The Fluorescence Detection of Phenolic Compounds in Plicosepalus curviflorus Extract Using Biosynthesized ZnO Nanoparticles and Their Biomedical Potential
Abstract
:1. Introduction
2. Materials and Methods
2.1. Apparatus
2.2. Chemicals and Reagents
2.3. Botanical Material
2.4. Microbial Pathogens and Cancer Cell Line
2.5. Extraction and Isolation Procedures
2.6. Spectral Analysis of Isolated Compounds
2.7. Green Preparation of ZnONPs Using Biomass of P. curviflorus
2.8. Spectroscopic and Microscopic Characterization of ZnONPs
2.9. Fluorescence Analysis
2.10. Constructed Calibration Graphs
2.11. Antibacterial Activity
2.12. Determination of Baterostatic and Bacteriocidal Concentration
2.13. Morphological Investigation of P. aeruginosa and S. aureus
2.14. Hemolytic Activity
2.15. Anticancer Activity
3. Results and Discussion
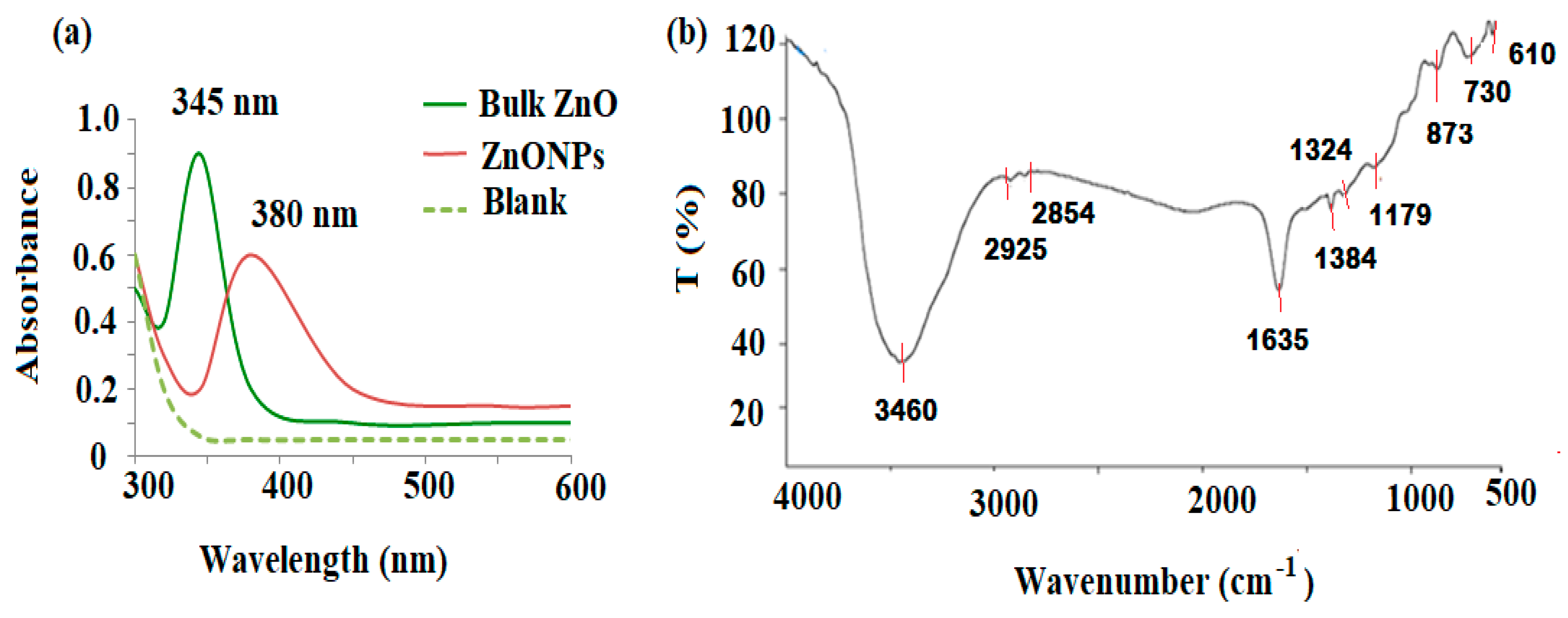
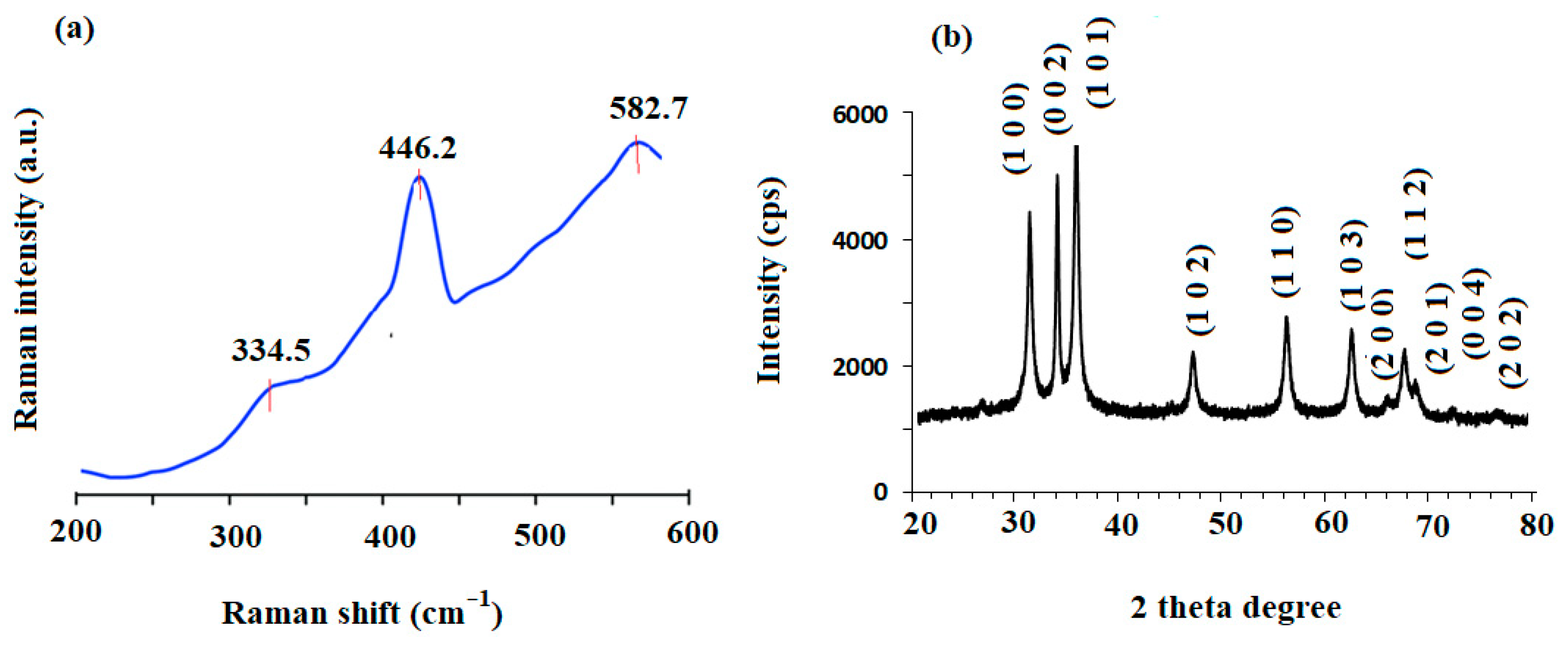
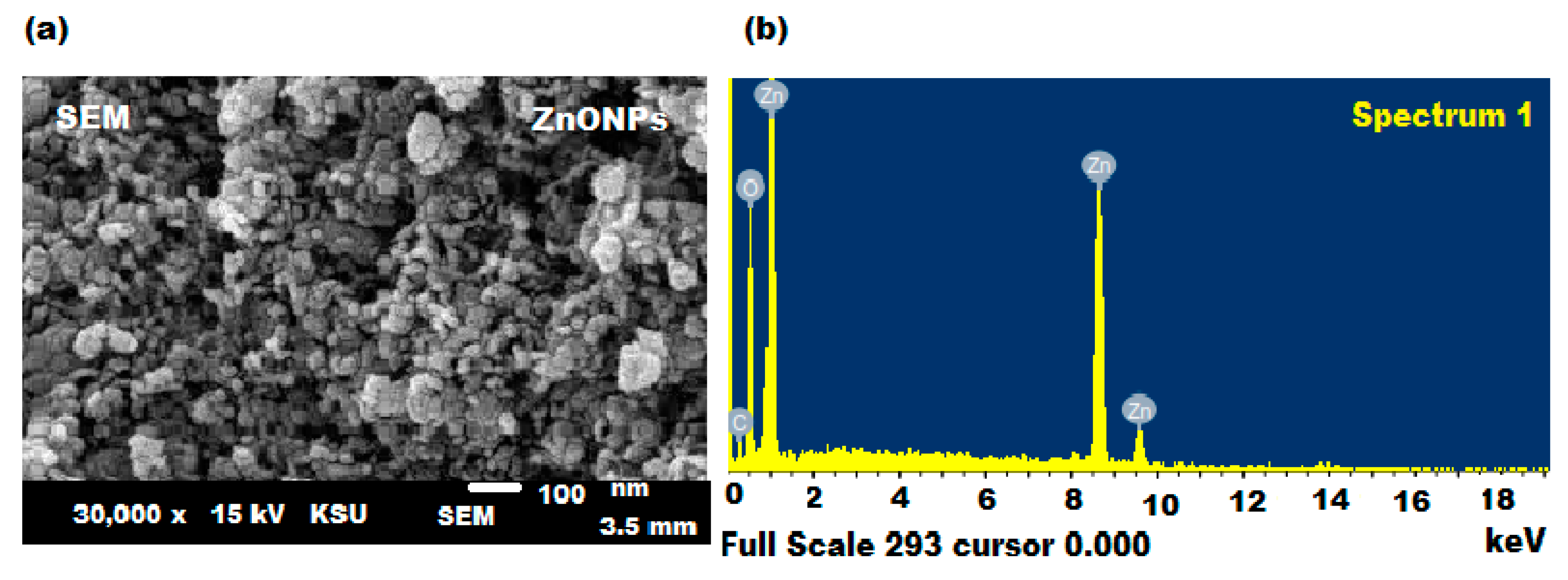
3.1. Spectroscopic Characterization of ZnONPs
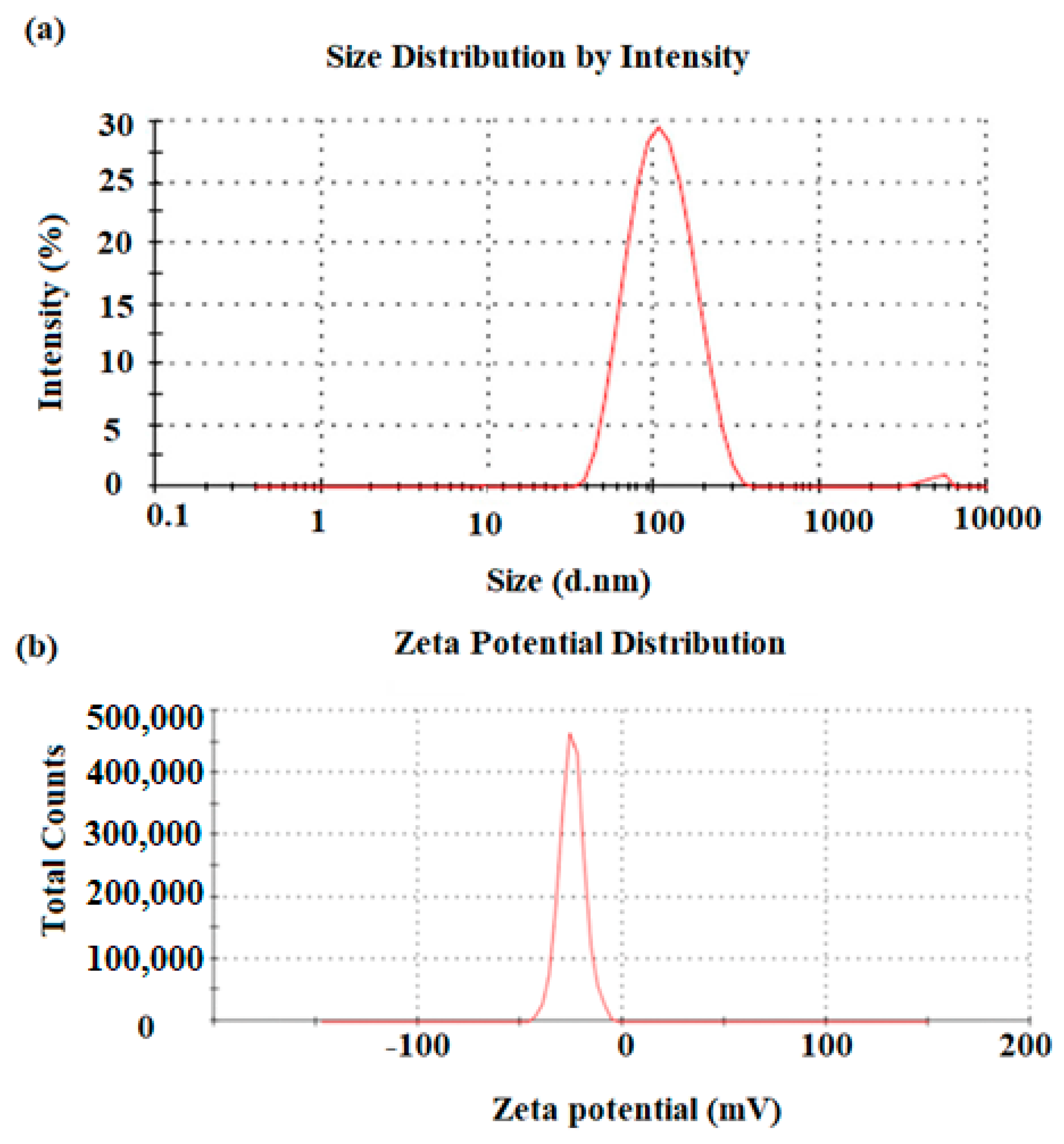
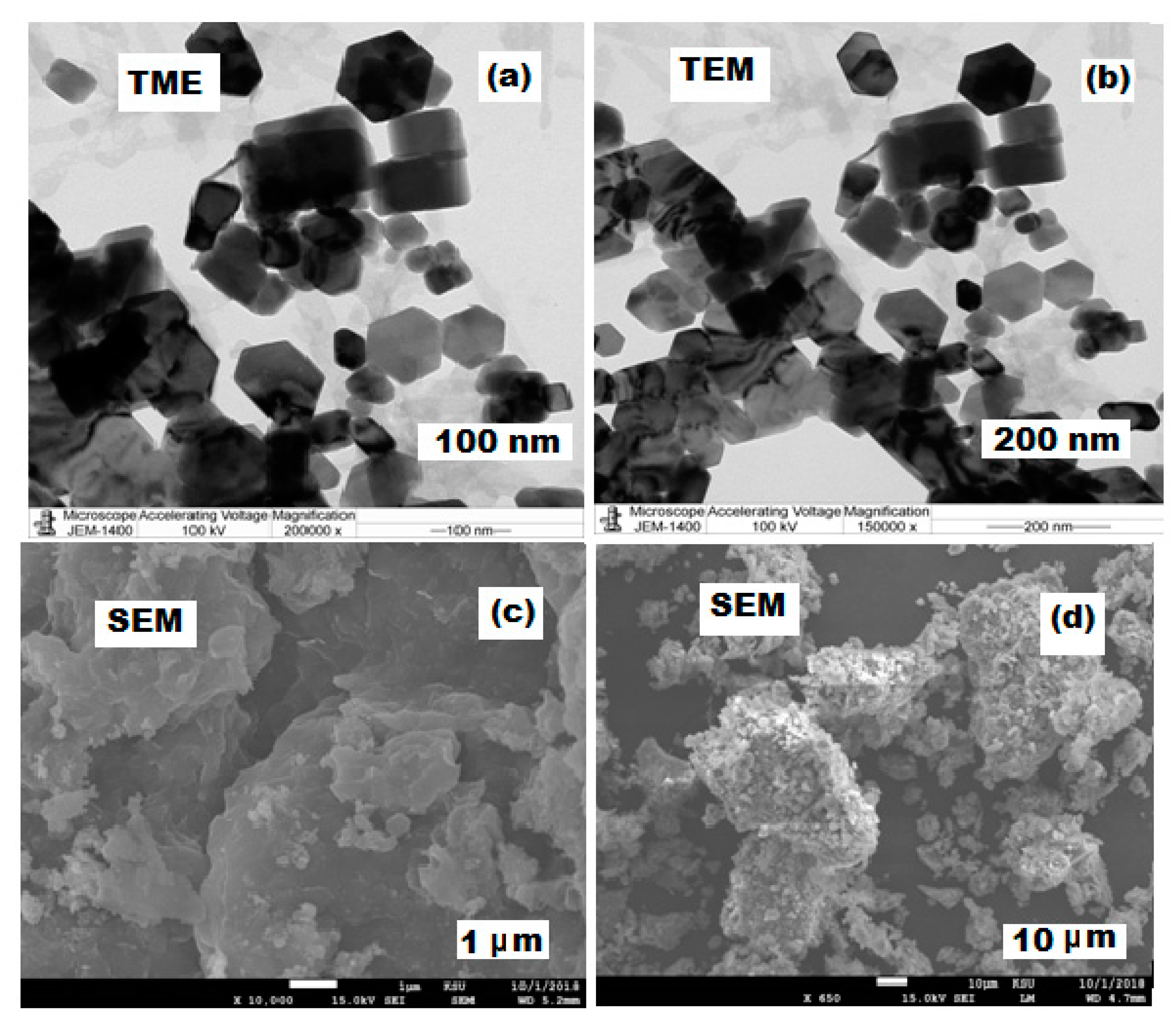
3.2. Microscopic Characterization
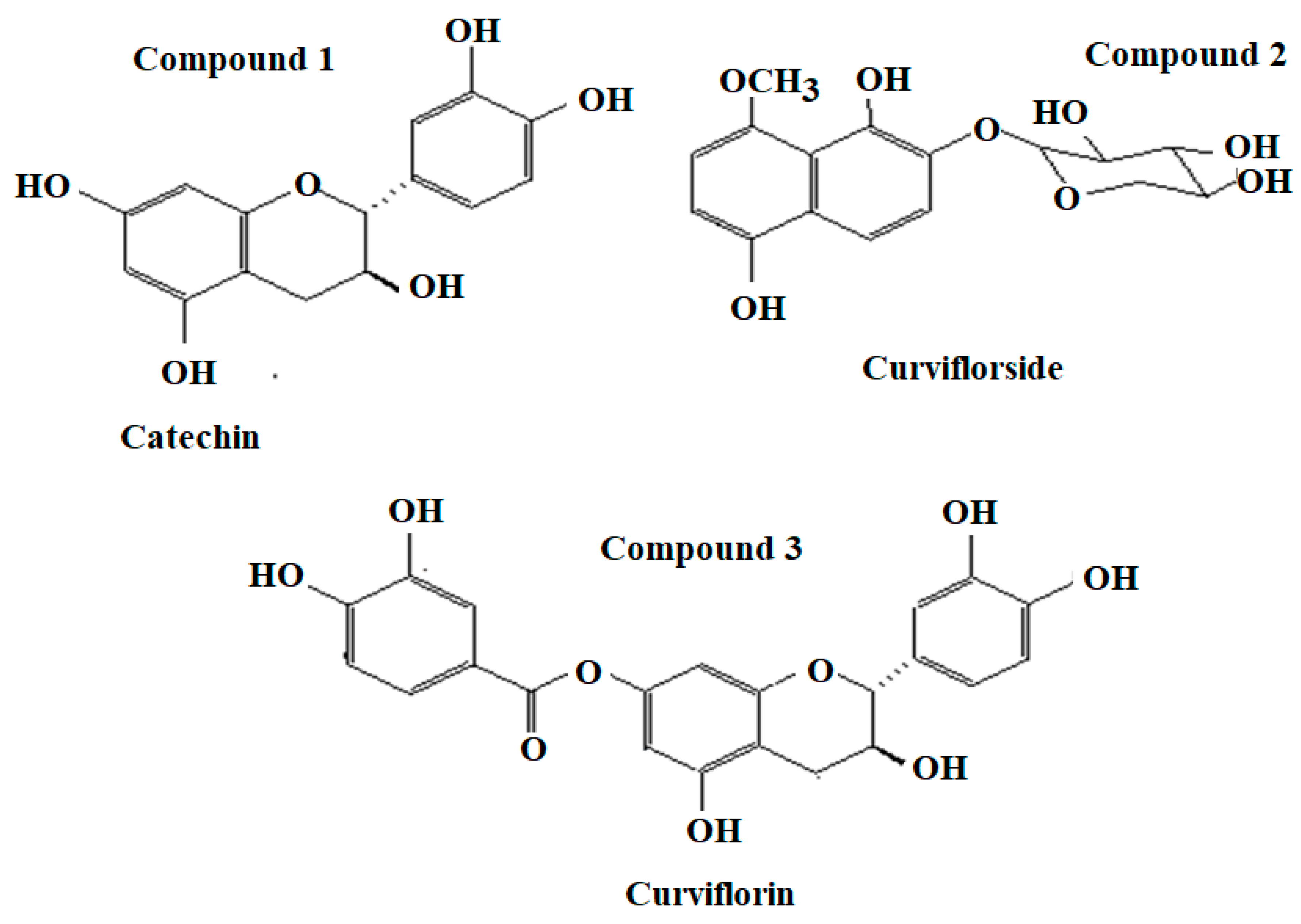
3.3. Spectral Characteristics of Isolated Compounds
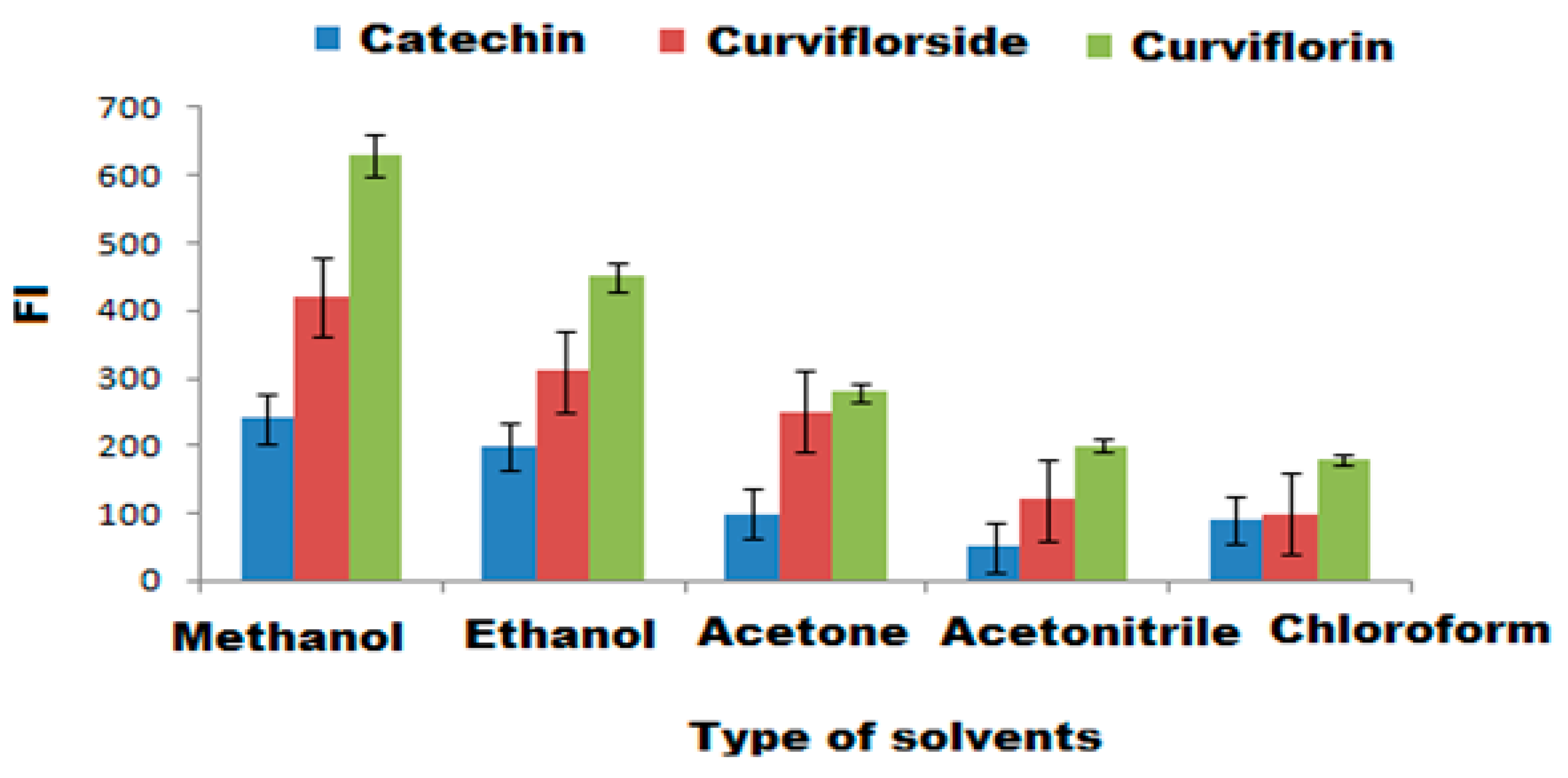
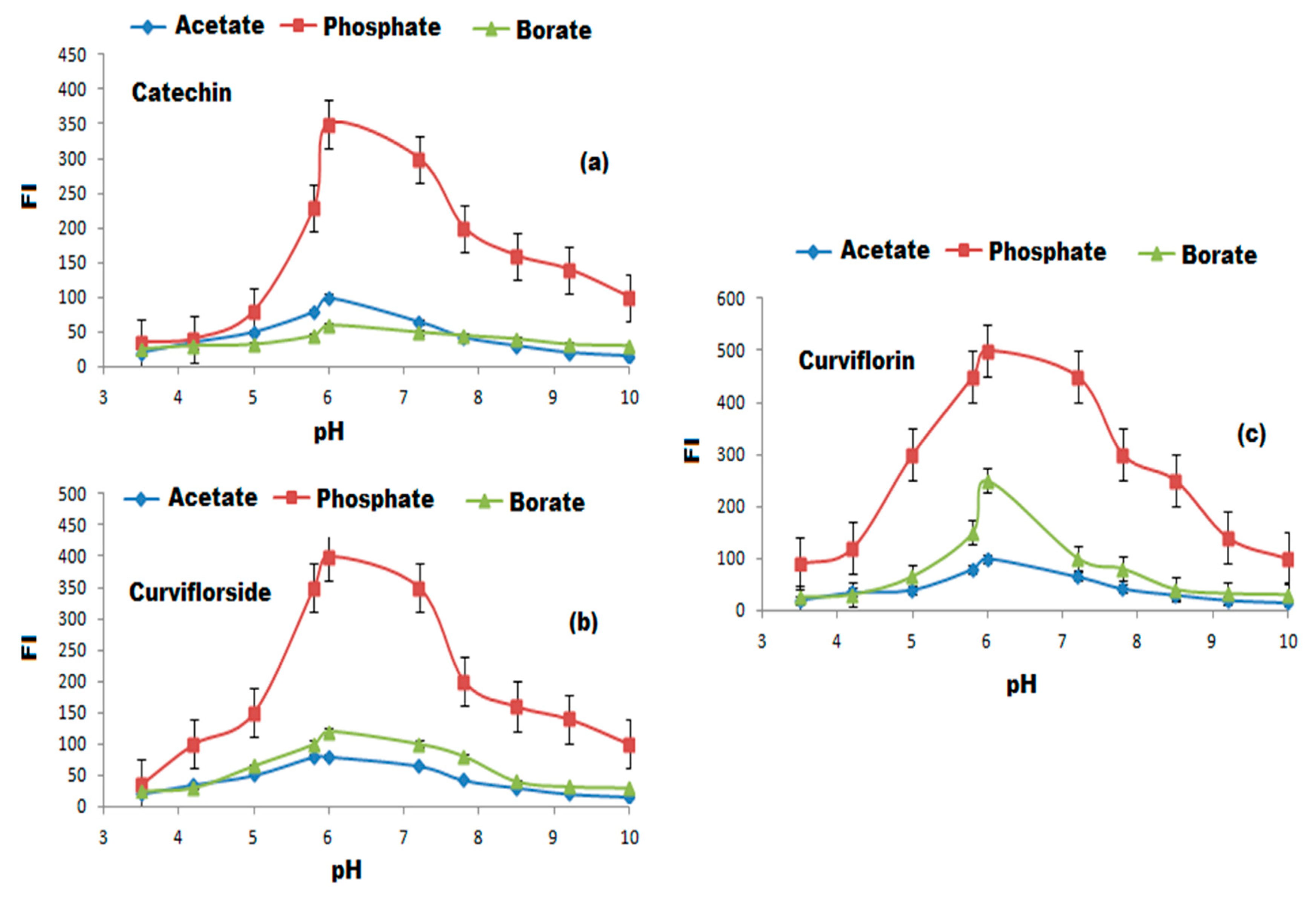
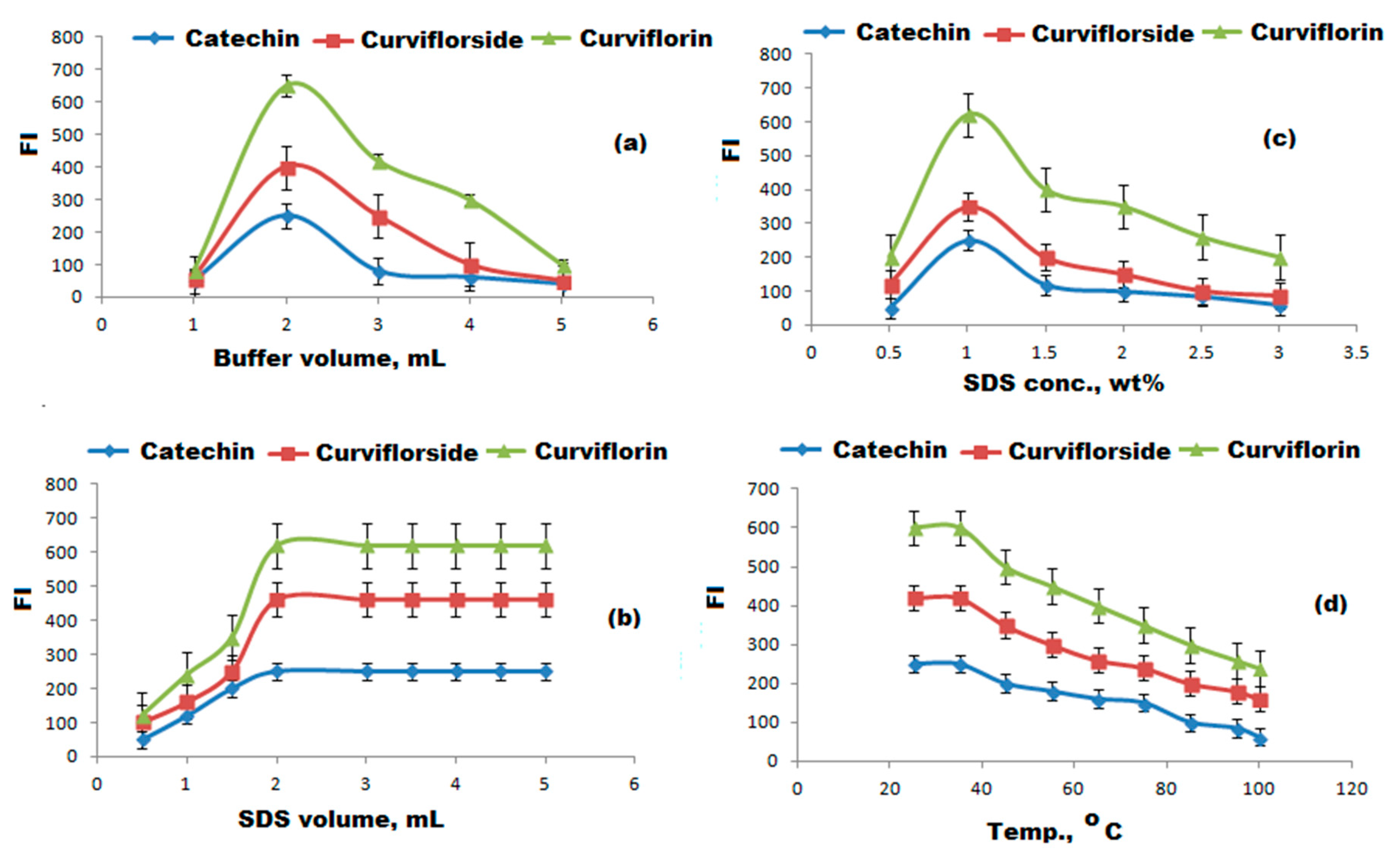
3.4. Selection of Optimum Experimental Conditions
3.5. Analysis of Isolated Catechin, Curviflorside, and Curviflorin in Real Extract
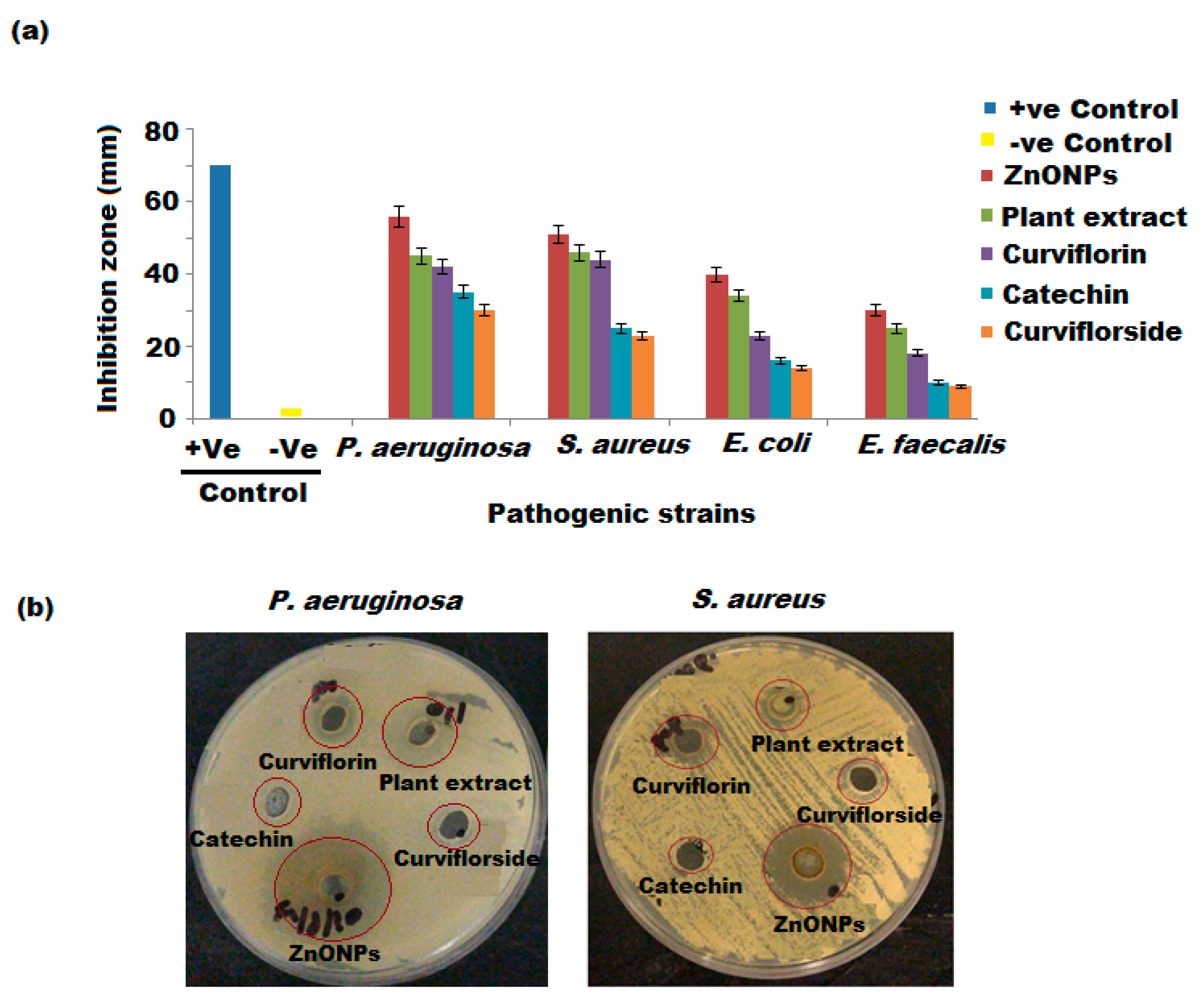
3.6. Antibacterial Activity
3.7. Morphological Study of P. aeruginosa and S. aureus (SEM)
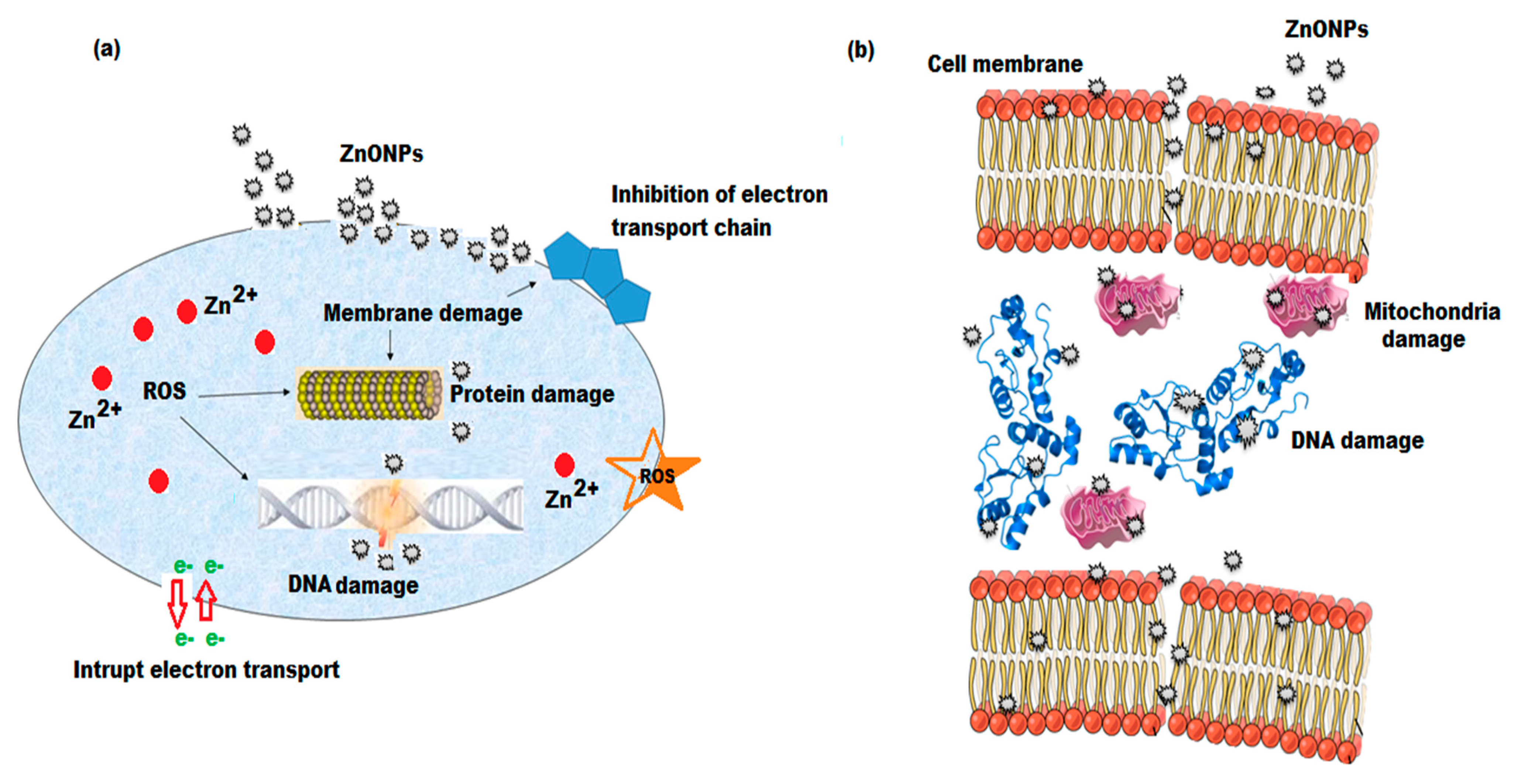
3.8. Anti-Hemolytic Effects
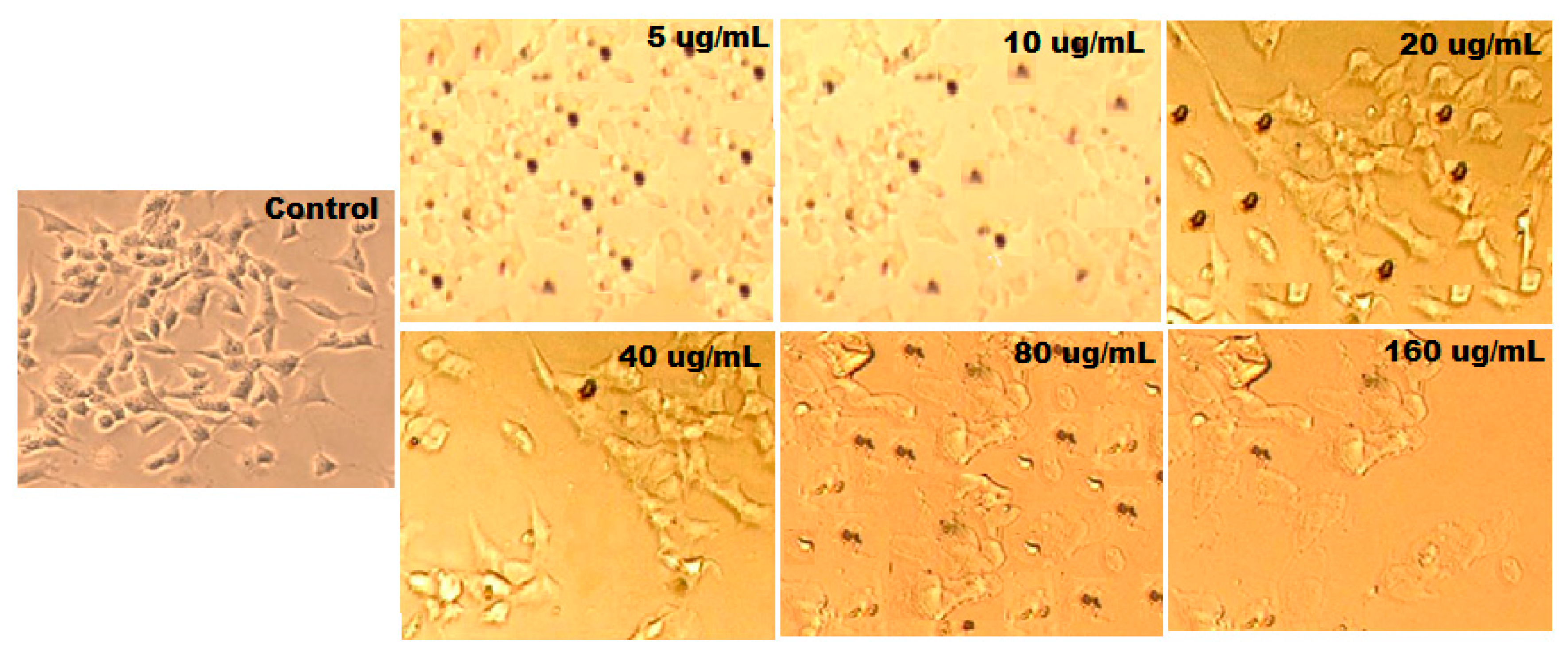
3.9. Anticancer Activity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fu, Y.; Luo, J.; Qin, J.; Yang, M. Screening techniques for the identification of bioactive compounds in natural products. J. Pharm. Biomed. Anal. 2019, 168, 189–200. [Google Scholar] [CrossRef]
- Ciesla, L.; Moaddel, R. Comparison of analytical techniques for the identification of bioactive compounds from natural products. Nat. Prod. Rep. 2016, 33, 1131–1145. [Google Scholar] [CrossRef] [Green Version]
- Ahmad Dar, A.; Sangwan, P.L.; Kumar, A. Chromatography: An important tool for drug discovery. J. Sep. Sci. 2020, 43, 105–119. [Google Scholar] [CrossRef]
- Fatemeh, F.; Mehdi, E.; Mehrabian, R.Z.; Golsefidi, M.A.; Moradi, A.V. Application of electrochemical techniques for determining and extracting natural product (EgCg) by the synthesized conductive polymer electrode (Ppy/Pan/rGO) impregnated with nano-Particles of TiO2. Sci. Rep. 2019, 9, 3940. [Google Scholar]
