Interference of Dihydrocoumarin with Hormone Transduction and Phenylpropanoid Biosynthesis Inhibits Barnyardgrass (Echinochloa crus-galli) Root Growth
Abstract
:1. Introduction
2. Results
2.1. Barnyardgrass and Rice Seedling Bioassay
2.2. Dihydrocoumarin Effects on Barnyardgrass Growth
2.3. Effect of Dihydrocoumarin on the Activity of Antioxidant Enzymes in Barnyardgrass Seedlings
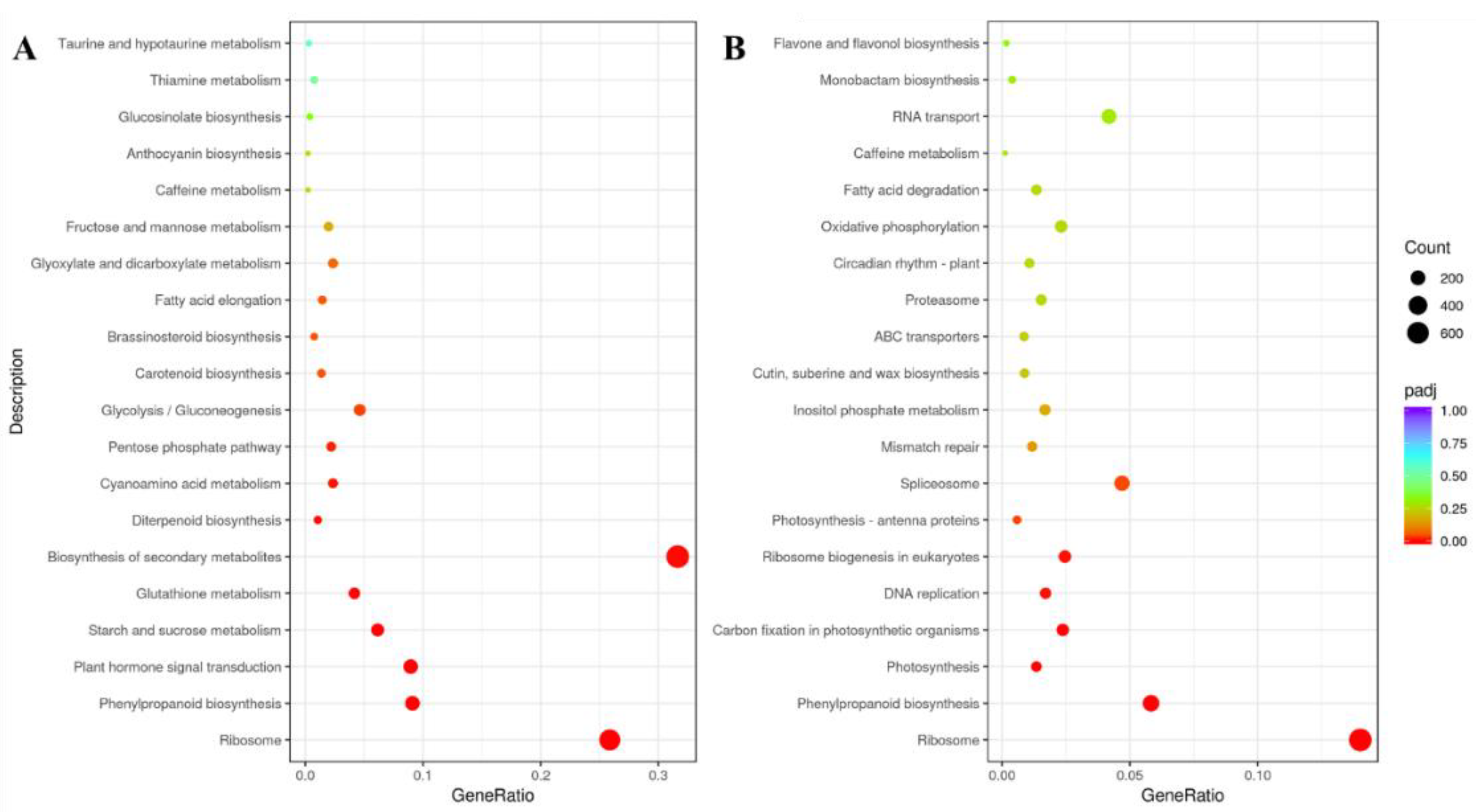
2.4. Transcriptomic Analysis
2.4.1. Plant Hormone Signal Transduction Pathway
2.4.2. Phenylpropanoid Biosynthesis Pathway
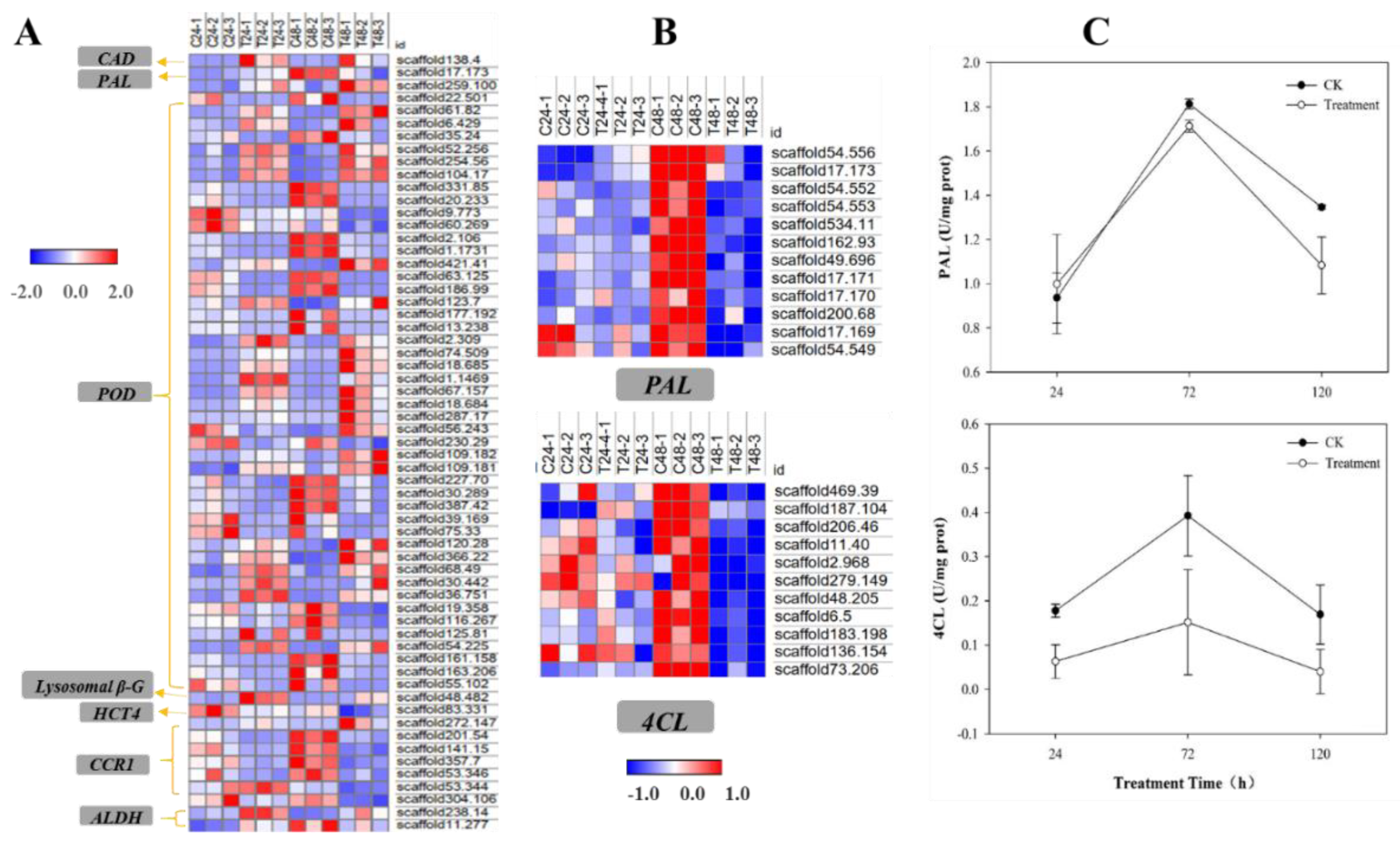
2.5. qRT-PCR Validation for Transcriptomic Analysis
2.6. PAL and 4CL Activities
3. Discussion
4. Materials and Methods
4.1. Materials and Plant Treatment
4.2. Bioassay for Barnyardgrass and Rice Seedlings
4.3. Cell Viability Analyses
4.4. ROS Production Analyses
4.5. Cell Membrane Permeability Analyses
4.6. Determination of the Activities of the Antioxidant Enzymes, PAL and 4CL
4.7. Transcriptome Sequencing
4.8. Differential Expression Analysis
4.9. Real-Time Quantitative Polymerase Chain Reaction Verification
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Keerthana, U.; Prabhukarthikeyan, S.R.; Baite, M.S.; Yadav, M.K.; Naveen Kumar, R.; Muthu Kumar, A.; Raghu, S.; Aravindan, S.; Rath, P.C. Fluorescent Pseudomonads: A Multifaceted Biocontrol Agent for Sustainable Agriculture. In New and Future Developments in Microbial Biotechnology and Bioengineering; Singh, H., Vaishnav, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 83–92. [Google Scholar]
- Shirkhan, F.; Mostafidi, M.; Tamaskani Zahedi, M.; Ziarati, P.; Vambol, V.; Vambol, S. Green technologies and environmental management: A new understanding and approach to the use of agricultural waste. Lett. Appl. NanoBioSci. 2022, 11, 3065–3075. [Google Scholar] [CrossRef]
