High-Density Linkage Mapping of Agronomic Trait QTLs in Wheat under Water Deficit Condition using Genotyping by Sequencing (GBS)
Abstract
:1. Introduction
2. Results
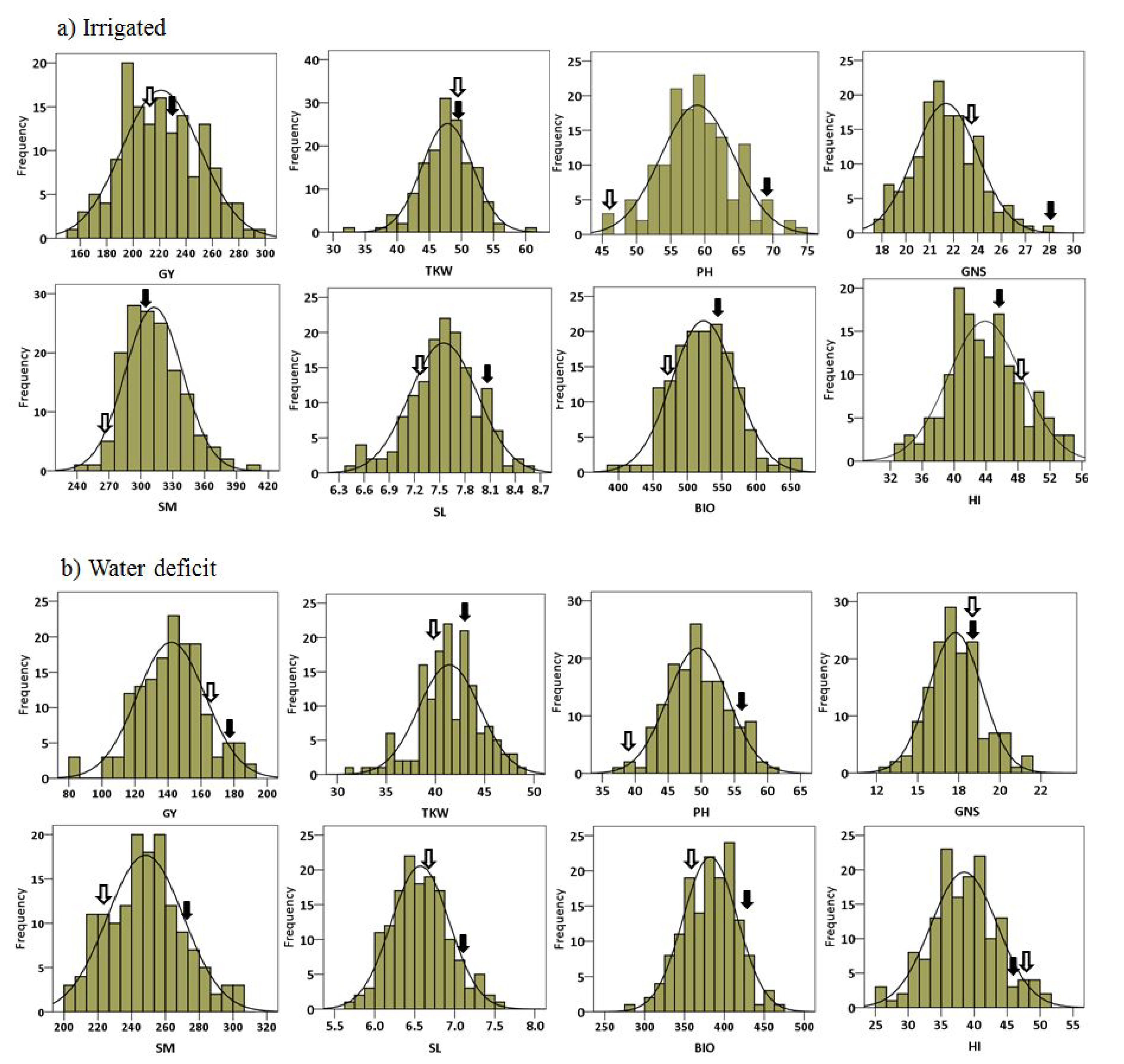
2.1. Phenotypic Data Analysis
2.2. Genome Wide SNP Discovery by GBS
2.3. Linkage Map Construction
2.4. QTL Mapping
2.4.1. Grain Yield (GY)
2.4.2. Plant Height (PH)
2.4.3. Number of Spikes per Square Meter (SM)
2.4.4. 1000. Kernel Weight (TKW)
2.4.5. Number of Grains per Spike (NGS)
2.4.6. Spike Length (SL)
2.4.7. Biomass (BIO)
2.4.8. Harvest Index (HI)
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Experimental Design and Phenotypic Evaluation
4.3. GBS Library Construction, Genotyping and SNP Calling
4.4. Linkage Map Construction and QTL Mapping
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zaynali Nezhad, K.; Weber, W.E.; Röder, M.S.; Sharma, S.; Lohwasser, U.; Meyer, R.C.; Saal, B.; Börner, A. QTL Analysis for Thousand-Grain Weight under Terminal Drought Stress in Bread Wheat (Triticumaestivum L.). Euphytica 2012, 186, 127–138. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Z.; Chang, X.; Jing, R. Genetic Dissection of the Developmental Behaviours of Plant Height in Wheat under Diverse Water Regimes. J. Exp. Bot. 2010, 61, 2923–2937. [Google Scholar] [CrossRef] [PubMed]
- Koua, A.P.; Oyiga, B.C.; Baig, M.M.; Léon, J.; Ballvora, A. Breeding Driven Enrichment of Genetic Variation for Key Yield Components and Grain Starch Content Under Drought Stress in Winter Wheat. Front. Plant Sci. 2021, 12, 1703. [Google Scholar] [CrossRef]
- Fleury, D.; Jefferies, S.; Kuchel, H.; Langridge, P. Genetic and Genomic Tools to Improve Drought Tolerance in Wheat. J. Exp. Bot. 2010, 61, 3211–3222. [Google Scholar] [CrossRef]
- Shatalina, M.; Wicker, T.; Buchmann, J.P.; Oberhaensli, S.; Šimková, H.; Doležel, J.; Keller, B. Genotype-Specific SNP Map Based on Whole Chromosome 3B Sequence Information from Wheat Cultivars Arina and Forno. Plant Biotechnol. J. 2013, 11, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Bhatta, M.; Morgounov, A.; Belamkar, V.; Baenziger, P.S. Genome-Wide Association Study Reveals Novel Genomic Regions for Grain Yield and Yield-Related Traits in Drought-Stressed Synthetic Hexaploid Wheat. Int. J. Mol. Sci. 2018, 19, 3011. [Google Scholar] [CrossRef]
- Edae, E.A.; Byrne, P.F.; Haley, S.D.; Lopes, M.S.; Reynolds, M.P. Genome-Wide Association Mapping of Yield and Yield Components of Spring Wheat under Contrasting Moisture Regimes. Theor. Appl. Genet. 2014, 127, 791–807. [Google Scholar] [CrossRef]
- Gahlaut, V.; Jaiswal, V.; Tyagi, B.S.; Singh, G.; Sareen, S.; Balyan, H.S.; Gupta, P.K. QTL Mapping for Nine Drought-Responsive Agronomic Traits in Bread Wheat under Irrigated and Rain-Fed Environments. PLoS ONE 2017, 12, e0182857. [Google Scholar] [CrossRef]
- Kirigwi, F.M.; Van Ginkel, M.; Brown-Guedira, G.; Gill, B.S.; Paulsen, G.M.; Fritz, A.K. Markers Associated with a QTL for Grain Yield in Wheat under Drought. Mol. Breed. 2007, 20, 401–413. [Google Scholar] [CrossRef]
- McIntyre, C.L.; Mathews, K.L.; Rattey, A.; Chapman, S.C.; Drenth, J.; Ghaderi, M.; Reynolds, M.; Shorter, R. Molecular Detection of Genomic Regions Associated with Grain Yield and Yield-Related Components in an Elite Bread Wheat Cross Evaluated under Irrigated and Rainfed Conditions. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2010, 120, 527–541. [Google Scholar] [CrossRef]
- Quarrie, S.A.; Steed, A.; Calestani, C.; Semikhodskii, A.; Lebreton, C.; Chinoy, C.; Steele, N.; Pljevljakusić, D.; Waterman, E.; Weyen, J.; et al. A High-Density Genetic Map of Hexaploid Wheat (Triticum Aestivum L.) from the Cross Chinese Spring × SQ1 and Its Use to Compare QTLs for Grain Yield across a Range of Environments. Theor. Appl. Genet. 2005, 110, 865–880. [Google Scholar] [CrossRef] [PubMed]
- Rufo, R.; López, A.; Lopes, M.S.; Bellvert, J.; Soriano, J.M. Identification of Quantitative Trait Loci Hotspots Affecting Agronomic Traits and High-Throughput Vegetation Indices in Rainfed Wheat. Front. Plant Sci. 2021, 12, 735192. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Singh, K.; Patil, R.V.; Kadam, S.; Bharti, S.; Prasad, P.; Singh, N.K.; Khanna-Chopra, R. Genomic Regions Associated with Grain Yield under Drought Stress in Wheat (Triticum Aestivum L.). Euphytica 2015, 203, 449–467. [Google Scholar] [CrossRef]