- Rojalin, T.; Phong, B.; Koster, H.J.; Carney, R.P. Nanoplasmonic approaches for sensitive detection and molecular characterization of extracellular vesicles. Front. Chem. 2019, 7, 279. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Li, P.; Yang, H.; Jiang, N.; Wang, D.; Sun, S.; Wei, W. Detection of 5-hydroxymethylfurfural Based on split-DNAzyme assisted signal amplification via quartz crystal microbalance. Sens. Actuators B Chem. 2021, 352, 131031. [Google Scholar] [CrossRef]
- Amina, M.; Alarfaj, N.A.; El-Tohamy, M.F.; Al-Musayeib, N.M.; Oraby, H.F. Sequential injection-chemiluminescence evaluation of stigmasterol glucoside and luteolin via green synthesis of silver nanoparticles using biomass of Plectranthus asirensis. Green Chem. Lett. Rev. 2018, 11, 523–533. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Zhu, J.; Ma, F.; Li, P.; Zhang, L.; Zhang, W.; Ding, X.; Zhang, Q. Quantitative determination of major capsaicinoids in serum by ELISA and time-resolved fluorescent immunoassay based on monoclonal antibodies. Biosens. Bioelectron. 2016, 81, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Geng, W.C.; Gao, J.; Wang, Y.Y.; Sun, H.; Guo, D.S. Ultrasensitive and specific fluorescence detection of a cancer biomarker via nanomolar binding to a guanidinium-modified calixarene. Chem. Sci. 2018, 9, 2087–2091. [Google Scholar] [CrossRef] [Green Version]
- Tagit, O.; Hildebrandt, N. Fluorescence sensing of circulating diagnostic biomarkers using molecular probes and nanoparticles. ACS Sens. 2017, 2, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Furlan, A.L.; Laurin, Y.; Botcazon, C.; Rodriguez-Moraga, N.; Rippa, S.; Deleu, M.; Lins, L.; Sarazin, C.; Buchoux, S. Contributions and limitations of biophysical approaches to study of the interactions between amphiphilic molecules and the plant plasma membrane. Plants 2020, 9, 648. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Kawasaki, A.; Karuo, Y.; Tarui, A.; Kawai, K.; Omote, M. Synthesis of new fluorescent molecules having an aggregation-induced emission property derived from 4-fluoroisoxazoles. Beilstein Arch. 2020, 1, 2020. [Google Scholar] [CrossRef] [PubMed]
- Galievsky, V.A.; Stasheuski, A.S.; Krylov, S.N. Improvement of LOD in fluorescence detection with spectrally nonuniform background by optimization of emission filtering. Anal. Chem. 2017, 89, 11122–11128. [Google Scholar] [CrossRef]
- Santos, D.R.; Soares, R.R.; Pinto, I.F.; Caneira, C.R.; Pinto, R.M.; Chu, V.; Conde, J.P. Label-free detection of biomolecules in microfluidic systems using on-chip UV and impedimetric sensors. IEEE Sens. J. 2019, 19, 7803–7812. [Google Scholar] [CrossRef]
- Gaviria-Arroyave, M.I.; Cano, J.B.; Penuela, G.A. Nanomaterial-based fluorescent biosensors for monitoring environmental pollutants: A critical review. Talanta Open 2020, 2, 100006. [Google Scholar] [CrossRef]