- Kong, C.H.; Dang, X.T.; Khanh, T.; Tran, H.D.; Nguyen Thanh, T. Allelochemicals and signaling chemicals in plants. Molecules 2019, 24, 2737. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, J.; Zhu, Y.; Zhao, T.; Guo, L.; Kang, L.; Yu, J.; Du, H.; Liu, D. Weed suppression and molecular mechanisms of isochlorogenic acid a isolated from Artemisia argyi extract via an activity-guided method. J. Agric. Food Chem. 2022, 70, 1494–1506. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.R.; Amist, N.; Bai, L.Y. Allelopathy in sustainable weeds management. Allelopath. J. 2019, 48, 109–138. [Google Scholar] [CrossRef]
- Macías, F.A.; Mejías, F.J.; Molinillo, J.M. Recent advances in allelopathy for weed control: From knowledge to applications. Pest Manag. Sci. 2019, 75, 2413–2436. [Google Scholar] [CrossRef]
- Lankau, R. Coevolution between invasive and native plants driven by chemical competition and soil biota. Proc. Natl. Acad. Sci. USA 2012, 109, 11240. [Google Scholar] [CrossRef]
- Kunz, C.; Sturm, D.J.; Varnholt, D.; Walker, F.; Gerhards, R. Allelopathic effects and weed suppressive ability of cover crops. Plant Soil Environ. 2016, 62, 60–66. [Google Scholar]
- Zhang, Z.; Liu, Y.; Lin, Y.; Weber, E.; Kleunen, M. Effect of allelopathy on plant performance: A meta-analysis. Ecol. Lett. 2020, 24, 348–362. [Google Scholar] [CrossRef]
- Otte, B.; Rice, C.; Davis, B.; Schomberg, H.; Mirsky, S.; Tully, K. Phenolic acids released to soil during cereal rye cover crop decomposition. Chemoecology 2020, 30, 25–34. [Google Scholar] [CrossRef]
- Yang, H.; Tang, Y.; Wang, L.; Guo, Y.; Luo, D.; Wu, L.; Deng, X.; Li, X.; Bai, L. The potential of Myosoton aquaticum extracts and compounds to control barnyardgrass (Echinochloa crus-galli). Weed Res. 2021, 61, 317–326. [Google Scholar] [CrossRef]
- Lu, D.; Li, Y.; Gong, Y. Organocatalytic asymmetric tandem michael addition-hemiacetalization: A route to chiral dihydrocoumarins, chromanes, and 4H-chromenes. J. Org. Chem. 2010, 75, 6900–6907. [Google Scholar] [CrossRef] [PubMed]
