A Survey of Enhanced Cold Tolerance and Low-Temperature-Induced Anthocyanin Accumulation in a Novel Zoysia japonica Biotype
Abstract
:1. Introduction
2. Results
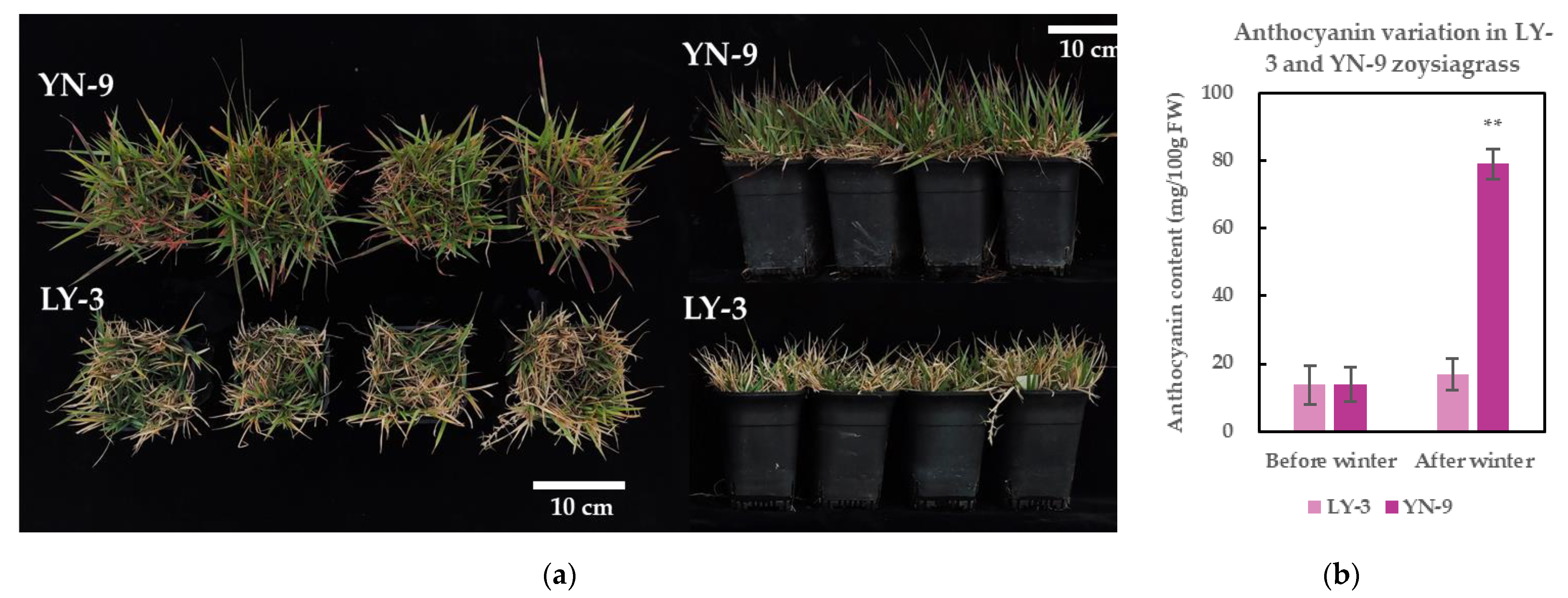
2.1. YN-9 Is a Zoysia japonica Biotype
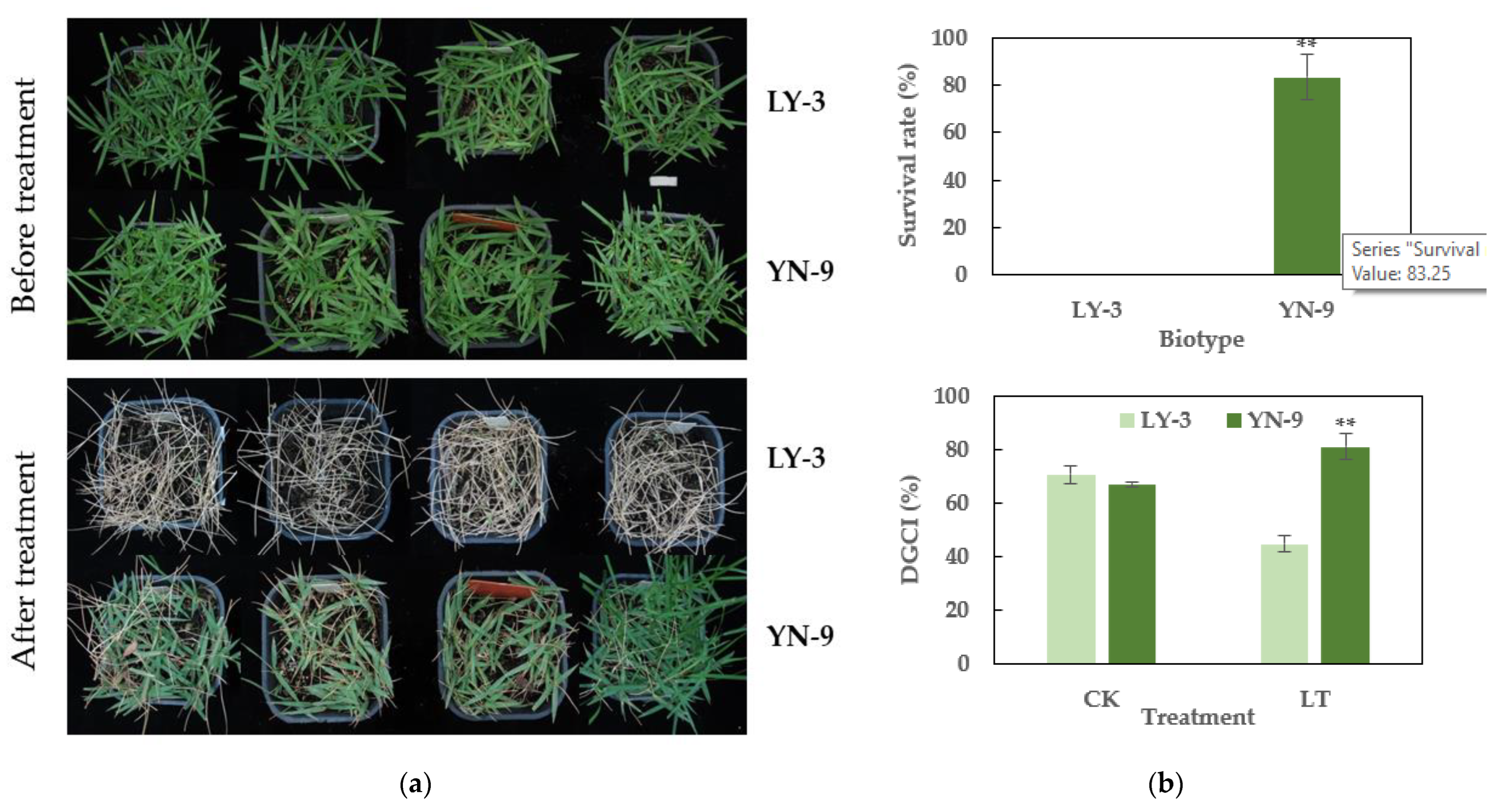
2.2. Enhanced Anthocyanin Accumulation and Cold Tolerance in YN-9
2.3. Read Trimming and Mapping
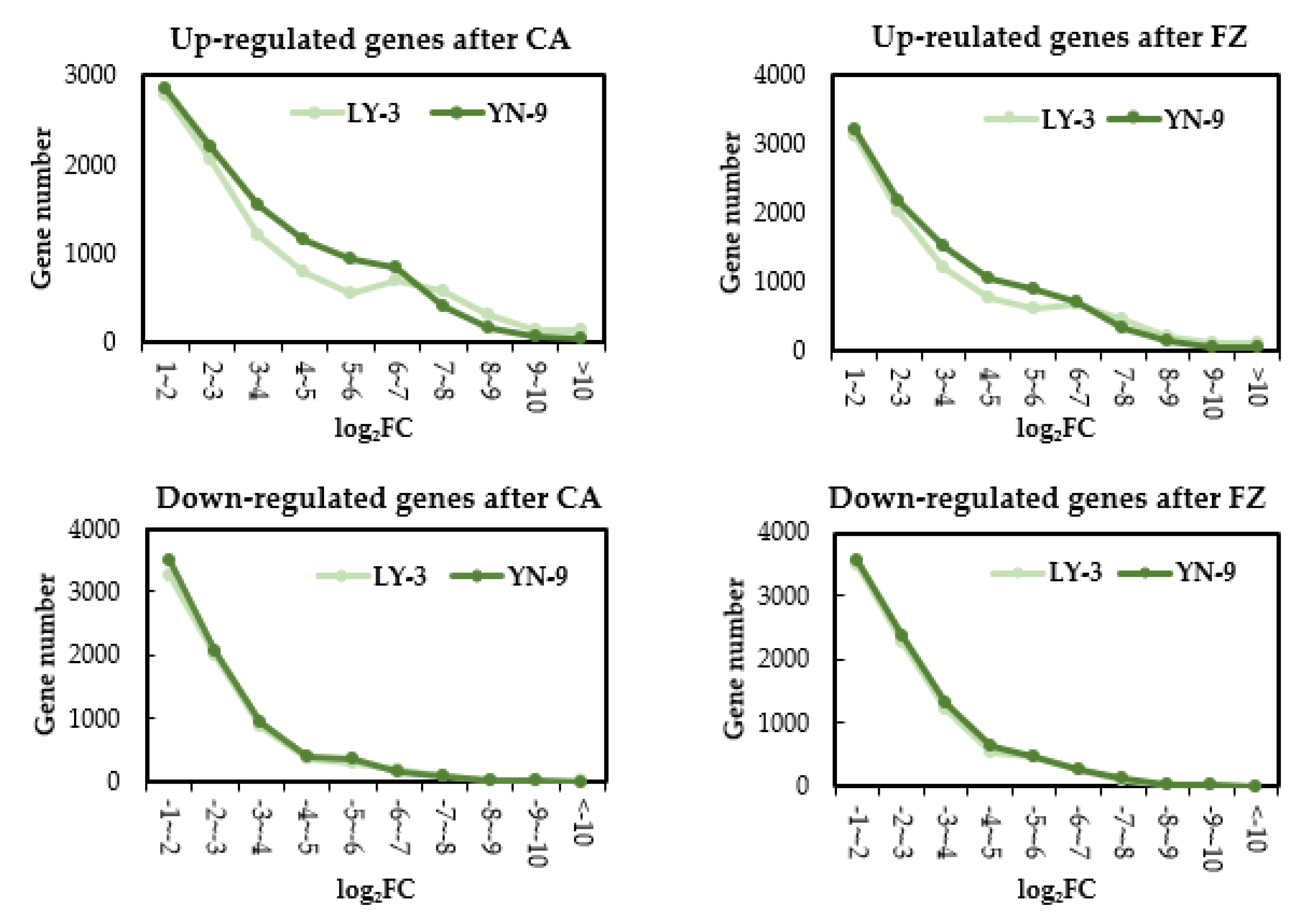
2.4. DEGs after Cold Treatment
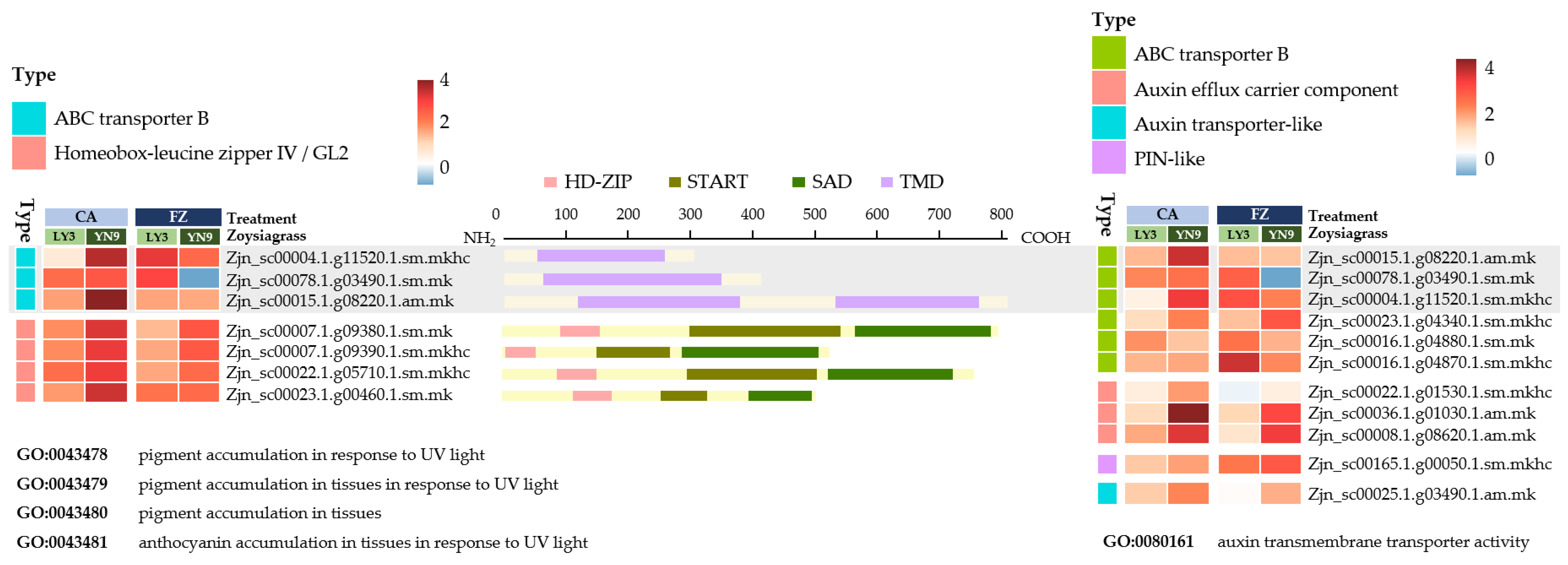
2.5. Genes Enriched in Pigmentation GOs
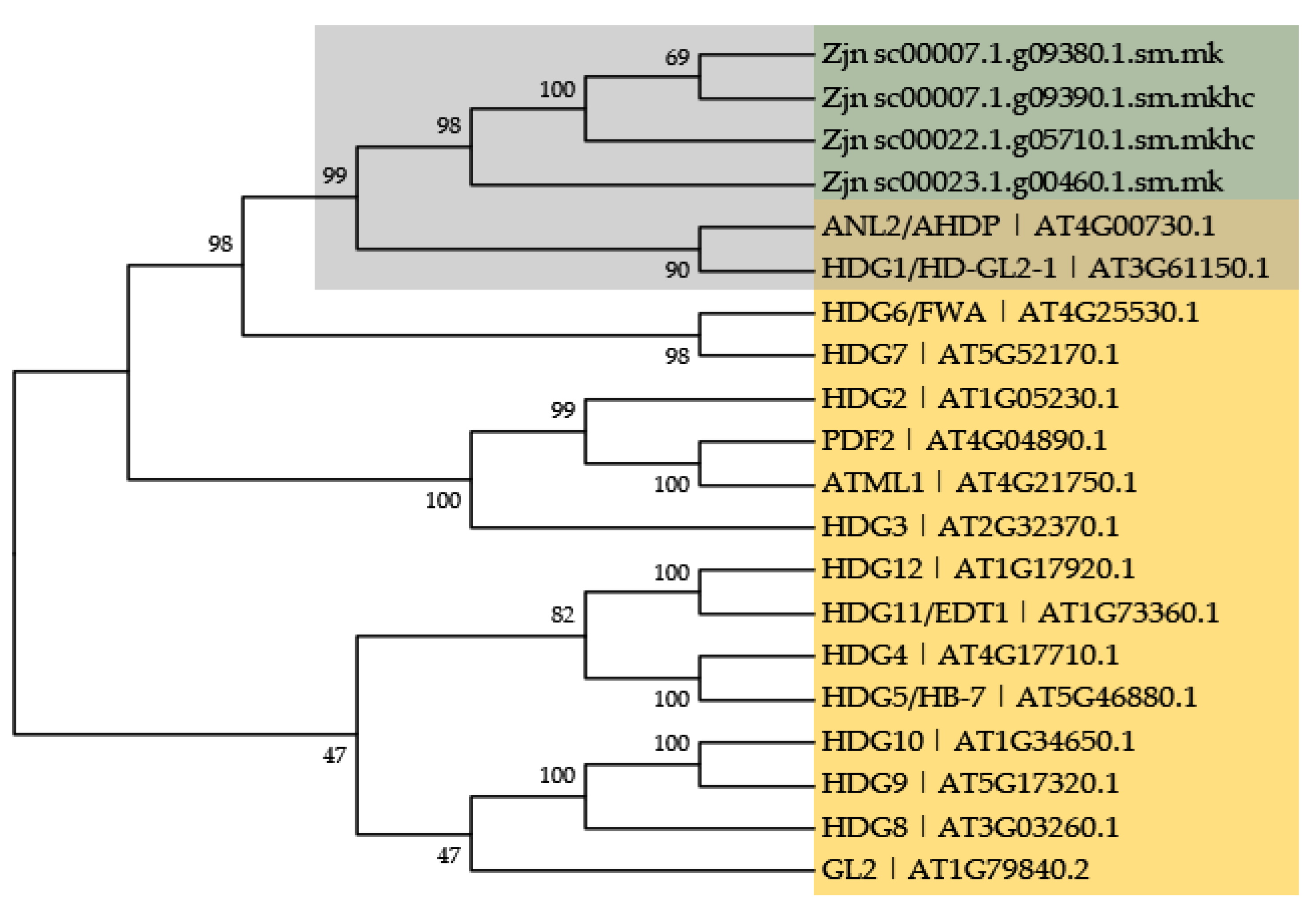
2.5.1. HD-Zop IV
2.5.2. ABCB Transmembrane Transporter
2.6. Expression Variations of Anthocyanin-Synthesis Genes
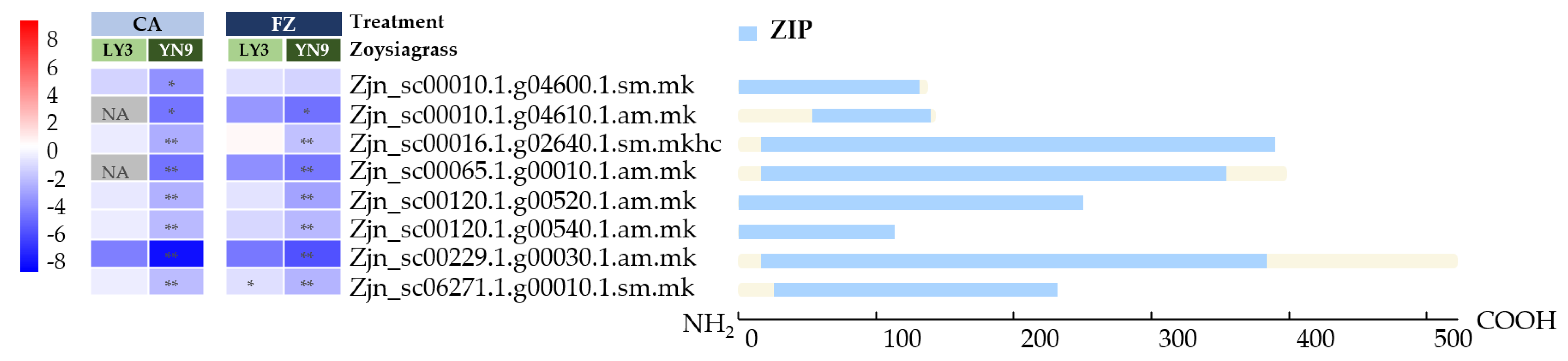
2.7. Down-Regulation of Zinc Transporters in YN-9
3. Discussion
3.1. Anthocyanin Accumulation and Cold-Tolerance in YN-9
3.2. Enhanced Expressions of Genes Associated with Anthocyanin Accumulation in YN-9
3.2.1. Anthocyanin-Biosynthesis Genes
3.2.2. HD-IV Transcription Factors
3.3. Enhanced Expressions of Anthocyanin Transport Genes in YN-9
3.4. Down-Regulation of Zinc Transporters Genes in YN-9
3.5. A Proposed Regulation Network of Anthocyanin Accumulation in YN-9
3.6. Future Outlook
4. Materials and Methods
4.1. Plant Materials
4.2. Anthocyanin Assay
4.3. Cold tolerance Assessment
4.4. RNA Extraction, cDNA Library Construction and Illumina Sequencing
4.5. RNA Seq Analysis
4.6. Anthocyanin-Synthesis Gene Profiling
4.7. Phylogenetic Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anderson, S.J. Taxonomy of Zoysia (Poaceae) Morphological and Molecular Variation; Texas A&M University: Dallas, TX, USA, 2000. [Google Scholar]
- Shouliang, C.; Phillips, S.M. Zoysia Willdenow, Ges. Naturf. Freunde Berlin Neue Schriften 3: 440. 1801, nom. cons. In Flora of China; Zhengyi, W., Raven, P.H., Eds.; Science Press and Missouri Botanical Garden Pre: Beijing, China; St. Louis, MO, USA, 2013; pp. 496–498. [Google Scholar]
- Braun, R.C.; Milla-Lewis, S.R.; Carbajal, E.M.; Schwartz, B.M.; Patton, A.J. Performance and playability of experimental low-input coarse-textured zoysiagrass in multiple climates. Grass Res. 2021, 1, 10. [Google Scholar] [CrossRef]
- Meeks, M.; Kong, S.T.; Genovesi, A.D.; Smith, B.; Chandra, A. Low-input golf course putting green performance of fine-textured inter- and intra-specific zoysiagrass (Zoysia spp.). Int. Turfgrass Soc. Res. J. 2021, 1–12. [Google Scholar] [CrossRef]
- Patton, A.J.; Schwartz, B.M.; Kenworthy, K.E. Zoysiagrass (Zoysia spp.) history, utilization, and improvement in the United States: A review. Crop Sci. 2017, 57, S-37–S-72. [Google Scholar] [CrossRef] [Green Version]
- Chandra, A.; Milla-Lewis, S.; Yu, Q. An overview of molecular advances in zoysiagrass. Crop Sci. 2017, 57, S-73. [Google Scholar] [CrossRef]