- Yogesh, G.K.; Shuaib, E.P.; Priya, A.K.; Rohini, P.; Anandhan, S.V.; Krishnan, U.M.; Kalyanavalli, V.; Shukla, S.; Sastikumar, D. Synthesis of water-soluble fluorescent carbon nanoparticles (CNPs) from nanosecond pulsed laser ablation in ethanol. Optics Laser Technol. 2021, 135, 106717. [Google Scholar] [CrossRef]
- Talebzadeh, S.; Queffelec, C.; Knight, D.A. Surface modification of plasmonic noble metal—Metal oxide core—Shell nanoparticles. Nanoscale Adv. 2019, 1, 4578–4591. [Google Scholar] [CrossRef] [Green Version]
- Abbandonato, G.; Hoffmann, K.; Resch-Genger, U. Determination of quantum yields of semiconductor nanocrystals at the single emitter level via fluorescence correlation spectroscopy. Nanoscale 2018, 10, 7147–7154. [Google Scholar] [CrossRef]
- Comini, E. Metal oxides nanowires chemical/gas sensors: Recent advances. Materials Today Adv. 2020, 7, 100099. [Google Scholar] [CrossRef]
- Du, L.; Chen, W.; Zhu, P.; Tian, Y.; Chen, Y.; Wu, C. Applications of functional metal-organic frameworks in biosensors. Biotechnol. J. 2021, 16, 1900424. [Google Scholar] [CrossRef]
- Purohit, B.; Vernekar, P.R.; Shetti, N.P.; Chandra, P. Biosensor nanoengineering: Design, operation, and implementation for biomolecular analysis. Sensor. Int. 2020, 1, 100040. [Google Scholar] [CrossRef]
- Ong, C.B.; Ng, L.Y.; Mohammad, A.W. A review of ZnO nanoparticles as solar photocatalysts: Synthesis, mechanisms and applications. Renew. Sust. Energ. Rev. 2018, 81, 536–551. [Google Scholar] [CrossRef]
- Vardhaman, B.A.; Amarnath, M.; Ramkumar, J.; Mondal, K. Enhanced tribological performances of zinc oxide/MWCNTs hybrid nanomaterials as the effective lubricant additive in engine oil. Mater. Chem. Phys. 2020, 253, 123447. [Google Scholar] [CrossRef]
- Alarfaj, N.A.; El-Tohamy, M.F.; Oraby, H.F. CA 19-9 pancreatic tumor marker fluorescence immunosensing detection via immobilized carbon quantum dots conjugated gold nanocomposite. Int. J. Mol. Sci. 2018, 19, 1162. [Google Scholar] [CrossRef] [Green Version]
- Soundharraj, P.; Dhinasekaran, D.; Rajendran, A.R.; Prakasarao, A.; Ganesan, S. N-Doped zinc oxide as an effective fluorescence sensor for urea detection. New J. Chem. 2021, 45, 6080–6090. [Google Scholar] [CrossRef]
- Hu, D.; Si, W.; Qin, W.; Jiao, J.; Li, X.; Gu, X.; Hao, Y. Cucurbita pepo leaf extract induced synthesis of zinc oxide nanoparticles, characterization for the treatment of femoral fracture. J. Photochem. Photobiol. B Biol. 2019, 195, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Viezcas, J.A.; Castillo-Michel, H.; Andrews, J.C.; Cotte, M.; Rico, C.; Peralta-Videa, J.R.; Ge, Y.; Priester, J.H.; Holden, P.A.; Gardea-Torresdey, J.L. In situ synchrotron X-ray fluorescence mapping and speciation of CeO2 and ZnO nanoparticles in soil cultivated soybean (Glycine max). ACS Nano 2013, 7, 1415–1423. [Google Scholar] [CrossRef]