- Wu, X.; Chang, X.; Jing, R. Genetic Insight into Yield-Associated Traits of Wheat Grown in Multiple Rain-Fed Environments. PLoS ONE 2012, 7, e31249. [Google Scholar] [CrossRef]
- Gao, F.; Wen, W.; Liu, J.; Rasheed, A.; Yin, G.; Xia, X.; Wu, X.; He, Z. Genome-Wide Linkage Mapping of QTL for Yield Components, Plant Height and Yield-Related Physiological Traits in the Chinese Wheat Cross Zhou 8425B/Chinese Spring. Front. Plant Sci. 2015, 6, 1099. [Google Scholar] [CrossRef]
- Li, C.; Bai, G.; Carver, B.F.; Chao, S.; Wang, Z. Single Nucleotide Polymorphism Markers Linked to QTL for Wheat Yield Traits. Euphytica 2015, 206, 89–101. [Google Scholar] [CrossRef]
- Li, F.; Wen, W.; He, Z.; Liu, J.; Jin, H.; Cao, S.; Geng, H.; Yan, J.; Zhang, P.; Wan, Y.; et al. Genome-Wide Linkage Mapping of Yield-Related Traits in Three Chinese Bread Wheat Populations Using High-Density SNP Markers. Theor. Appl. Genet. 2018, 131, 1903–1924. [Google Scholar] [CrossRef]
- Liu, G.; Jia, L.; Lu, L.; Qin, D.; Zhang, J.; Guan, P.; Ni, Z.; Yao, Y.; Sun, Q.; Peng, H. Mapping QTLs of Yield-Related Traits Using RIL Population Derived from Common Wheat and Tibetan Semi-Wild Wheat. Theor. Appl. Genet. 2014, 127, 2415–2432. [Google Scholar] [CrossRef]
- Qu, P.; Wang, J.; Wen, W.; Gao, F.; Liu, J.; Xia, X.; Peng, H.; Zhang, L. Construction of Consensus Genetic Map With Applications in Gene Mapping of Wheat (Triticum aestivum L.) Using 90K SNP Array. Front. Plant Sci. 2021, 12, 727077. [Google Scholar] [CrossRef]
- Qu, X.; Li, C.; Liu, H.; Liu, J.; Luo, W.; Xu, Q.; Tang, H.; Mu, Y.; Deng, M.; Pu, Z.; et al. Quick Mapping and Characterization of a Co-Located Kernel Length and Thousand-Kernel Weight-Related QTL in Wheat. Theor. Appl. Genet. 2022, 135, 2849–2860. [Google Scholar] [CrossRef]
- Sehgal, D.; Autrique, E.; Singh, R.; Ellis, M.; Singh, S.; Dreisigacker, S. Identification of Genomic Regions for Grain Yield and Yield Stability and Their Epistatic Interactions. Sci. Rep. 2017, 7, 41578. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Zhang, X.; Zhang, W.; Zhang, N.; Song, L.; Liu, L.; Xue, X.; Liu, G.; Liu, J.; Meng, D.; et al. QTL Detection for Kernel Size and Weight in Bread Wheat (Triticum Aestivum L.) Using a High-Density SNP and SSR-Based Linkage Map. Front. Plant Sci. 2018, 9, 1484. [Google Scholar] [CrossRef]
- Sun, C.; Zhang, F.; Yan, X.; Zhang, X.; Dong, Z.; Cui, D.; Chen, F. Genome-Wide Association Study for 13 Agronomic Traits Reveals Distribution of Superior Alleles in Bread Wheat from the Yellow and Huai Valley of China. Plant Biotechnol. J. 2017, 15, 953–969. [Google Scholar] [CrossRef] [PubMed]
- Vitale, P.; Fania, F.; Esposito, S.; Pecorella, I.; Pecchioni, N.; Palombieri, S.; Sestili, F.; Lafiandra, D.; Taranto, F.; De Vita, P. QTL Analysis of Five Morpho-Physiological Traits in Bread Wheat Using Two Mapping Populations Derived from Common Parents. Genes 2021, 12, 604. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hu, H.; Jiang, X.; Tao, Y.; Lin, Y.; Wu, F.; Hou, S.; Liu, S.; Li, C.; Chen, G.; et al. Identification and Validation of a Novel Major Quantitative Trait Locus for Plant Height in Common Wheat (Triticum aestivum L.). Front. Genet. 2020, 11, 602495. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Li, S.; Liu, H.; Xu, Q.; Tang, H.; Mu, Y.; Deng, M.; Jiang, Q.; Chen, G.; Qi, P.; et al. Identification and Validation of a Major QTL for Kernel Length in Bread Wheat Based on Two F3 Biparental Populations. BMC Genomics 2022, 23, 386. [Google Scholar] [CrossRef]
- Yang, L.; Zhao, D.; Meng, Z.; Xu, K.; Yan, J.; Xia, X.; Cao, S.; Tian, Y.; He, Z.; Zhang, Y. QTL Mapping for Grain Yield-Related Traits in Bread Wheat via SNP-Based Selective Genotyping. Theor. Appl. Genet. 2020, 133, 857–872. [Google Scholar] [CrossRef]
- Yang, Y.; Dhakal, S.; Chu, C.; Wang, S.; Xue, Q.; Rudd, J.C.; Ibrahim, A.M.H.; Jessup, K.; Baker, J.; Fuentealba, M.P.; et al. Genome Wide Identification of QTL Associated with Yield and Yield Components in Two Popular Wheat Cultivars TAM 111 and TAM 112. PLoS ONE 2020, 15, e0237293. [Google Scholar] [CrossRef]