- Long, S.; Yan, F.; Yang, L.; Sun, Z.; Wei, S. Responses of Manila grass (Zoysia matrella) to chilling stress: From transcriptomics to physiology. PLoS ONE 2020, 15, e0235972. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Zhou, R.; Bi, B.; Song, L.; Chai, M.; Wang, Q.; Song, G. Analysis of a radiation-induced dwarf mutant of a warm-season turf grass reveals potential mechanisms involved in the dwarfing mutant. Sci. Rep. 2020, 10, 18913. [Google Scholar] [CrossRef]
- Wang, R.; Wang, X.; Liu, K.; Zhang, X.J.; Zhang, L.Y.; Fan, S.J. Comparative transcriptome analysis of halophyte Zoysia macrostachya in response to salinity stress. Plants 2020, 9, 458. [Google Scholar] [CrossRef] [Green Version]
- Ahn, J.H.; Kim, J.-S.; Kim, S.; Soh, H.Y.; Shin, H.; Jang, H.; Ryu, J.H.; Kim, A.; Yun, K.-Y.; Kim, S.; et al. De novo transcriptome analysis to identify anthocyanin biosynthesis genes responsible for tissue-specific pigmentation in zoysiagrass (Zoysia japonica Steud.). PLoS ONE 2015, 10, e0124497. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Ai, L.; Wang, L.; Yin, P.; Liu, C.; Li, S.; Zeng, H. De novo transcriptome analysis of Rhizoctonia solani AG1 IA strain early invasion in Zoysia japonica root. Front. Microbiol. 2016, 7, 708. [Google Scholar] [CrossRef]
- Li, L.; He, X.; Zhao, F.; Zhu, C.; Zeng, H. WUS and PIN1-related genes undergo dynamic expressional change during organ regeneration in response to wounding in Zoysia japonica. Mol. Biol. Rep. 2018, 45, 1733–1744. [Google Scholar] [CrossRef]
- Wang, J.; An, C.; Guo, H.; Yang, X.; Chen, J.; Zong, J.; Li, J.; Liu, J. Physiological and transcriptomic analyses reveal the mechanisms underlying the salt tolerance of Zoysia japonica Steud. BMC Plant Biol. 2020, 20, 114. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Niu, J.; Xu, X.; Xu, L.; Zhang, Y.; Fan, B.; Liang, X.; Zhang, L.; Yin, S.; Han, L. De novo assembly of the Japanese lawngrass (Zoysia japonica Steud.) root transcriptome and identification of candidate unigenes related to early responses under salt stress. Front. Plant Sci. 2015, 6, 610. [Google Scholar] [CrossRef] [Green Version]
- Wei, S.; Du, Z.; Gao, F.; Ke, X.; Li, J.; Liu, J.; Zhou, Y. Global transcriptome profiles of “Meyer” zoysiagrass in response to cold stress. PLoS ONE 2015, 10, e0131153. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.; Zhu, J. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Agarwal, P.K.; Agarwal, P.; Sopory, M.K.R.S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006, 25, 1263–1274. [Google Scholar] [CrossRef] [PubMed]
- Lan, S.Y.S.; Ming, F.D. Effects of Calcium and calmodulin antagonists on chilling stress-induced proline accumulation in Jatropha curcas L. J. Plant Growth Regul. 2016, 35, 815–826. [Google Scholar] [CrossRef]