- Ambika, S.; Sundrarajan, M. Green biosynthesis of ZnO nanoparticles using Vitex negundo L. extract: Spectroscopic investigation of interaction between ZnO nanoparticles and human serum albumin. J. Photochem. Photobiol. B Biol. 2015, 149, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Aly, S.M.; Eita, M.; Khan, J.I.; Alarousu, E.; Mohammed, O.F. Remarkable fluorescence enhancement versus complex formation of cationic porphyrins on the surface of ZnO nanoparticles. J. Phys. Chem. C 2014, 118, 12154–12161. [Google Scholar] [CrossRef]
- Jiang, J.; Pi, J.; Cai, J. The advancing of zinc oxide nanoparticles for biomedical applications. Bioinorg. Chem. Appl. 2018, 2018, 1062562. [Google Scholar] [CrossRef]
- Fawzy, G.A.; Al-Taweel, A.M.; Perveen, S. Anticancer activity of flavane gallates isolated from Plicosepalus curviflorus. Pharmacogn. Mag. 2014, 10, S519. [Google Scholar] [CrossRef] [Green Version]
- Aldawsari, H.M.; Hanafy, A.; Labib, G.S.; Badr, J.M. Antihyperglycemic activities of extracts of the mistletoes Plicosepalus acaciae and P. curviflorus in comparison to their solid lipid nanoparticle suspension formulations. Z. Naturforsch. C 2014, 69, 391–398. [Google Scholar] [CrossRef]
- Waly, N.M.; El-Din, A.A.; Jrais, R.N. Botanical and Biological studies of six parasitic species of family Loranthaceae growing in Kingdom of Saudi Arabia. Int. J. Environ. Sci. 2012, 4, 196–205. [Google Scholar]
- Sher, H.; Alyemeni, M.N. Pharmaceutically important plants used in traditional system of Arab medicine for the treatment of livestock ailments in the kingdom of Saudi Arabia. Afr. J. Biotechnol. 2011, 10, 9153–9159. [Google Scholar]
- Mossa, J.S. A study on the crude antidiabetic drugs used in Arabian folk medicine. Int. J. Crude Drug Res. 1985, 23, 137–145. [Google Scholar] [CrossRef]
- Al-Fatimi, M.; Wurster, M.; Schroder, G.; Lindequist, U. Antioxidant, antimicrobial and cytotoxic activities of selected medicinal plants from Yemen. J. Ethnopharmacol. 2007, 111, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Al-Taweel, A.M.; Perveen, S.; Fawzy, G.A.; Alqasoumi, S.I.; El Tahir, K.E. New flavane gallates isolated from the leaves of Plicosepalus curviflorus and their hypoglycemic activity. Fitoterapia 2012, 83, 610–1615. [Google Scholar] [CrossRef]
- Badr, J.M.; Ibrahim, S.R.; Abou-Hussein, D.R. Plicosepalin A, a new antioxidant catechin–gallic acid derivative of inositol from the mistletoe Plicosepalus curviflorus. Z. Naturforsch. C 2016, 711, 375–380. [Google Scholar] [CrossRef]
- Al-Musayeib, N.M.; Ibrahim, S.R.; Amina, M.; Al-Hamoud, G.A.; Mohamed, G.A. Curviflorside and curviflorin, new naphthalene glycoside and flavanol from Plicosepalus curviflorus. Z. Naturforsch. C 2017, 72, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Akbar, S.; Tauseef, I.; Subhan, F.; Sultana, N.; Khan, I.; Ahmed, U.; Haleem, K.S. An overview of the plant-mediated synthesis of zinc oxide nanoparticles and their antimicrobial potential. Inorg. Nano Metal Chem. 2020, 50, 257–271. [Google Scholar] [CrossRef]