- Zhai, H.; Feng, Z.; Li, J.; Liu, X.; Xiao, S.; Ni, Z.; Sun, Q. QTL Analysis of Spike Morphological Traits and Plant Height in Winter Wheat (Triticum aestivum L.) Using a High-Density SNP and SSR-Based Linkage Map. Front. Plant Sci. 2016, 7, 1617. [Google Scholar] [CrossRef]
- Berkman, P.J.; Lai, K.; Lorenc, M.T.; Edwards, D. Next-Generation Sequencing Applications for Wheat Crop Improvement. Am. J. Bot. 2012, 99, 365–371. [Google Scholar] [CrossRef]
- Poland, J.A.; Rife, T.W. Genotyping-by-Sequencing for Plant Breeding and Genetics. Plant Genome 2012, 5, 92–102. [Google Scholar] [CrossRef]
- Zhou, Y.; Conway, B.; Miller, D.; Marshall, D.; Cooper, A.; Murphy, P.; Chao, S.; Brown-Guedira, G.; Costa, J. Quantitative Trait Loci Mapping for Spike Characteristics in Hexaploid Wheat. Plant Genome 2017, 10, plantgenome2016.10.0101. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Liu, C.; Wang, D.; St. Amand, P.; Bernardo, A.; Li, W.; He, F.; Li, L.; Wang, L.; Yuan, X.; et al. High-Resolution Genome-Wide Association Study Identifies Genomic Regions and Candidate Genes for Important Agronomic Traits in Wheat. Mol. Plant 2020, 13, 1311–1327. [Google Scholar] [CrossRef]
- Juliana, P.; Poland, J.; Huerta-Espino, J.; Shrestha, S.; Crossa, J.; Crespo-Herrera, L.; Toledo, F.H.; Govindan, V.; Mondal, S.; Kumar, U.; et al. Improving Grain Yield, Stress Resilience and Quality of Bread Wheat Using Large-Scale Genomics. Nat. Genet. 2019, 51, 1530–1539. [Google Scholar] [CrossRef] [PubMed]
- Ehdaie, B.; Mohammadi, S.A.; Nouraein, M. QTLs for Root Traits at Mid-Tillering and for Root and Shoot Traits at Maturity in a RIL Population of Spring Bread Wheat Grown under Well-Watered Conditions. Euphytica 2016, 211, 17–38. [Google Scholar] [CrossRef]
- The International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the Limits in Wheat Research and Breeding Using a Fully Annotated Reference Genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Spindel, J.; Wright, M.; Chen, C.; Cobb, J.; Gage, J.; Harrington, S.; Lorieux, M.; Ahmadi, N.; McCouch, S. Bridging the Genotyping Gap: Using Genotyping by Sequencing (GBS) to Add High-Density SNP Markers and New Value to Traditional Bi-Parental Mapping and Breeding Populations. Theor. Appl. Genet. 2013, 126, 2699–2716. [Google Scholar] [CrossRef]
- Hussain, W.; Baenziger, P.S.; Belamkar, V.; Guttieri, M.J.; Venegas, J.P.; Easterly, A.; Sallam, A.; Poland, J. Genotyping-by-Sequencing Derived High-Density Linkage Map and Its Application to QTL Mapping of Flag Leaf Traits in Bread Wheat. Sci. Rep. 2017, 7, 16394. [Google Scholar] [CrossRef]
- Verma, S.; Gupta, S.; Bandhiwal, N.; Kumar, T.; Bharadwaj, C.; Bhatia, S. High-Density Linkage Map Construction and Mapping of Seed Trait QTLs in Chickpea (Cicer arietinum L.) Using Genotyping-by-Sequencing (GBS). Sci. Rep. 2015, 5, 17512. [Google Scholar] [CrossRef]
- Zanke, C.D.; Ling, J.; Plieske, J.; Kollers, S.; Ebmeyer, E.; Korzun, V.; Argillier, O.; Stiewe, G.; Hinze, M.; Neumann, F.; et al. Analysis of Main Effect QTL for Thousand Grain Weight in European Winter Wheat (Triticum aestivum L.) by Genome-Wide Association Mapping. Front. Plant Sci. 2015, 6, 644. [Google Scholar] [CrossRef] [Green Version]
- Griffiths, S.; Simmonds, J.; Leverington, M.; Wang, Y.; Fish, L.; Sayers, L.; Alibert, L.; Orford, S.; Wingen, L.; Snape, J. Meta-QTL Analysis of the Genetic Control of Crop Height in Elite European Winter Wheat Germplasm. Mol. Breed. 2012, 29, 159–171. [Google Scholar] [CrossRef]
- Zanke, C.D.; Ling, J.; Plieske, J.; Kollers, S.; Ebmeyer, E.; Korzun, V.; Argillier, O.; Stiewe, G.; Hinze, M.; Neumann, K.; et al. Whole Genome Association Mapping of Plant Height in Winter Wheat (Triticum aestivum L.). PLoS ONE 2014, 9, e113287. [Google Scholar] [CrossRef] [PubMed]
- Mathews, K.L.; Malosetti, M.; Chapman, S.; McIntyre, L.; Reynolds, M.; Shorter, R.; van Eeuwijk, F. Multi-Environment QTL Mixed Models for Drought Stress Adaptation in Wheat. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2008, 117, 1077–1091. [Google Scholar] [CrossRef] [PubMed]