- Lupini, A.; Araniti, F.; Sunseri, F.; Abenavoli, M.R. Coumarin interacts with auxin polar transport to modify root system architecture in Arabidopsis thaliana. Plant Growth Regul. 2014, 74, 23–31. [Google Scholar] [CrossRef]
- Bruno, L.; Talarico, E.; Cabeiras-Freijanes, L.; Madeo, M.; Muto, A.; Minervino, M.; Lucini, L.; Miras-Moreno, B.; Sofo, A.; Araniti, F. Coumarin interferes with polar auxin transport altering microtubule cortical array organization in Arabidopsis thaliana (L.) Heynh. root apical meristem. Int. J. Mol. Sci. 2021, 22, 7305. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.M.; Abu El-Soud, W. Evidence for “gibberellin-like” activity of coumarin. S. Afr. J. Bot. 2015, 100, 51–57. [Google Scholar] [CrossRef]
- Chen, B.X.; Peng, Y.X.; Yang, X.Q.; Liu, J. Delayed germination of Brassica parachinensis seeds by coumarin involves decreased GA 4 production and a consequent reduction of ROS accumulation. Seed Sci. Res. 2021, 31, 1–12. [Google Scholar] [CrossRef]
- Chen, B.X.; Peng, Y.X.; Gao, J.D.; Zhang, Q.; Liu, Q.J.; Fu, H.; Liu, J. Coumarin-induced delay of rice seed germination is mediated by suppression of abscisic acid catabolism and reactive oxygen species production. Front. Plant Sci. 2019, 10, 828. [Google Scholar] [CrossRef]
- Serra, S.; Castagna, A.; Valentino, M. Biocatalytic synthesis of natural dihydrocoumarin by microbial reduction of coumarin. Catalysts 2019, 9, 665. [Google Scholar] [CrossRef]
- Api, A.M.; Belsito, D.; Botelho, D.; Bruze, M.; Burton, G.A.; Buschmann, J.; Cancellieri, M.A.; Dagli, M.L.; Date, M.; Dekant, W.; et al. RIFM fragrance ingredient safety assessment, dihydrocoumarin, CAS registry number 119-84-6. Food Chem. Toxicol. 2021, 156, 112557. [Google Scholar] [CrossRef]