- Singh, T.A.; Das, J.; Sil, P.C. Zinc oxide nanoparticles: A comprehensive review on its synthesis, anticancer and drug delivery applications as well as health risks. Adv. Coll. Inter. Sci. 2020, 286, 102317. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Mitra, M.D.; Hussain, A.; Kaul, G. Exploration of immunomodulatory and protective effect of Withania somnifera on trace metal oxide (zinc oxide nanoparticles) induced toxicity in Balb/c mice. Mol. Biol. Rep. 2019, 46, 2447–2459. [Google Scholar] [CrossRef]
- Brahmkshatriya, P.P.; Brahmkshatriya, P.S. Terpenes: Chemistry, biological role, and therapeutic applications. Nat. Prod. 2013, 12, 2665–2691. [Google Scholar]
- Vuong, Q.V.; Golding, J.B.; Nguyen, M.; Roach, P.D. Extraction and isolation of catechins from tea. J. Sep. Sci. 2010, 33, 3415–3428. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Huang, Y.; Zhu, S.; Abbes, N.; Jing, X.; Zhang, L. A review of the green synthesis of ZnO nanoparticles using plant extracts and their prospects for application in antibacterial textiles. J. Eng. Fibers Fabr. 2021, 16, 15589250211046242. [Google Scholar] [CrossRef]
- Liu, Y.J.; Li, J.F.; Chen, Z.L. Research progress of green synthesis of iron nanoparticle and its application on contaminants removal from water. Technol. Water Treat. 2019, 45, 6–11. [Google Scholar]
- Agarwal, H.; Kumar, S.V.; Rajeshkumar, S. A review on green synthesis of zinc oxide nanoparticles—An eco-friendly approach. Resour. Eff. Technol. 2017, 3, 406–413. [Google Scholar] [CrossRef]
- Matinise, N.; Kaviyarasu, K.; Mongwaketsi, N.; Khamlich, S.; Kotsedi, L.; Mayedwa, N.; Maaza, M. Green synthesis of novel zinc iron oxide (ZnFe2O4) nanocomposite via Moringa Oleifera natural extract for electrochemical applications. Appl. Surf. Sci. 2018, 446, 66–73. [Google Scholar] [CrossRef]
- Osuntokun, J.; Onwudiwe, D.C.; Ebenso, E.E. Green synthesis of ZnO nanoparticles using aqueous Brassica oleracea L. var. italica and the photocatalytic activity. Green Chem. Lett. Rev. 2019, 12, 444–457. [Google Scholar] [CrossRef] [Green Version]
- Karnan, T.; Selvakumar, S.A.S. Biosynthesis of ZnO nanoparticles using rambutan (Nephelium lappaceum L.) peel extract and their photocatalytic activity on methyl orange dye. J. Mol. Struct. 2016, 1125, 358–365. [Google Scholar] [CrossRef]
- Mayedwa, N.; Mongwaketsi, N.; Khamlich, S.; Kaviyarasu, K.; Matinise, N.; Maaza, M. Green synthesis of nickel oxide, palladium and palladium oxide synthesized via Aspalathus linearis natural extracts: Physical properties & mechanism of formation. Appl. Surf. Sci. 2018, 446, 266–272. [Google Scholar]