- Isham, K.; Wang, R.; Zhao, W.; Wheeler, J.; Klassen, N.; Akhunov, E.; Chen, J. QTL Mapping for Grain Yield and Three Yield Components in a Population Derived from Two High-Yielding Spring Wheat Cultivars. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2021, 134, 2079–2095. [Google Scholar] [CrossRef]
- Zhang, K.; Tian, J.; Zhao, L.; Wang, S. Mapping QTLs with Epistatic Effects and QTL × Environment Interactions for Plant Height Using a Doubled Haploid Population in Cultivated Wheat. J. Genet. Genomics 2008, 35, 119–127. [Google Scholar] [CrossRef]
- Ain, Q.-U.; Rasheed, A.; Anwar, A.; Mahmood, T.; Imtiaz, M.; Mahmood, T.; Xia, X.; He, Z.; Quraishi, U.M. Genome-Wide Association for Grain Yield under Rainfed Conditions in Historical Wheat Cultivars from Pakistan. Front. Plant Sci. 2015, 6, 743. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, J.; Li, R.; Deng, Z.; Zhang, K.; Liu, B.; Tian, J. Conditional QTL Mapping of Three Yield Components in Common Wheat (Triticum aestivum L.). Crop J. 2016, 4, 220–228. [Google Scholar] [CrossRef]
- Rizza, F.; Badeck, F.W.; Cattivelli, L.; Lidestri, O.; Di Fonzo, N.; Stanca, A.M. Use of a Water Stress Index to Identify Barley Genotypes Adapted to Rainfed and Irrigated Conditions. Crop Sci. 2004, 44, 2127–2137. [Google Scholar] [CrossRef]
- Sukumaran, S.; Dreisigacker, S.; Lopes, M.; Chavez, P.; Reynolds, M.P. Genome-Wide Association Study for Grain Yield and Related Traits in an Elite Spring Wheat Population Grown in Temperate Irrigated Environments. Theor. Appl. Genet. 2015, 128, 353–363. [Google Scholar] [CrossRef]
- Perez-Lara, E.; Semagn, K.; Chen, H.; Iqbal, M.; N’Diaye, A.; Kamran, A.; Navabi, A.; Pozniak, C.; Spaner, D. QTLs Associated with Agronomic Traits in the Cutler × AC Barrie Spring Wheat Mapping Population Using Single Nucleotide Polymorphic Markers. PLoS ONE 2016, 11, e0160623. [Google Scholar] [CrossRef] [Green Version]
- Acuña-Galindo, M.A.; Mason, R.E.; Subramanian, N.K.; Hays, D.B. Meta-Analysis of Wheat QTL Regions Associated with Adaptation to Drought and Heat Stress. Crop Sci. 2015, 55, 477–492. [Google Scholar] [CrossRef]
- Wang, S.-X.; Zhu, Y.-L.; Zhang, D.-X.; Shao, H.; Liu, P.; Hu, J.-B.; Zhang, H.; Zhang, H.-P.; Chang, C.; Lu, J.; et al. Genome-Wide Association Study for Grain Yield and Related Traits in Elite Wheat Varieties and Advanced Lines Using SNP Markers. PLoS ONE 2017, 12, e0188662. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Q.; Kempf, H.; Ganal, M.W.; Röder, M.S. Advanced Backcross QTL Analysis in Progenies Derived from a Cross between a German Elite Winter Wheat Variety and a Synthetic Wheat (Triticum Aestivum L.). TAG Theor. Appl. Genet. Theor. Angew. Genet. 2004, 109, 933–943. [Google Scholar] [CrossRef]
- Gagne, J.M.; Smalle, J.; Gingerich, D.J.; Walker, J.M.; Yoo, S.-D.; Yanagisawa, S.; Vierstra, R.D. Arabidopsis EIN3-Binding F-Box 1 and 2 Form Ubiquitin-Protein Ligases That Repress Ethylene Action and Promote Growth by Directing EIN3 Degradation. Proc. Natl. Acad. Sci. USA 2004, 101, 6803–6808. [Google Scholar] [CrossRef]
- Strader, L.C.; Ritchie, S.; Soule, J.D.; McGinnis, K.M.; Steber, C.M. Recessive-Interfering Mutations in the Gibberellin Signaling Gene SLEEPY1 Are Rescued by Overexpression of Its Homologue, SNEEZY. Proc. Natl. Acad. Sci. USA 2004, 101, 12771–12776. [Google Scholar] [CrossRef]
- Lechner, E.; Achard, P.; Vansiri, A.; Potuschak, T.; Genschik, P. F-Box Proteins Everywhere. Curr. Opin. Plant Biol. 2006, 9, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Nijhawan, A.; Arora, R.; Agarwal, P.; Ray, S.; Sharma, P.; Kapoor, S.; Tyagi, A.K.; Khurana, J.P. F-Box Proteins in Rice. Genome-Wide Analysis, Classification, Temporal and Spatial Gene Expression during Panicle and Seed Development, and Regulation by Light and Abiotic Stress. Plant Physiol. 2007, 143, 1467–1483. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Zhang, H.; Tian, S.; Chang, X.; Jing, R. TaSnRK2.4, an SNF1-Type Serine/Threonine Protein Kinase of Wheat (Triticum aestivum L.), Confers Enhanced Multistress Tolerance in Arabidopsis. J. Exp. Bot. 2010, 61, 683–696. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of Plant Heat-Shock Proteins and Molecular Chaperones in the Abiotic Stress Response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Feki, K.; Brini, F. Role of Proteins in Alleviating Drought Stress in Plants. In Water Stress and Crop Plants; Ahmad, P., Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2016; pp. 165–176. ISBN 978-1-119-05445-0. [Google Scholar]
