, 520–528. [Google Scholar] [CrossRef]
- Zainuddin, I.M.; Fathoni, A.; Sudarmonowati, E.; Beeching, J.R.; Gruissem, W.; Vanderschuren, H. Cassava post-harvest physiological deterioration: From triggers to symptoms. Postharvest Biol. Technol. 2018, 142, 115–123. [Google Scholar] [CrossRef]
- Rao, G.P.; Alvarez, E.; Yadav, A. Phytoplasma Diseases of Industrial Crops. In Phytoplasmas: Plant Pathogenic Bacteria; Rao, G., Bertaccini, A., Fiore, N., Liefting, L., Eds.; Springer: Singapore, 2018; Volume 1, pp. 91–121. [Google Scholar]
- Thomas-Sharma, S.; Abdurahman, A.; Ali, S.; Andrade-Piedra, J.L.; Bao, S.; Charkowski, A.O.; Crook, D.; Kadian, M.; Kromann, P.; Struik, P.C.; et al. Seed degeneration in potato: The need for an integrated seed health strategy to mitigate the problem in developing countries. Plant Pathol. 2016, 65, 3–16. [Google Scholar] [CrossRef] [Green Version]
- McCallum, E.J.; Anjanappa, R.B.; Gruissem, W. Tackling agriculturally relevant diseases in the staple crop cassava (Manihot esculenta). Curr. Opin. Plant Biol. 2017, 38, 50–58. [Google Scholar] [CrossRef]


| Treatment | Seed type a | Average Severity of CFSD Root Symptoms (% Infected Plants) | ||
|---|---|---|---|---|
| 1st Crop Cycle (2004–2005) b | 2nd Crop Cycle (2005–2006) b | 3rd Crop Cycle (2006–2007) b | ||
| Open field, no fumigation | CFSD | 3.78 (77.3%) | 3.72 (75.2%) | 3.90 (79.3%) |
| Open field, no fumigation | Disease-free | 1.00 (0.0%) | 2.55 (34.3%) | 2.60 (38.2%) |
| Open field + fumigation | CFSD | 3.22 (57.7%) | 3.77 (77.0%) | 3.90 (81.0%) |
| Open field + fumigation | Disease-free | 1.00 (0.0%) | 1.60 (9.0%) | 1.70 (15%) |
| Screen house + fumigation | CFSD | 2.67 (38.5%) | 2.93 (47.6%) | 2.20 (33.3%) |
| Screen house + fumigation | Disease-free | 1.00 (0.0%) | 1.00 (0.0%) | 1.00 (0.0%) |
| Family | Pathogen | Primers (5′ to 3′) | Test | Ref |
|---|---|---|---|---|
| Reoviridae | CsFSaV | CsFSaV-F: TGG CCG GGA GAA CAA TAA TA CsFSaV-R: GCG AAG TAA GTT CCG TCG TT | RT-PCR | [14] |
| Secoviridae | CsTLV | CsTLV-1F: GAC TCA ATG AAG GAG GAG GAT AGA CsTLV-1R: ACC AGA GCT TGT CCT AAT AGC AAC | RT-PCR | [15,20] |
| Luteoviridae | CsPLV | CsPLV-F2: TTG CAT TCA AAG ATC AGT TCT CTC CsPLV-R3: TGG TTG ACA GCT GTT TCA GAG G | RT-PCR | |
| Alphaflexiviridae | CsVX | CsVX-RdRp-F1: GCR TTG ACC AGG CAG TCA CCW GAC CsVX-RdRp-R1: TAG CCC TCT ATC ACG TCC TCA | RT-PCR | [21,23] |
| CsCMV | CsCMV_3269F: GAG GCT CTT CTC TGG GAA AC CsCMV_3896R: CTT GAG TCC AGT TTG ATG TC | RT-PCR | ||
| CsNAV | CsNAV-RdRp-F: TGA GAG CAA TYT RAA GGA AA CsNAV-RdRp-R: GAT GAT ATC GTC AGG AAG AC | RT-PCR | ||
| Acholeplasmataceae | Cassava frogskin Phytoplasma (16SrIII) | rpIII-PF: GAG AAG CAC AAG CAA TTT TGA TG rpIII-PR: CAG CGT TGG CAA CAG CAC Probe: FAM/ACC CCA AAA GCA GCT TCT CCA ATC G/BHQ | qPCR | [24] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pardo, J.M.; Alvarez, E.; Becerra Lopez-Lavalle, L.A.; Olaya, C.; Leiva, A.M.; Cuellar, W.J. Cassava Frogskin Disease: Current Knowledge on a Re-Emerging Disease in the Americas. Plants 2022, 11, 1841. https://doi.org/10.3390/plants11141841
Pardo JM, Alvarez E, Becerra Lopez-Lavalle LA, Olaya C, Leiva AM, Cuellar WJ. Cassava Frogskin Disease: Current Knowledge on a Re-Emerging Disease in the Americas. Plants. 2022; 11(14):1841. https://doi.org/10.3390/plants11141841
Chicago/Turabian StylePardo, Juan Manuel, Elizabeth Alvarez, Luis Augusto Becerra Lopez-Lavalle, Cristian Olaya, Ana Maria Leiva, and Wilmer Jose Cuellar. 2022. "Cassava Frogskin Disease: Current Knowledge on a Re-Emerging Disease in the Americas" Plants 11, no. 14: 1841. https://doi.org/10.3390/plants11141841
APA StylePardo, J. M., Alvarez, E., Becerra Lopez-Lavalle, L. A., Olaya, C., Leiva, A. M., & Cuellar, W. J. (2022). Cassava Frogskin Disease: Current Knowledge on a Re-Emerging Disease in the Americas. Plants, 11(14), 1841. https://doi.org/10.3390/plants11141841