- Sahariah, P.; Gaware, V.S.; Lieder, R.; Jonsdottir, S.; Hjalmarsdottir, M.A.; Sigurjonsson, O.E.; Masson, M. The effect of substituent, degree of acetylation and positioning of the cationic charge on the antibacterial activity of quaternary chitosan derivatives. Mar. Drugs. 2014, 12, 4635–4658. [Google Scholar] [CrossRef]
- Khan, M.; Wei, C.; Chen, M.; Tao, J.; Huang, N.; Qi, Z.; Li, L. CTAB-mediated synthesis and characterization of ZnO/Ag core–shell nanocomposites. J. Alloys Compd. 2014, 6, 306–314. [Google Scholar] [CrossRef]
- Decremps, F.; Pellicer-Porres, J.; Saitta, A.M.; Chervin, J.C.; Polian, A. High-pressure Raman spectroscopy study of wurtzite ZnO. Phys. Rev. B 2002, 65, 092101. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, J.; Murmu, P.P.; Manikandan, E.; Lee, S.Y. Investigation of structural and photoluminescence properties of gas and metal ions doped zinc oxide single crystals. J. Alloys Compd. 2014, 616, 614–617. [Google Scholar] [CrossRef]
- Ghimbeu, C.M.; Schoonman, J.; Lumbreras, M.; Siadat, M. Electrostatic spray deposited zinc oxide films for gas sensor applications. Appl. Surf. Sci. 2007, 253, 7483–7489. [Google Scholar] [CrossRef]
- Arefi, M.R.; Rezaei-Zarchi, S. Synthesis of zinc oxide nanoparticles and their effect on the compressive strength and setting time of self-compacted concrete paste as cementitious composites. Int. J. Mol. Sci. 2012, 13, 4340–4350. [Google Scholar] [CrossRef]
- Sundrarajan, M.; Ambika, S.; Bharathi, K. Plant-extract mediated synthesis of ZnO nanoparticles using Pongamia pinnata and their activity against pathogenic bacteria. Adv. Powder Technol. 2015, 26, 1294–1299. [Google Scholar] [CrossRef]
- Vidya, C.; Hiremath, S.; Chandraprabha, M.N.; Antonyraj, M.L.; Gopal, I.V.; Jain, A.; Bansal, K. Green synthesis of ZnO nanoparticles by Calotropis gigantea. Int. J. Curr. Eng. Technol. 2013, 1, 118–120. [Google Scholar]
- Kumar, S.S.; Venkateswarlu, P.; Rao, V.R.; Rao, G.N. Synthesis, characterization and optical properties of zinc oxide nanoparticles. Int. Nano Lett. 2013, 3, 30. [Google Scholar] [CrossRef] [Green Version]
- Oostenbrink, C.; van Gunsteren, W.F. Free energies of ligand binding for structurally diverse compounds. Proc. Natl. Acad. Sci. USA 2005, 102, 6750–6754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malicka, J.; Gryczynski, I.; Fang, J.; Kusba, J.; Lakowicz, J.R. Increased resonance energy transfer between fluorophores bound to DNA in proximity to metallic silver particles. Anal. Biochem. 2003, 315, 160–169. [Google Scholar] [CrossRef] [Green Version]
- Guo, B.L.; Han, P.; Guo, L.C.; Cao, Y.Q.; Li, A.D.; Kong, J.Z.; Zhai, H.F.; Wu, D. The antibacterial activity of Ta-doped ZnO nanoparticles. Nanoscale Res. Lett. 2015, 10, 336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, G.; Korenkov, V.; Judy, J.D.; Bertsch, P.N. Nanoparticles composed of Zn and ZnO inhibit Peronospora tabacina spore germination in vitro and P. tabacina infectivity on tobacco leaves. Nanomaterials 2016, 6, 50. [Google Scholar] [CrossRef] [Green Version]