- Chini, A.; Grant, J.J.; Seki, M.; Shinozaki, K.; Loake, G.J. Drought Tolerance Established by Enhanced Expression of the CC-NBS-LRR Gene, ADR1, Requires Salicylic Acid, EDS1 and ABI1. Plant J. 2004, 38, 810–822. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two Transcription Factors, DREB1 and DREB2, with an EREBP/AP2 DNA Binding Domain Separate Two Cellular Signal Transduction Pathways in Drought- and Low-Temperature-Responsive Gene Expression, Respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Song, C.-P.; Agarwal, M.; Ohta, M.; Guo, Y.; Halfter, U.; Wang, P.; Zhu, J.-K. Role of an Arabidopsis AP2/EREBP-Type Transcriptional Repressor in Abscisic Acid and Drought Stress Responses. Plant Cell 2005, 17, 2384–2396. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xu, C.; Li-Beisson, Y.; Philippar, K. Fatty Acid and Lipid Transport in Plant Cells. Trends Plant Sci. 2016, 21, 145–158. [Google Scholar] [CrossRef]
- Siegel, D.; Gustafson, D.L.; Dehn, D.L.; Han, J.Y.; Boonchoong, P.; Berliner, L.J.; Ross, D. NAD(P)H:Quinone Oxidoreductase 1: Role as a Superoxide Scavenger. Mol. Pharmacol. 2004, 65, 1238–1247. [Google Scholar] [CrossRef]
- Simova-Stoilova, L.; Vaseva, I.; Grigorova, B.; Demirevska, K.; Feller, U. Proteolytic Activity and Cysteine Protease Expression in Wheat Leaves under Severe Soil Drought and Recovery. Plant Physiol. Biochem. PPB 2010, 48, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, S.; Takano, T. Two Cysteine Proteinase Inhibitors from Arabidopsis Thaliana, AtCYSa and AtCYSb, Increasing the Salt, Drought, Oxidation and Cold Tolerance. Plant Mol. Biol. 2008, 68, 131–143. [Google Scholar] [CrossRef]
- Tamiru, M.; Undan, J.R.; Takagi, H.; Abe, A.; Yoshida, K.; Undan, J.Q.; Natsume, S.; Uemura, A.; Saitoh, H.; Matsumura, H.; et al. A Cytochrome P450, OsDSS1, Is Involved in Growth and Drought Stress Responses in Rice (Oryza sativa L.). Plant Mol. Biol. 2015, 88, 85–99. [Google Scholar] [CrossRef]
- Wan, X.; Mo, A.; Liu, S.; Yang, L.; Li, L. Constitutive Expression of a Peanut Ubiquitin-Conjugating Enzyme Gene in Arabidopsis Confers Improved Water-Stress Tolerance through Regulation of Stress-Responsive Gene Expression. J. Biosci. Bioeng. 2011, 111, 478–484. [Google Scholar] [CrossRef]
- Poland, J.A.; Brown, P.J.; Sorrells, M.E.; Jannink, J.-L. Development of High-Density Genetic Maps for Barley and Wheat Using a Novel Two-Enzyme Genotyping-by-Sequencing Approach. PLoS ONE 2012, 7, e32253. [Google Scholar] [CrossRef]
- Mascher, M.; Wu, S.; Amand, P.S.; Stein, N.; Poland, J. Application of Genotyping-by-Sequencing on Semiconductor Sequencing Platforms: A Comparison of Genetic and Reference-Based Marker Ordering in Barley. PLoS ONE 2013, 8, e76925. [Google Scholar] [CrossRef] [Green Version]
- Ooijen, J.; Ooijen, J.; van ’t Verlaat, J.; Ooijen, J.; Tol, J.; Dalén, J.; Buren, J.; Meer, J.M.; Krieken, J.V.; Ooijen, J.; et al. JoinMap® 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma BV: Wageningen, The Netherlands, 2006. [Google Scholar]
- Kosambi, D.D. The Estimation of Map Distances from Recombination Values. Ann. Eugen. 1943, 12, 172–175. [Google Scholar] [CrossRef]
- WinQTLCart. Available online: https://brcwebportal.cos.ncsu.edu/qtlcart/WQTLCart.htm (accessed on 19 August 2022).
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Water Condition | PH | SL | SM | GNS | TKW | BIO | GY | HI | |
|---|---|---|---|---|---|---|---|---|---|
| PH | irrigated | 1 | |||||||
| water deficit | 1 | ||||||||
| SL | irrigated | 0.269 ** | 1 | ||||||
| water deficit | 0.358 ** | 1 | |||||||
| SM | irrigated | −0.197 * | −0.238 ** | 1 | |||||
| water deficit | −0.088 | −0.141 | 1 | ||||||
| GNS | irrigated | 0.321 ** | 0.517 ** | −0.174 * | 1 | ||||
| water deficit | 0.242 ** | 0.486 ** | −0.103 | 1 | |||||