- Naing, A.H.; Ai, T.N.; Lim, K.B.; Lee, I.J.; Kim, C.K.; Allen, R.D. Overexpression of rosea1 from snapdragon enhances anthocyanin accumulation and abiotic stress tolerance in transgenic tobacco. Front. Plant Sci. 2018, 9, 1070. [Google Scholar] [CrossRef] [PubMed]
- Christie, P.J.; Alfenito, M.R.; Walbot, V. Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: Enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta 1994, 194, 541–549. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhai, J.; Shao, L.; Lin, W.; Peng, C. Accumulation of anthocyanins: An adaptation strategy of Mikania micrantha to low temperature in winter. Front. Plant Sci. 2019, 10, 1049. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, N.U.; Park, J.I.; Jung, H.J.; Hur, Y.; Nou, I.S. Anthocyanin biosynthesis for cold and freezing stress tolerance and desirable color in Brassica rapa. Funct. Integr. Genom. 2015, 15, 383–394. [Google Scholar] [CrossRef]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: A review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- Naing, A.H.; Kim, C.K. Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant. 2021, 172, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.F.; Gyu, H.; Quan, K.; Hong, C.; Young, M.; Hyeon, P.; Sun, J.; Kim, J. A novel basic helix-loop-helix transcription factor, ZjICE2 from Zoysia japonica confers abiotic stress tolerance to transgenic plants via activating the DREB/CBF regulon and enhancing ROS scavenging. Plant Mol. Biol. 2020, 102, 447–462. [Google Scholar] [CrossRef]
- Sun, X.; Li, X.; Zhu, J.; Huang, N.; Bian, X.; Li, H.; Wang, L.; Han, L. Polyamines and ethylene metabolism during cold acclimation in zoysiagrass (Zoysia japonica Steud.). Acta Physiol. Plant 2020, 42, 1–10. [Google Scholar] [CrossRef]
- Brown, J.M.; Dacosta, M.; Patton, A.J.; Livingston, D.P.; Bernstein, R.P.; Lu, J.; Dunne, J.C.; Arellano, C.; Milla-lewis, S.R. Differences in proteome response to cold acclimation in Zoysia japonica cultivars with different levels of freeze tolerance. Crop Sci. 2020, 60, 2744–2756. [Google Scholar] [CrossRef]
- Zuo, Z.; Sun, H.; Lee, H.; Kang, H.; Zjice, Z. Identification of bHLH genes through genome-wide association study and antisense expression of ZjbHLH076/ZjICE1 influence tolerance to low temperature and salinity in Zoysia japonica. Plant Sci. 2021, 313, 111088. [Google Scholar] [CrossRef] [PubMed]
- Okeyo, D.O.; Fry, J.D.; Bremer, D.; Rajashekar, C.B.; Kennelly, M.; Chandra, A.; Genovesi, D.A.; Engelke, M.C. Freezing tolerance and seasonal color of experimental zoysiagrasses. Crop Sci. 2011, 51, 2858–2863. [Google Scholar] [CrossRef]
- Hinton, J.D.; Livingston, D.P.; Miller, G.L.; Peacock, C.H.; Tuong, T. Freeze tolerance of nine zoysiagrass cultivars using natural cold acclimation and freeze chambers. HortScience 2012, 47, 112–115. [Google Scholar] [CrossRef] [Green Version]