- Krishnamoorthy, V.; Hiller, D.B.; Ripper, R.; Lin, B.; Vogel, S.M.; Feinstein, D.L.; Oswald, S.; Rothschild, L.; Hensel, P.; Rubinstein, I.; et al. Epinephrine induces rapid deterioration in pulmonary oxygen exchange in intact, anesthetized RatsA flow and pulmonary capillary pressure-dependent phenomenon. Anesthesiology 2012, 117, 745–754. [Google Scholar] [CrossRef] [Green Version]
- Stoimenov, P.K.; Klinger, R.L.; Marchin, G.L.; Klabunde, K.J. Metal oxide nanoparticles as bactericidal agents. Langmuir 2002, 18, 6679–6686. [Google Scholar] [CrossRef]
- Zhang, L.; Ding, Y.; Povey, M.; York, D. ZnO nanofluids—A potential antibacterial agent. Prog. Nat. Sci. 2008, 18, 939–944. [Google Scholar] [CrossRef]
- Slavin, Y.N.; Asnis, J.; Hafeli, U.O.; Bach, H. Metal nanoparticles: Understanding the mechanisms behind antibacterial activity. J. Nanobiotechnol. 2017, 15, 65. [Google Scholar] [CrossRef]
- Clemens, M.R.; Waller, H.D. Lipid peroxidation in erythrocytes. Chem. Phys. Lipids. 1987, 45, 251–268. [Google Scholar] [CrossRef]
- Niki, E. Interaction of ascorbate and alpha-tocopherol. Ann. N. Y. Acad. Sci. 1987, 498, 186–199. [Google Scholar] [CrossRef]
- Iwatsuki, M.; Komuro, E.; Niki, E. Antioxidant activities of aminophenols against oxidation of methyl linoleate in solution. Bull. Chem. Soc. Jpn. 1995, 68, 620–624. [Google Scholar] [CrossRef]
- Murugesan, B.; Sonamuthu, J.; Samayanan, S.; Arumugam, S.; Mahalingam, S. Highly biological active antibiofilm, anticancer and osteoblast adhesion efficacy from MWCNT/PPy/Pd nanocomposite. Appl. Surf. Sci. 2018, 434, 400–411. [Google Scholar] [CrossRef]
- Gowdhami, B.; Jaabir, M.; Archunan, G.; Suganthy, N. Anticancer potential of zinc oxide nanoparticles against cervical carcinoma cells synthesized via biogenic route using aqueous extract of Gracilaria edulis. Mater. Sci. Eng. C 2019, 103, 109840. [Google Scholar]
- Majeed, S.; Danish, M.; Ismail, M.H.; Ansari, M.T.; Ibrahim, M.N. Anticancer and apoptotic activity of biologically synthesized zinc oxide nanoparticles against human colon cancer HCT-116 cell line-in vitro study. Sustain. Chem. Pharm. 2019, 14, 100179. [Google Scholar] [CrossRef]



| Samples | Concentration (µg mL−1) | Zone of Inhibition (ZOI) | |||
|---|---|---|---|---|---|
| S. aureus | E. faecalis | E. coli | P. aeruginosa | ||
| P. curiflorus extract | 120 | 18.7 ± 0.5 | 9.2 ± 0.8 | 15.0 ± 1.2 | 20.6 ± 0.5 |
| Catechin | 120 | 14.2 ± 0.6 | 11.3 ± 0.5 | 14.2 ± 1.1 | 16.9 ± 0.5 |
| Curviflorside | 120 | 16.3 ± 0.5 | 10.2 ± 0.8 | 13.5 ± 0.4 | 18.6 ± 0.7 |
| Curviflorin | 120 | 19.2 ± 0.5 | 9.8 ± 1.2 | 15.4 ± 0.8 | 21.2 ± 0.9 |
| ZnONPs | 120 | 28.2 ± 1.1 | 15.1 ± 0.8 | 17.2 ± 0.52 | 30.1 ± 0.48 |
| Positive control (Ciprofloxacin) | 120 | 32.6 ± 2.1 | 17.9 ± 1.5 | 21.3 ± 1.2 | 33.4 ± 1.4 |