| TKW | irrigated | 0.165 * | 0.141 | 0.057 | 0.170 * | 1 | |||
| water deficit | 0.112 | 0.027 | −0.038 | 0.073 | 1 | ||||
| BIO | irrigated | 0.439 ** | 0.354 ** | 0.253 ** | 0.336 ** | 0.346 ** | 1 | ||
| water deficit | 0.344 ** | 0.290 ** | 0.351 ** | 0.322 ** | 0.244 ** | 1 | |||
| GY | irrigated | 0.276 ** | 0.261 ** | 0.330 ** | 0.451 ** | 0.423 ** | 0.640 ** | 1 | |
| water deficit | 0.149 | 0.207 * | 0.295 ** | 0.385 ** | 0.421 ** | 0.561 ** | 1 | ||
| HI | irrigated | 0.008 | 0.1 | 0.198 * | 0.314 ** | 0.281 ** | 0.019 | 0.755 ** | 1 |
| water deficit | −0.086 | 0.058 | 0.082 | 0.217 ** | 0.292 ** | −0.102 | 0.742 ** | 1 |
| Chromosome | No. SNP | No. SNP after Filtering | Mapped Loci to Linkage Group | Length of Linkage Group (cM) | Distance between Two Adjacent Markers | SNP per cM |
|---|---|---|---|---|---|---|
| 1A | 2080 | 476 | 399 | 189.22 | 0.47 | 2.11 |
| 2A | 1868 | 451 | 356 | 231.32 | 0.65 | 1.54 |
| 3A | 1447 | 342 | 267 | 208.69 | 0.78 | 1.28 |
| 4A | 2135 | 396 | 335 | 220.39 | 0.66 | 1.52 |
| 5A | 1767 | 424 | 343 | 276.68 | 0.81 | 1.24 |
| 6A | 1305 | 323 | 254 | 179.48 | 0.71 | 1.42 |
| 7A | 2730 | 630 | 500 | 260.88 | 0.52 | 1.92 |
| A genome | 13,332 | 3042 | 2454 | 1566.64 | ||
| 1B | 2520 | 548 | 419 | 190.72 | 0.46 | 2.20 |
| 2B | 3384 | 785 | 570 | 268.49 | 0.47 | 2.12 |
| 3B | 3516 | 707 | 540 | 257.19 | 0.48 | 2.10 |
| 4B | 1164 | 222 | 180 | 131.50 | 0.73 | 1.37 |
| 5B | 2499 | 492 | 380 | 281.99 | 0.74 | 1.35 |
| 6B | 2926 | 636 | 422 | 162.63 | 0.39 | 2.59 |
| 7B | 2828 | 587 | 422 | 227.19 | 0.54 | 1.86 |
| B genome | 18,837 | 3977 | 2933 | 1519.70 | ||
| 1D | 601 | 159 | 101 | 115.38 | 1.14 | 0.88 |
| 2D | 837 | 237 | 126 | 152.11 | 1.21 | 0.83 |
| 3D | 268 | 43 | 29 | 83.70 | 2.89 | 0.35 |
| 4D | 134 | 27 | 15 | 12.89 | 0.86 | 1.16 |
| 5D | 276 | 49 | 29 | 18.17 | 0.63 | 1.60 |
| 6D | 498 | 128 | 49 | 9.19 | 0.19 | 5.33 |
| 7D | 622 | 126 | 95 | 164.36 | 1.73 | 0.58 |
| D genome | 3236 | 769 | 444 | 555.80 | ||
| Total | 35,405 | 7788 | 5831 | 3642.14 | 0.62 | 1.60 |
| Trait | Water Condition | Environment | QTL | Closest Marker | Position (cM) | LOD | R2 (%) a | Add b |
|---|---|---|---|---|---|---|---|---|
| GY | Irrigated | Mahabad | Qgy-Irr.mah-4A | chr4A_614700608 | 82 | 3.92 | 8.57 | 12.94 |
| Qgy1-Irr.mah-6B | chr6B_33629751 | 43 | 3.80 | 8.87 | 11.76 | |||
| Qgy2-Irr.mah-6B | chr6B_34082536 | 49 | 3.65 | 8.50 | 11.51 | |||
| Miandoab | Qgy-Irr.mia-2A | chr2A_30568266 | 43 | 3.98 | 9.21 | −12.01 | ||
| Mean of 3 Places | Qgy1-Irr.3P-6B | chr6B_33629751 | 43 | 4.22 | 9.90 | 9.63 | ||
| Qgy2-Irr.3P-6B | chr6B_34340678 | 49 | 4.06 | 9.50 | 9.39 | |||
| Water deficit | Miandoab | Qgy-Wd.mia-2A | chr2A_24214196 | 34 | 4.21 | 10.50 | −9.10 | |
| Mean of 3 Places | Qgy-Wd.3P-2A | chr2A_24214196 | 34 | 3.49 | 8.30 | −5.97 | ||
| PH | Irrigated | Mahabad | Qph-Irr.mah-3D | chr3D_453986741 | 24 | 3.59 | 9.78 | 2.42 |
| Miandoab | Qph1-Irr.mia-6B | chr6B_567765886 | 109 | 3.64 | 8.6 | −3.62 | ||
| Qph2-Irr.mia-6B | chr6B_659589376 | 129 | 4.61 | 11.42 | 4.43 | |||
| Tabriz | Qph1-Irr.tab-4B | chr4B_28735878 | 44 | 7.21 | 14.00 | −2.96 | ||
| Qph2-Irr.tab-4B | chr4B_37347202 | 50 | 10.60 | 19.00 | −3.45 | |||
| Qph-Irr.tab-4D | chr4D_120094414 | 5 | 10.22 | 18.60 | −3.40 | |||
| Qph-Irr.tab-7A | chr7A_83776793 | 113 | 3.79 | 6.20 | −1.97 | |||
| Mean of 3 Places | Qph-Irr.3P-2B | chr2B_770220840 | 239 | 3.82 | 9.30 | 1.61 | ||
| Qph-Irr.3P-3A | chr3A_35421232 | 62 | 3.60 | 8.30 | −1.51 | |||
| Qph-Irr.3P-7B | chr7B_650619590 | 140 | 3.65 | 8.30 | 1.54 | |||
| Water deficit | Mahabad | Qph-Wd.mah-7D | chr7D_485517060 | 110 | 4.24 | 14.00 | −2.65 | |
| Miandoab | Qph1-Wd.mia-7A | chr7A_16302867 | 16 | 3.88 | 9.10 | −1.86 | ||
| Qph2-Wd.mia-7A | chr7A_22919790 | 26 | 3.81 | 8.90 | −1.83 | |||
| Tabriz | Qph-Wd.tab-4B | chr4B_28735878 | 44 | 6.13 | 12.70 | −2.60 | ||
| Qph-Wd.tab-4D | chr4D_120094414 | 2 | 4.67 | 9.04 | −2.20 | |||
| Mean of 3 Places | Qph-Wd.3P-4A | chr4A_614700608 | 76 | 4.06 | 9.30 | 1.48 | ||
| Qph1-Wd.3P-7A | chr7A_16302867 | 16 | 4.27 | 10.00 | −1.42 | |||
| Qph2-Wd.3P-7A | chr7A_18775675 | 22 | 4.90 | 11.40 | −1.50 | |||