- Paul, J.; Choudhary, A.K.; Suri, V.K.; Sharma, A.K.; Kumar, V.; Shobhna. Bioresource nutrient recycling and its relationship with biofertility indicators of soil health and nutrient dynamics in rice–wheat cropping system. Commun. Soil Sci. Plant Anal. 2014, 45, 912–924. [Google Scholar] [CrossRef]
- Chauhan, B.S. Shade reduces growth and seed production of Echinochloa colona, Echinochloa crus-galli, and Echinochloa glabrescens. Crop Prot. 2013, 43, 241–245. [Google Scholar] [CrossRef]
- Motte, H.; Vanneste, S.; Beeckman, T. Molecular and environmental regulation of root development. Annu. Rev. Plant Biol. 2019, 70, 465–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, G.L.B.; Scortecci, K.C. Auxin and its role in plant development: Structure, signalling, regulation and response mechanisms. Plant Biol. 2021, 23, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Martínez, O.; Pernas, M.; Carol, R.J.; Dolan, L. Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 2007, 317, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, S.; Rosado, A.; Vaughan-Hirsch, J.; Bishopp, A.; Chini, A. Molecular locks and keys: The role of small molecules in phytohormone research. Front. Plant Sci. 2014, 5, 709. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant–environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef] [PubMed]
- Mase, K.; Tsukagoshi, H. Reactive oxygen species link gene regulatory networks during Arabidopsis root development. Front. Plant Sci. 2021, 12, 660274. [Google Scholar] [CrossRef]
- Pasternak, T.; Potters, G.; Caubergs, R.; Jansen, M.A.K. Complementary interactions between oxidative stress and auxins control plant growth responses at plant, organ, and cellular level. J. Exp. Bot. 2005, 56, 1991–2001. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, H.; Wang, H.; Yang, X.; Xu, Y.; Yang, Z.; Xu, C.; Li, P. Integrating transcriptome, co-expression and QTL-seq reveal metabolism of flavonoids and auxin associated with the primary root development in maize. J. Exp. Bot. 2021, 72, 4773–4795. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Mittler, R. ROS-induced ROS release in plant and animal cells. Free Radic. Biol. Med. 2018, 122, 21–27. [Google Scholar] [CrossRef]