- Zuo, Z.; Kang, H.; Park, M.; Jeong, H.; Sun, H. Plant Science Zoysia japonica MYC type transcription factor ZjICE1 regulates cold tolerance in transgenic Arabidopsis. Plant Sci. 2019, 289, 110254. [Google Scholar] [CrossRef]
- Ji, Y.; Dae, K.; Yang, H.; Young, M.; Hyeon, P.; Sun, J.; Soon, P.; Hong, S.; Kang, G.; Gai, A.I.; et al. Overexpression of Zoysia ZjCIGR1 gene confers cold stress resistance to zoysiagrass. Plant Biotechnol. Rep. 2020, 14, 21–31. [Google Scholar] [CrossRef]
- Xuan, J.; Song, Y.; Zhang, H.; Liu, J.; Guo, Z.; Hua, Y. Comparative proteomic analysis of the stolon cold stress response between the C4 perennial grass species Zoysia japonica and Zoysia matrella. PLoS ONE 2013, 8, e75705. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Z.M.; Zhang, Q.F.; Li, J.L. Enhanced chilling tolerance in Zoysia matrella by pre-treatment with salicylic acid, calcium chloride, hydrogen peroxide or 6-benzylaminopurine. Biol. Plant. 2009, 53, 179–182. [Google Scholar] [CrossRef]
- Brown, J.M.; Yu, X.; Patton, A.J.; Holloway, H.M.P.; Arellano, C.; Tuong, T.D.; Livingston, D.P.; Schwartz, B.M.; Milla-lewis, S.R. Identification of QTL associated with cold acclimation and freezing tolerance in Zoysia japonica. Crop Sci. 2021, 61, 3044–3055. [Google Scholar] [CrossRef]
- Zuo, Z.F.; Kang, H.G.; Park, M.Y.; Jeong, H.; Sun, H.J.; Yang, D.H.; Lee, Y.E.; Song, P.S.; Lee, H.Y. Overexpression of ICE1, a regulator of cold-induced transcriptome, confers cold tolerance to transgenic Zoysia japonica. J. Plant Biol. 2019, 62, 137–146. [Google Scholar] [CrossRef]
- Andolfo, G.; Ruocco, M.; Di Donato, A.; Frusciante, L.; Lorito, M.; Scala, F.; Ercolano, M.R. Genetic variability and evolutionary diversification of membrane ABC transporters in plants. BMC Plant Biol. 2015, 15, 51. [Google Scholar] [CrossRef] [Green Version]
- Dean, M.; Annilo, T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu. Rev. Genomics Hum. Genet. 2005, 6, 123–142. [Google Scholar] [CrossRef]
- Theodoulou, F.L. Plant ABC transporters. Biochim. Biophys. Acta—Biomembr. 2000, 1465, 79–103. [Google Scholar] [CrossRef] [Green Version]
- Lane, T.S.; Rempe, C.S.; Davitt, J.; Staton, M.E.; Peng, Y.; Soltis, D.E.; Melkonian, M.; Deyholos, M.; Leebens-Mack, J.H.; Chase, M.; et al. Diversity of ABC transporter genes across the plant kingdom and their potential utility in biotechnology. BMC Biotechnol. 2016, 16, 47. [Google Scholar] [CrossRef] [Green Version]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [Green Version]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Moustaka, J.; Tanou, G.; Giannakoula, A.; Adamakis, I.S.; Panteris, E.; Eleftheriou, E.P.; Moustakas, M. Anthocyanin accumulation in poinsettia leaves and its functional role in photo-oxidative stress. Environ. Exp. Bot. 2020, 175, 104065. [Google Scholar] [CrossRef]