| Qph3-Wd.3P-7A | chr7A_22919790 | 27 | 4.24 | 10.50 | −1.40 | |||
| SM | Irrigated | Mahabad | Qsm1-Irr.mah-6B | chr6B_33629751 | 43 | 3.81 | 9.46 | 9.49 |
| Qsm2-Irr.mah-6B | chr6B_34340678 | 49 | 3.81 | 9.46 | 9.47 | |||
| Tabriz | Qsm-Irr.tab-5A | chr5A_589287461 | 166 | 8.01 | 17.60 | 24.26 | ||
| Mean of 3 Places | Qsm1-Irr.3P-2B | chr2B_777297032 | 247 | 3.68 | 8.80 | −8.68 | ||
| Qsm2-Irr.3P-2B | chr2B_780590240 | 252 | 4.02 | 9.50 | −8.50 | |||
| Qsm-Irr.3P-5A | chr5A_581488651 | 165 | 5.81 | 13.80 | 10.45 | |||
| Water deficit | Tabriz | Qsm-Wd.tab-2B | chr2B_513284737 | 128 | 3.97 | 8.70 | −14.64 | |
| TKW | Irrigated | Mahabad | Qtkw1-Irr.mah-1B | chr1B_640559774 | 145 | 5.17 | 14.61 | −3.91 |
| Qtkw2-Irr.mah-1B | chr1B_642697116 | 150 | 4.78 | 12.04 | −3.42 | |||
| Miandoab | Qtkw-Irr.mia-1B | chr1B_642697116 | 150 | 4.62 | 11.57 | −3.67 | ||
| Tabriz | Qtkw1-Irr.tab-5B | chr5B_339425357 | 64 | 5.73 | 13.00 | 1.29 | ||
| Qtkw2-Irr.tab-5B | chr5B_378853109 | 74 | 4.44 | 10.00 | 1.09 | |||
| Qtkw-Irr.tab-6B | chr6B_668966788 | 139 | 3.40 | 7.90 | 0.93 | |||
| Water deficit | Mahabad | Qtkw-Wd.mah-3B | chr3B_21770957 | 44 | 4.18 | 11.59 | 2.51 | |
| Miandoab | Qtkw-Wd.mia-3B | chr3B_20721650 | 42 | 3.39 | 8.60 | 1.79 | ||
| Tabriz | Qtkw1-Wd.tab-4B | chr4B_21573529 | 37 | 4.35 | 11.60 | −1.08 | ||
| Qtkw2-Wd.tab-4B | chr4B_37347202 | 50 | 3.79 | 8.40 | −0.90 | |||
| Qtkw1-Wd.tab-5B | chr5B_412238630 | 77 | 4.48 | 11.00 | 1.08 | |||
| Qtkw2-Wd.tab-5B | chr5B_430752407 | 85 | 3.32 | 8.30 | 0.92 | |||
| Qtkw-Wd.tab-7B | chr7B_701649237 | 173 | 3.90 | 8.80 | −0.91 | |||
| GNS | Irrigated | Mahabad | Qgns-Irr.mah-3A | chr3A_695662512 | 150 | 4.19 | 8.13 | −0.65 |
| Tabriz | Qgns1-Irr.tab-4A | chr4A_632236000 | 122 | 7.58 | 19.25 | 1.61 | ||
| Qgns2-Irr.tab-4A | chr4A_683874492 | 129 | 6.27 | 14.60 | 1.40 | |||
| Water deficit | Mahabad | Qgns1-Wd.mah-7D | chr7D_94715776 | 85 | 2.82 | 6.60 | 0.65 | |
| Qgns2-Wd.mah-7D | chr7D_180083703 | 98 | 4.04 | 10.00 | −0.77 | |||
| Miandoab | Qgns-Wd.mia-1A | chr1A_11934211 | 22 | 4.29 | 11.10 | −0.68 | ||
| Mean of 3 Places | Qgns-Wd.3P-4A | chr4A_683874492 | 130 | 3.64 | 9.10 | 0.51 | ||
| SL | Irrigated | Tabriz | Qsl-Irr.tab-3A | chr3A_697615272 | 158 | 4.74 | 11.92 | −0.26 |
| Qsl1-Irr.tab-4A | chr4A_639994434 | 124 | 3.45 | 7.40 | 0.21 | |||
| Qsl2-Irr.tab-4A | chr4A_681683160 | 132 | 4.61 | 10.43 | 0.25 | |||
| Qsl1-Irr.tab-7D | chr7D_89732435 | 80 | 4.01 | 9.10 | −0.23 | |||
| Qsl2-Irr.tab-7D | chr7D_104889647 | 89 | 4.61 | 9.70 | −0.24 | |||
| Water deficit | Mahabad | Qsl-Wd.mah-3B | chr3B_43100457 | 76 | 3.80 | 8.80 | −0.21 | |
| Qsl-Wd.mah-7A | chr7A_66641918 | 94 | 7.15 | 19.18 | −0.25 | |||
| Miandoab | Qsl-Wd.mia-2B | chr2B_26987136 | 46 | 3.97 | 11.64 | 0.17 | ||
| Mean of 3 Places | Qsl-Wd.3P-2A | chr2A_24111229 | 25 | 3.27 | 8.20 | −0.11 | ||
| BIO | Irrigated | Mahabad | Qbio-Irr.mah-4A | chr4A_614700608 | 77 | 3.77 | 9.42 | 17.03 |
| Miandoab | Qbio-Irr.mia-1D | chr1D_34011022 | 55 | 3.65 | 10.78 | 18.86 | ||
| Mean of 3 Places | Qbio-Irr.3P-4A | chr4A_614700608 | 78 | 3.98 | 10.70 | 15.66 | ||
| Water deficit | Mahabad | Qbio-Wd.mah-5A | chr5A_552523257 | 128 | 3.84 | 10.38 | −16.00 | |
| Miandoab | Qbio1-Wd.mia-1B | chr1B_9691095 | 9 | 3.67 | 9.30 | 13.95 | ||
| Qbio2-Wd.mia-1B | chr1B_15368052 | 17 | 3.84 | 9.60 | 14.41 | |||
| Mean of 3 Places | Qbio-Wd.3P-5A | chr5A_552523257 | 131 | 4.43 | 10.95 | −11.43 | ||
| HI | Irrigated | Mahabad | Qhi-Irr.mah-3D | chr3D_197992771 | 23 | 3.72 | 9.70 | 2.14 |
| Miandoab | Qhi1-Irr.mia-2A | chr2A_3975444 | 5 | 3.67 | 8.10 | −2.20 | ||
| Qhi2-Irr.mia-2A | chr2A_6269139 | 14 | 5.02 | 11.77 | −2.61 | |||
| Qhi-Irr.mia-6A | chr6A_591883833 | 122 | 6.43 | 14.20 | −2.91 | |||
| Tabriz | Qhi1-Irr.tab-1B | chr1B_420591625 | 77 | 4.87 | 11.60 | −2.71 | ||
| Qhi2-Irr.tab-1B | chr1B_498063961 | 87 | 5.09 | 12.10 | −2.64 | |||
| Mean of 3 Places | Qhi-Irr.3P-6A | chr6A_591883833 | 122 | 4.42 | 9.70 | −1.47 | ||
| Water deficit | Mahabad | Qhi-Wd.mah-2D | chr2D_618150012 | 142 | 2.55 | 6.06 | 1.96 | |
| Qhi1-Wd.mah-3B | chr3B_178266388 | 109 | 2.86 | 7.25 | 2.32 | |||
| Qhi2-Wd.mah-3B | chr3B_419560745 | 117 | 4.57 | 11.54 | 2.98 | |||