- Petrov, V.; Hille, J.; Mueller-Roeber, B.; Gechev, T. ROS-mediated abiotic stress-induced programmed cell death in plants. Front. Plant Sci. 2015, 6, 69. [Google Scholar] [CrossRef]
- Zhu, Y.F.; Wu, Y.X.; Hu, Y.; Jia, X.M.; Zhao, T.; Cheng, L.; Wang, Y.X. Tolerance of two apple rootstocks to short-term salt stress: Focus on chlorophyll degradation, photosynthesis, hormone and leaf ultrastructures. Acta Physiol. Plant. 2019, 41, 87. [Google Scholar] [CrossRef]
- Nawaz, M.A.; Jiao, Y.; Chen, C.; Shireen, F.; Zheng, Z.; Imtiaz, M.; Bie, Z.; Huang, Y. Melatonin pretreatment improves vanadium stress tolerance of watermelon seedlings by reducing vanadium concentration in the leaves and regulating melatonin biosynthesis and antioxidant-related gene expression. J. Plant Physiol. 2018, 220, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Gniazdowska, A.; Bogatek, R. Allelopathic interactions between plants. Multi site action of allelochemicals. Acta Physiol. Plant. 2005, 27, 395–407. [Google Scholar] [CrossRef]
- Araniti, F.; Miras-Moreno, B.; Lucini, L.; Landi, M.; Abenavoli, M.R. Metabolomic, proteomic and physiological insights into the potential mode of action of thymol, a phytotoxic natural monoterpenoid phenol. Plant Physiol. Biochem. 2020, 153, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zou, J.J.; Zheng, H.; Xu, M.; Zong, X.; Wang, L. RNA-seq transcriptome analysis of rice primary roots reveals the role of flavonoids in regulating the rice primary root growth. Genes 2019, 10, 213. [Google Scholar] [CrossRef]
- Li, Z.; Xiong, F.; Guo, W.; Mo, C.; Wu, H.; Du, L. The root transcriptome analyses of peanut wild species Arachis correntina (Burkart) Krapov. & W.C. Gregory and cultivated variety Xiaobaisha in response to benzoic acid and p-cumaric acid stress. Genet. Resour. Crop Evolut. 2020, 67, 9–20. [Google Scholar] [CrossRef]