- Li, X.; Ahammed, G.J.; Zhang, X.-N.; Zhang, L.; Yan, P.; Zhang, L.; Fu, J.; Han, W. Melatonin-mediated regulation of anthocyanin biosynthesis and antioxidant defense confer tolerance to arsenic stress in Camellia sinensis L. J. Hazard. Mater. 2021, 403, 123922. [Google Scholar] [CrossRef] [PubMed]
- Teng, H.; Mi, Y.; Cao, H.; Chen, L. Enzymatic acylation of raspberry anthocyanin: Evaluations on its stability and oxidative stress prevention. Food Chem. 2022, 372, 130766. [Google Scholar] [CrossRef] [PubMed]
- Hinojosa-Gómez, J.; Martín-Hernández, C.S.; Heredia, J.B.; León-Félix, J.; Osuna-Enciso, T.; Muy-Rangel, M.D. Anthocyanin induction by drought stress in the calyx of Roselle cultivars. Molecules 2020, 25, 1555. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Zhang, A.; Wu, X.; Zhu, Z.; Yang, Z.; Zhu, Y.; Zha, D. Transcriptome analysis revealed expression of genes related to anthocyanin biosynthesis in eggplant (Solanum melongena L.) under high-temperature stress. BMC Plant Biol. 2019, 19, 387. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Chen, B.; Lin, H.; Liu, Y.; Wei, Y.; Chen, F.; Li, W. Identification and characterization of the glutathione S-Transferase (GST) family in radish reveals a likely role in anthocyanin biosynthesis and heavy metal stress tolerance. Gene 2020, 743, 144484. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef]
- Kaplan, F.; Kopka, J.; Sung, D.Y.; Zhao, W.; Popp, M.; Porat, R.; Guy, C.L. Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J. 2007, 50, 967–981. [Google Scholar] [CrossRef]
- Crifò, T.; Puglisi, I.; Petrone, G.; Recupero, G.R.; Lo Piero, A.R. Expression analysis in response to low temperature stress in blood oranges: Implication of the flavonoid biosynthetic pathway. Gene 2011, 476, 1–9. [Google Scholar] [CrossRef]
- Becker, C.; Klaering, H.P.; Kroh, L.W.; Krumbein, A. Cool-cultivated red leaf lettuce accumulates cyanidin-3-O-(6″-O- malonyl)-glucoside and caffeoylmalic acid. Food Chem. 2014, 146, 404–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Teng, S.; Yin, H.; Zhang, S.; Tuo, X.; Tran, L.S.P. Transcriptome analysis reveals roles of anthocyanin-and jasmonic acid-biosynthetic pathways in rapeseed in response to high light stress. Int. J. Mol. Sci. 2021, 22, 13027. [Google Scholar] [CrossRef] [PubMed]
- Schulz, E.; Tohge, T.; Zuther, E.; Fernie, A.R.; Hincha, D.K. Natural variation in flavonol and anthocyanin metabolism during cold acclimation in Arabidopsis thaliana accessions. Plant Cell Environ. 2015, 38, 1658–1672. [Google Scholar] [CrossRef] [PubMed]