| Qhi3-Wd.mah-3B | chr3B_492105670 | 125 | 5.06 | 12.47 | 3.19 | |||
| Qhi4-Wd.mah-3B | chr3B_728922412 | 159 | 3.57 | 8.90 | −2.54 | |||
| Miandoab | Qhi-Wd.mia-5A | chr5A_52357876 | 63 | 5.33 | 13.00 | −3.85 | ||
| Tabriz | Qhi1-Wd.tab-1B | chr1B_420591625 | 77 | 4.89 | 11.50 | −2.74 | ||
| Qhi2-Wd.tab-1B | chr1B_427069393 | 81 | 4.89 | 11.50 | −2.74 |
| QTL Name | Flanking Markers a | Gene-ID | Annotation |
|---|---|---|---|
| Qtkw1-Irr.mah-1B | chr1B_637810003-chr1B_641627897 | TraesCS1B01G415500.1 | Ubiquitin-conjugating enzyme E2 |
| Qtkw-Irr.mia-1B | chr1B_639448207-chr1B_642697116 | ||
| Qtkw-Wd.mia-3B | chr3B_20439595-chr3B_22054094 | TraesCS3B01G041700.1 | Alpha-glucosidase |
| Qtkw-Wd.mah-3B | chr3B_20439595-chr3B_22054094 | TraesCS3B01G042400.1 | AP2-EREBP transcription factor |
| Qtkw-Wd.tab-7B | chr7B_692926289-chr7B_702176728 | TraesCS7B01G434600.1 | FBD-associated F-box protein |
| Qgy-Wd.mia-2A | chr2A_20237446-chr2A_33006222 | TraesCS2A01G057700.1 | NBS-LRR disease resistance protein |
| Qgy-Wd.3P-2A | |||
| Qgy-Irr.mia-2A | |||
| Qgy2-Irr.3P-6B | chr6B_28012560-chr6B_35789585 | TraesCS6B01G053900.1 | Cysteine proteinase |
| Qsm2-Irr.mah-6B | |||
| Qgy2-Irr.mah-6B | |||
| Qgns2-Irr.tab-4A | chr4A_681683121-chr4A_684909655 | TraesCS4A01G411500.1 | NAD(P)H-quinone oxidoreductase subunit H |
| Qgns-Wd.3P-4A | |||
| Qgns2-Irr.tab-4A | |||
| Qsl1-Irr.tab-4A | chr4A_632236000-chr4A_640906743 | TraesCS4A01G366600.1 | F-box family protein |
| Qsl-Wd.3P-2A | chr2A_15816001-chr2A_21812312 | TraesCS2A01G056900.1 | Multidrug resistance protein, ABC transporter family protein |
| Qsl-Wd.mia-2B | chr2B_25477352-chr2B_26987136 | TraesCS2B01G055300.1 | NBS-LRR disease resistance protein-like |
| Qsm-Wd.tab-2B | chr2B_485443263-chr2B_565076231 | TraesCS2B01G359800.1 | Mitochondrial inner membrane protease ATP23 |
| Qsm2-Irr.3P-2B | chr2B_779912793-chr2B_785208751 | TraesCS2B01G596900.1 | Acyl-CoA-binding domain-containing protein |
| Qsm-Irr.3P-5A | chr5A_581488651-chr5A_589302806 | TraesCS5A01G383800.1 | Heat shock transcription factor |
| Qph2-Irr.mia-6B | chr6B_659809861-chr6B_662562269 | TraesCS6B01G384600.1 | Stigma-specific protein Stig1 |
| Qph1-Irr.tab-4B | chr4B_21573518-chr4B_37529724 | TraesCS4B01G042200.2 | Serine/threonine-protein kinase |
| Qph-Wd.tab-4B | |||
| Qph1-Irr.mia-6B | chr6B_569936355-chr6B_623369219 | TraesCS6B01G320500.1 | F-box family protein |
| Qph-Irr.tab-7A | chr7A_82949085-chr7A_85912154 | TraesCS7A01G130300.1 | Protein DETOXIFICATION |
| Qph1-Wd.mia-7A | chr7A_13905540-chr7A_19958972 | TraesCS7A01G036200.1 | 60 kDa chaperonin |
| Qph1-Wd.3P-7A | |||
| Qph-Wd.mah-7D | chr7D_451622823-chr7D_554279150 | TraesCS7D01G375100.1 | Phosphatidylcholine:diacylglycerol cholinephosphotransferase 1 |
| Qbio-Wd.3P-5A | chr5A_549192141-chr5A_555596100 | TraesCS5A01G349500.1 | Plant regulator RWP-RK family protein |
| Qbio-Wd.mah-5A | |||
| Qhi2-Irr.mia-2A | chr2A_2810448-chr2A_13880947 | TraesCS2A01G015400.1 | Cytochrome P450 |
| Qhi2-Irr.tab-1B | chr1B_402043386-chr1B_516951252 | TraesCS1B01G286300.1 | Cytochrome P450 |
| Qhi1-Wd.mah-3B | chr3B_122826152-chr3B_190473903 | TraesCS3B01G156900.1 | B-box zinc finger family protein |
| Qhi2-Wd.mah-3B | chr3B_411269889-chr3B_452490772 | TraesCS3B01G264100.1 | Protein CHUP1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdollahi Sisi, N.; Stein, N.; Himmelbach, A.; Mohammadi, S.A. High-Density Linkage Mapping of Agronomic Trait QTLs in Wheat under Water Deficit Condition using Genotyping by Sequencing (GBS). Plants 2022, 11, 2533. https://doi.org/10.3390/plants11192533
Abdollahi Sisi N, Stein N, Himmelbach A, Mohammadi SA. High-Density Linkage Mapping of Agronomic Trait QTLs in Wheat under Water Deficit Condition using Genotyping by Sequencing (GBS). Plants. 2022; 11(19):2533. https://doi.org/10.3390/plants11192533
Chicago/Turabian StyleAbdollahi Sisi, Nayyer, Nils Stein, Axel Himmelbach, and Seyed Abolghasem Mohammadi. 2022. "High-Density Linkage Mapping of Agronomic Trait QTLs in Wheat under Water Deficit Condition using Genotyping by Sequencing (GBS)" Plants 11, no. 19: 2533. https://doi.org/10.3390/plants11192533