- Chi, W.C.; Chen, Y.A.; Hsiung, Y.C.; Fu, S.F.; Chou, C.H.; Trinh, N.N.; Chen, Y.C.; Huang, H.J. Autotoxicity mechanism of Oryza sativa: Transcriptome response in rice roots exposed to ferulic acid. BMC Genom. 2013, 14, 351. [Google Scholar] [CrossRef]
- Liscum, E.; Reed, J.W. Genetics of AUX/IAA and ARF action in plant growth and development. Plant Mol. Biol. 2002, 49, 387–400. [Google Scholar] [CrossRef]
- Hannah, M.; Heyer, A.; Hincha, D. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet. 2005, 1, e26. [Google Scholar] [CrossRef]
- Ljung, K.; Bhalerao, R.P.; Sandberg, G. Sites and homeostatic control of auxin biosynthesis in arabidopsis during vegetative growth. Plant J. 2001, 28, 465–474. [Google Scholar] [CrossRef]
- Song, M.; Fan, X.; Chen, J.; Qu, H.; Luo, L.; Xu, G. OsNAR2.1 interaction with OsNIT1 and OsNIT2 functions in root-growth responses to nitrate and ammonium. Plant Physiol. 2020, 183, 289–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Hou, L.; Meng, J.; You, H.; Li, Z.; Gong, Z.; Yang, S.; Shi, Y. The antagonistic action of abscisic acid and cytokinin signaling mediates drought stress response in Arabidopsis. Mol. Plant 2018, 11, 970–982. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tian, S.; Hou, L.; Huang, X.; Zhang, X.; Guo, H.; Yang, S. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 2012, 24, 2578–2595. [Google Scholar] [CrossRef]
- Yang, Z.B.; Liu, G.; Liu, J.; Zhang, B.; Meng, W.; Müller, B.; Hayashi, K.I.; Zhang, X.; Zhao, Z.; De Smet, I.; et al. Synergistic action of auxin and cytokinin mediates aluminum-induced root growth inhibition in Arabidopsis. EMBO Rep. 2017, 18, 1213–1230. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, Z.; Gao, J.; Wang, P.; Hu, T.; Wang, Z.; Hou, Y.J.; Wan, Y.; Liu, W.; Xie, S.; et al. Arabidopsis duodecuple mutant of PYL ABA receptors revealsPYL repression of ABA-independent SnRK2 activity. Cell Rep. 2018, 23, 3340–3351.e3345. [Google Scholar] [CrossRef]