| Negative control (Dist. Water) | - | 0.00 | 0.00 | 0.00 | 0.00 |
| Sample | S. aureus | E. faecalis | E. coli | P. aeruginosa | ||||
|---|---|---|---|---|---|---|---|---|
| MIC (μg/mL) | MLC (μg/mL) | MIC (μg/mL) | MLC (μg/mL) | MIC (μg/mL) | MLC (μg/mL) | MIC (μg/mL) | MLC (μg/mL) | |
| Plant extract | 512 | 1024 | 1024 | 2048 | 1024 | 2048 | 510 | 1024 |
| Catechin | 2048 | 4096 | 1024 | ≥8192 | 1024 | 2048 | 1024 | 2048 |
| Curvilforside | 2048 | 4096 | 2048 | ≥8192 | 2048 | 4096 | 1024 | 2048 |
| Curviflorin | 512 | 1024 | 1024 | 4096 | 2048 | 2048 | 510 | 2048 |
| ZnONPs | 250 | 625 | 823 | 1525 | 815 | 1510 | 230 | 560 |
| Gentamicin | 8 | 10 | 8 | 10 | 5 | 8 | 5 | 5 |
| Erythromycin | 1 | 60 | 130 | 540 | 78 | 500 | 1 | 30 |
| Negative control (Dist. Water) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Sample | Hemolytic Activity HC50 (μg/mL) | Selectivity (HC50/MIC) | |||
|---|---|---|---|---|---|
| S. aureus | E. faecalis | E. coli | P. aeruginosa | ||
| Plant extract | 52 | 0.8 | 0.6 | 0.3 | 0.1 |
| Catechin | 1500 | 0.7 | 1.5 | 1.5 | 1.5 |
| Curviflorside | ≥8192 | ≥4 | ≥4 | ≥4 | ≥8 |
| Curviflorin | 1500 | 3 | 1.5 | 0.7 | 3 |
| ZnONPs | 28 | 0.1 | 0.02 | 0.02 | 0.05 |
| Concentration of ZnONPs (µg/mL) | Absorbance (nm) | % Inhibition of HeLa Cell Line |
|---|---|---|
| 160 | 0.250 | 64.06 |
| 80 | 0.305 | 54.35 |
| 40 | 0.382 | 41.85 |
| 20 | 0.430 | 32.82 |
| 10 | 0.485 | 22.06 |
| 5 | 0.492 | 12.02 |
| Control | 160 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amina, M.; Al Musayeib, N.M.; Alarfaj, N.A.; El-Tohamy, M.F.; Al-Hamoud, G.A.; Alqenaei, M.K.M. The Fluorescence Detection of Phenolic Compounds in Plicosepalus curviflorus Extract Using Biosynthesized ZnO Nanoparticles and Their Biomedical Potential. Plants 2022, 11, 361. https://doi.org/10.3390/plants11030361
Amina M, Al Musayeib NM, Alarfaj NA, El-Tohamy MF, Al-Hamoud GA, Alqenaei MKM. The Fluorescence Detection of Phenolic Compounds in Plicosepalus curviflorus Extract Using Biosynthesized ZnO Nanoparticles and Their Biomedical Potential. Plants. 2022; 11(3):361. https://doi.org/10.3390/plants11030361
Chicago/Turabian StyleAmina, Musarat, Nawal M. Al Musayeib, Nawal A. Alarfaj, Maha F. El-Tohamy, Gadah A. Al-Hamoud, and Muneerah K. M. Alqenaei. 2022. "The Fluorescence Detection of Phenolic Compounds in Plicosepalus curviflorus Extract Using Biosynthesized ZnO Nanoparticles and Their Biomedical Potential" Plants 11, no. 3: 361. https://doi.org/10.3390/plants11030361
APA StyleAmina, M., Al Musayeib, N. M., Alarfaj, N. A., El-Tohamy, M. F., Al-Hamoud, G. A., & Alqenaei, M. K. M. (2022). The Fluorescence Detection of Phenolic Compounds in Plicosepalus curviflorus Extract Using Biosynthesized ZnO Nanoparticles and Their Biomedical Potential. Plants, 11(3), 361. https://doi.org/10.3390/plants11030361