- Schulz, E.; Tohge, T.; Zuther, E.; Fernie, A.R.; Hincha, D.K. Flavonoids are determinants of freezing tolerance and cold acclimation in Arabidopsis thaliana. Sci. Rep. 2016, 6, 34027. [Google Scholar] [CrossRef] [PubMed]
- Quattrocchio, F.; Wing, J.F.; Leppen, H.T.C.; Mol, J.N.M.; Koes, R.E. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 1993, 5, 1497–1512. [Google Scholar] [CrossRef]
- Sun, C.; Deng, L.; Du, M.; Zhao, J.; Chen, Q.; Huang, T.; Jiang, H.; Li, C.-B.; Li, C. A transcriptional network promotes anthocyanin biosynthesis in tomato flesh. Mol. Plant 2020, 13, 42–58. [Google Scholar] [CrossRef] [PubMed]
- Karppinen, K.; Lafferty, D.J.; Albert, N.W.; Mikkola, N.; McGhie, T.; Allan, A.C.; Afzal, B.M.; Häggman, H.; Espley, R.V.; Jaakola, L. MYBA and MYBPA transcription factors co-regulate anthocyanin biosynthesis in blue-coloured berries. New Phytol. 2021, 232, 1350–1367. [Google Scholar] [CrossRef]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Li, Y.; Liu, Z.; Zhou, P.; Zeng, L.; Zhang, X.; Zhang, J.; et al. StMYB44 negatively regulates anthocyanin biosynthesis at high temperatures in tuber flesh of potato. J. Exp. Bot. 2019, 70, 3809–3824. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Z.; Wang, H.; Li, D.; Yu, B.; Hui, Q.; Yan, S.; Huang, Z.; Cui, X.; Cao, B. Identification of candidate HY5-dependent and -independent regulators of anthocyanin biosynthesis in tomato. Plant Cell Physiol. 2019, 60, 643–656. [Google Scholar] [CrossRef]
- Chen, L.; Hu, B.; Qin, Y.; Hu, G.; Zhao, J. Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Biochem. 2019, 136, 178–187. [Google Scholar] [CrossRef]
- Xu, Z.S.; Yang, Q.Q.; Feng, K.; Yu, X.; Xiong, A.S. DcMYB113, a root-specific R2R3-MYB, conditions anthocyanin biosynthesis and modification in carrot. Plant Biotechnol. J. 2020, 18, 1585–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Wu, H.; Zhu, H.; Huang, C.; Liu, C.; Chang, Y.; Kong, Z.; Zhou, Z.; Wang, G.; Lin, Y.; et al. Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves. New Phytol. 2019, 223, 705–721. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Cavagnaro, P.F.; Bostan, H.; Zhao, Y.; Zhang, J.; Simon, P.W. A cluster of MYB transcription factors regulates anthocyanin biosynthesis in Carrot (Daucus carota L.) root and petiole. Front. Plant Sci. Front. Plant Sci. 2019, 9, 1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Tang, W.; Hu, Y.; Zhang, Y.; Sun, J.; Guo, X.; Lu, H.; Yang, Y.; Fang, C.; Niu, X.; et al. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 2019, 99, 359–378. [Google Scholar] [CrossRef]
- Huang, D.; Tang, Z.; Fu, J.; Yuan, Y.; Deng, X.; Xu, Q. CsMYB3 and CsRuby1 form an ‘activator-and-repressor’ loop for the regulation of anthocyanin biosynthesis in Citrus. Plant Cell Physiol. 2020, 61, 318–330. [Google Scholar] [CrossRef]