- Deng, Y.; Lu, S. Biosynthesis and regulation of phenylpropanoids in plants. Crit. Rev. Plant Sci. 2017, 36, 257–290. [Google Scholar] [CrossRef]
- Li, Y.; Yu, T.; Wu, T.; Wang, R.; Wang, H.; Du, H.; Xu, X.; Xie, D.; Xu, X. The dynamic transcriptome of pepper (Capsicum annuum) whole roots reveals an important role for the phenylpropanoid biosynthesis pathway in root resistance to Phytophthora capsici. Gene 2019, 728, 144288. [Google Scholar] [CrossRef]
- Pan, Q.H.; Ji, C.Z.; Liu, H.; Zhang, J.; Chen, J.; Wen, P.; Huang, W. Salicylic acid synthesized by benzoic acid 2-hydroxylase participates in the development of thermotolerance in pea plants. Plant Sci. 2006, 171, 226–233. [Google Scholar] [CrossRef]
- Wu, L.; Fang, Y.; Yang, H.; Bai, L. Effects of drought-stress on seed germination and growth physiology of quinclorac-resistant Echinochloa crusgalli. PLoS ONE 2019, 14, e0214480. [Google Scholar] [CrossRef]
- Pan, J.W.; Zhu, M.Y.; Chen, H. Aluminum-induced cell death in root-tip cells of barley. Environ. Exp. Bot. 2001, 46, 71–79. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kobayashi, Y.; Devi, S.; Rikiishi, S.; Matsumoto, H. Aluminum toxicity is associated with mitochondrial dysfunction and the production of reactive oxygen species in plant cells. Plant Physiol. 2002, 128, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Zabalza, A.; Gaston, S.; Sandalio, L.; Del Río, L.; Royuela, M. Oxidative stress is not related to the mode of action of herbicides that inhibit acetolactate synthase. Environ. Exp. Bot. 2007, 59, 150–159. [Google Scholar] [CrossRef]
- Wang, J.; Xu, C.; Cheng, Q.; Zhao, J.; Wu, S.; Li, W.; Ma, W.; Liu, C.; Jiang, X. RNA sequencing revealed signals of evolution from gallbladder stone to gallbladder carcinoma. Front. Oncol. 2020, 10, 00823. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Tang, Y.; Pan, L.; Wang, C.; Guo, Y.; Yang, H.; Li, Z.; Bai, L.; Wang, L. Effect of fulvic acid on barnyardgrass (Echinochloa crus-galli ) seedling growth under flooding conditions. Weed Sci. 2021, 69, 192–202. [Google Scholar] [CrossRef]
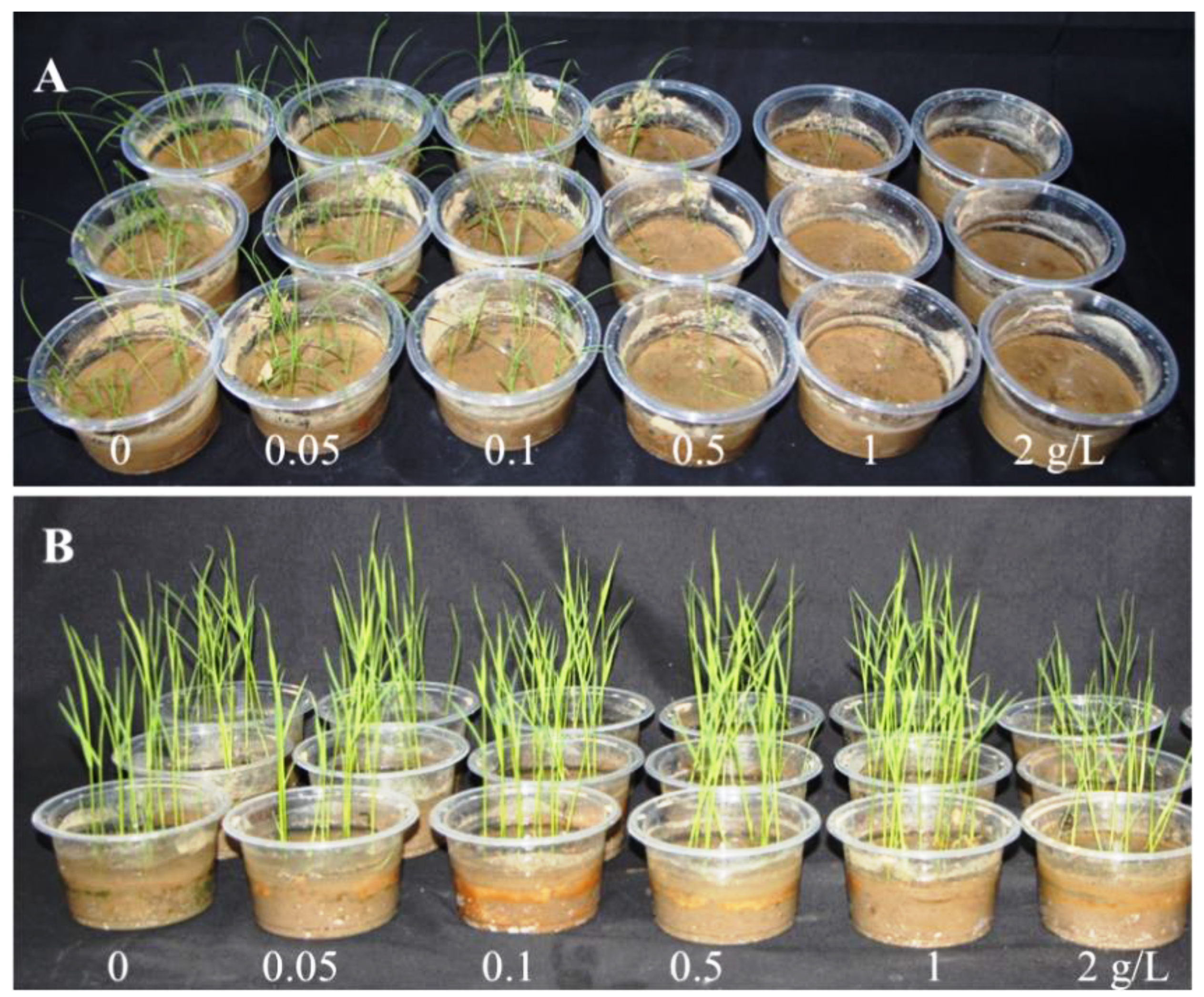
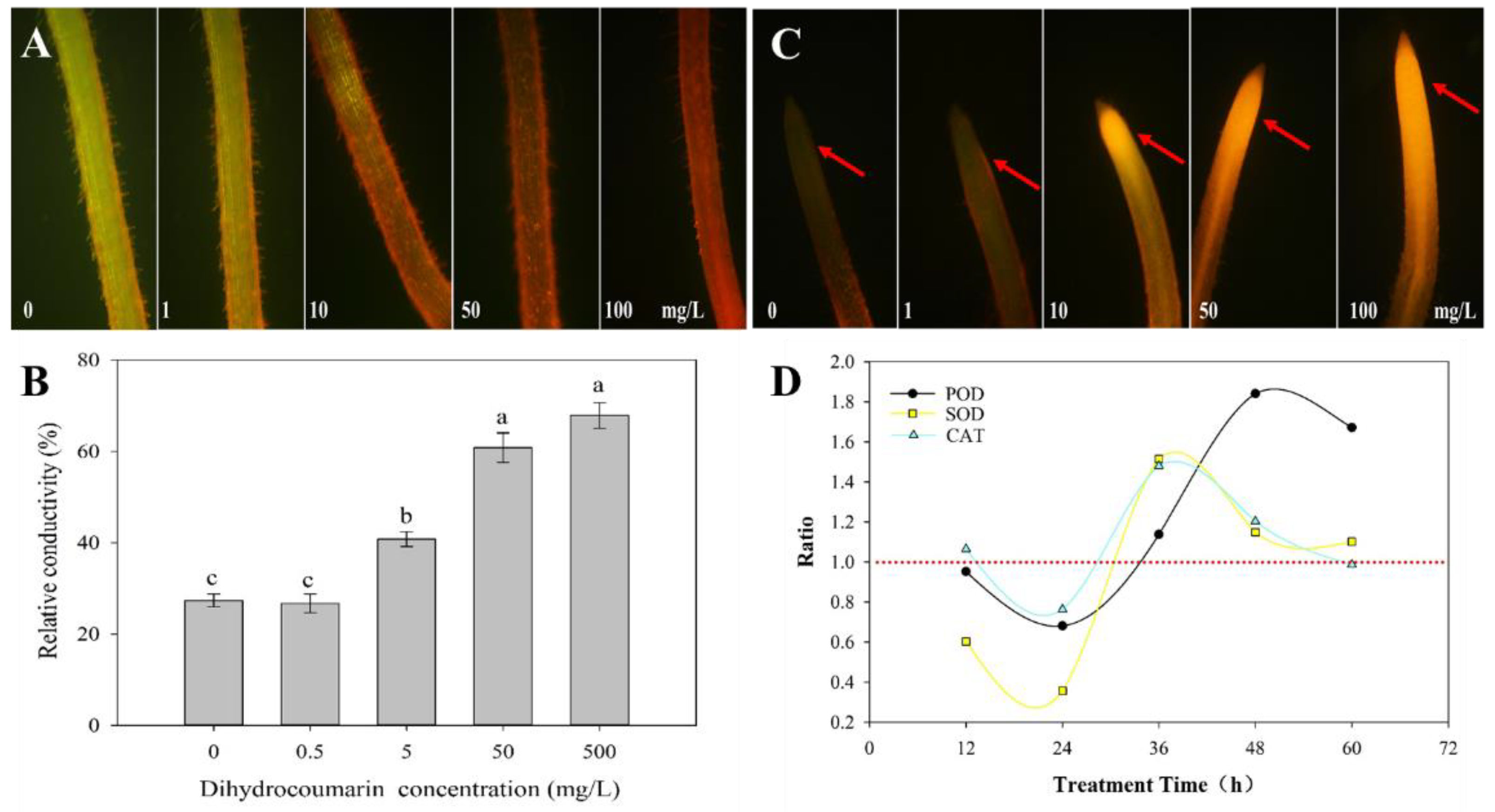

| Concentration (g/L) | Fresh Weight (g) | Shoot Length (cm) |
|---|---|---|
| 0 | 0.79 ± 0.08 a | 15.41 ± 0.85 a |
| 0.05 | 0.81 ± 0.04 a | 15.81 ± 1.13 a |
| 0.1 | 0.78 ± 0.06 a | 14.16 ± 0.82 a |
| 0.5 | 0.78 ± 0.07 a | 15.04 ± 0.51 a |
| 1 | 0.70 ± 0.09 ab | 13.93 ± 0.44 a |
| 2 | 0.55 ± 0.04 b | 10.76 ± 0.87 b |
| Concentration (g/L) | Fresh Weight (mg) | Shoot Length (cm) |
|---|---|---|
| 0 | 38.00 ± 3.21 a | 8.67 ± 0.24 a |
| 0.05 | 38.33 ± 3.28 a | 8.43 ± 0.20 a |
| 0.1 | 30.00 ± 1.00 b | 8.15 ± 0.61 a |
| 0.5 | 12.33 ± 0.88 c | 4.37 ± 0.62 b |
| 1 | 5.33 ± 2.40 d | 1.53 ± 0.69 c |
| 2 | 0.00 ± 0.00 d | 0.00 ± 0.00 d |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Zhou, S.; Wu, L.; Wang, L. Interference of Dihydrocoumarin with Hormone Transduction and Phenylpropanoid Biosynthesis Inhibits Barnyardgrass (Echinochloa crus-galli) Root Growth. Plants 2022, 11, 2505. https://doi.org/10.3390/plants11192505
Yang H, Zhou S, Wu L, Wang L. Interference of Dihydrocoumarin with Hormone Transduction and Phenylpropanoid Biosynthesis Inhibits Barnyardgrass (Echinochloa crus-galli) Root Growth. Plants. 2022; 11(19):2505. https://doi.org/10.3390/plants11192505
Chicago/Turabian StyleYang, Haona, Shangfeng Zhou, Lamei Wu, and Lifeng Wang. 2022. "Interference of Dihydrocoumarin with Hormone Transduction and Phenylpropanoid Biosynthesis Inhibits Barnyardgrass (Echinochloa crus-galli) Root Growth" Plants 11, no. 19: 2505. https://doi.org/10.3390/plants11192505