- Yang, L.; Pengbo, X.; Chen, G.; Wu, J.; Liu, Z.; Lian, H. FvbHLH9 functions as a positive regulator of anthocyanin biosynthesis by forming a HY5-bHLH9 transcription complex in strawberry fruits. Plant Cell Physiol. 2020, 61, 826–837. [Google Scholar] [CrossRef]
- Ni, J.; Bai, S.; Zhao, Y.; Qian, M.; Tao, R.; Yin, L.; Gao, L.; Teng, Y. Ethylene response factors Pp4ERF24 and Pp12ERF96 regulate blue light-induced anthocyanin biosynthesis in ‘Red Zaosu’ pear fruits by interacting with MYB114. Plant Mol. Biol. 2019, 99, 67–78. [Google Scholar] [CrossRef]
- Li, C.; Wu, J.; Hu, K.-D.; Wei, S.-W.; Sun, H.-Y.; Hu, L.-Y.; Han, Z.; Yao, G.-F.; Zhang, H. PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Hortic. Res. 2020, 7, 37. [Google Scholar] [CrossRef] [Green Version]
- Tang, B.; Li, L.; Hu, Z.; Chen, Y.; Tan, T.; Jia, Y.; Xie, Q.; Chen, G. Anthocyanin accumulation and transcriptional regulation of anthocyanin biosynthesis in Purple Pepper. J. Agric. Food Chem. 2020, 68, 12152–12163. [Google Scholar] [CrossRef]
- Han, M.; Yang, C.; Zhou, J.; Zhu, J.; Meng, J.; Shen, T.; Xin, Z.; Li, H. Analysis of flavonoids and anthocyanin biosynthesis-related genes expression reveals the mechanism of petal color fading of Malus hupehensis (Rosaceae). Braz. J. Bot. 2020, 43, 81–89. [Google Scholar] [CrossRef]
- Li, W.-F.; Ning, G.-X.; Mao, J.; Guo, Z.-G.; Zhou, Q.; Chen, B.-H. Whole-genome DNA methylation patterns and complex associations with gene expression associated with anthocyanin biosynthesis in apple fruit skin. Planta 2019, 250, 1833–1847. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, P.; Gu, M.; Hou, B.; Zhang, C.; Zheng, Y.; Sun, Y.; Jin, S.; Ye, N. Identification of PAL genes related to anthocyanin synthesis in tea plants and its correlation with anthocyanin content. Hortic. Plant J. 2021. [Google Scholar] [CrossRef]
- Li, J.; An, Y.; Wang, L. Transcriptomic analysis of Ficus carica peels with a focus on the key genes for anthocyanin biosynthesis. Int. J. Mol. Sci. 2020, 21, 1245. [Google Scholar] [CrossRef] [Green Version]
- Sicilia, A.; Catara, V.; Scialò, E.; Piero, A.R. Lo Fungal infection induces anthocyanin biosynthesis and changes in dna methylation configuration of blood orange [Citrus sinensis L. (osbeck)]. Plants 2021, 10, 244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sun, F.; Zhang, C.; Zhang, M.; Wang, W.; Zhang, C.; Xi, Y. Anthocyanin biosynthesis and a regulatory network of different-colored wheat grains revealed by multiomics analysis. J. Agric. Food Chem. 2022. [Google Scholar] [CrossRef]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [Green Version]
- Olsen, K.M.; Lea, U.S.; Slimestad, R.; Verheul, M.; Lillo, C. Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J. Plant Physiol. 2008, 165, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.W.; Li, C.; Jiang, Q.T.; Wei, Y.; Yao, H.; Chen, H.; Wu, Q. Cloning and characterization of a cold inducible Pal promoter from Fagopyrum tataricum. Cent. Eur. J. Biol. 2014, 9, 290–297. [Google Scholar] [CrossRef] [Green Version]
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef]
- Chen, S.; Wang, S. GLABRA2, a common regulator for epidermal cell fate determination and anthocyanin biosynthesis in Arabidopsis. Int. J. Mol. Sci. 2019, 20, 4997. [Google Scholar] [CrossRef] [Green Version]
- Rerie, W.G.; Feldmann, K.A.; Marks, M.D. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 1994, 8, 1388–1399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Cristina, M.; Sessa, G.; Dolan, L.; Linstead, P.; Baima, S.; Ruberti, I.; Morelli, G. The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 1996, 10, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Katsumata, H.; Abe, M.; Yabe, N.; Komeda, Y.; Yamamoto, K.T.; Takahashi, T. Characterization of the class IV homeodomain-Leucine zipper gene family in Arabidopsis. Plant Physiol. 2006, 141, 1363–1375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubo, H.; Peeters, A.J.M.; Aarts, M.G.M.; Pereira, A.; Koornneef, M. ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell 1999, 11, 1217–1226. [Google Scholar] [CrossRef] [Green Version]
- Kubo, H.; Hayashi, K. Characterization of root cells of anl2 mutant in Arabidopsis thaliana. Plant Sci. 2011, 180, 679–685. [Google Scholar] [CrossRef]
- Braidot, E.; Zancani, M.; Petrussa, E.; Peresson, C.; Bertolini, A.; Patui, S.; Macrì, F.; Vianello, A. Transport and accumulation of flavonoids in grapevine (Vitis vinifera L.). Plant Signal. Behav. 2008, 3, 626–632. [Google Scholar] [CrossRef] [Green Version]
- Kaur, S.; Sharma, N.; Kapoor, P.; Chunduri, V.; Pandey, A.K.; Garg, M. Spotlight on the overlapping routes and partners for anthocyanin transport in plants. Physiol. Plant 2021, 171, 868–881. [Google Scholar] [CrossRef]
- Mackon, E.; Jeazet Dongho Epse Mackon, G.C.; Ma, Y.; Haneef Kashif, M.; Ali, N.; Usman, B.; Liu, P. Recent insights into anthocyanin pigmentation, synthesis, trafficking, and regulatory mechanisms in Rice (Oryza sativa L.) Caryopsis. Biomolecules 2021, 11, 394. [Google Scholar] [CrossRef]
- Bannoud, F.; Carvajal, S.; Ellison, S.; Senalik, D.; Gomez Talquenca, S.; Iorizzo, M.; Simon, P.W.; Cavagnaro, P.F. Genetic and transcription profile analysis of tissue-specific anthocyanin pigmentation in carrot root phloem. Genes 2021, 12, 1464. [Google Scholar] [CrossRef]
- Jiang, T.; Zhang, M.; Wen, C.; Xie, X.; Tian, W.; Wen, S.; Lu, R.; Liu, L. Integrated metabolomic and transcriptomic analysis of the anthocyanin regulatory networks in Salvia miltiorrhiza Bge. flowers. BMC Plant Biol. 2020, 20, 349. [Google Scholar] [CrossRef]
- Huang, B.; Rong, H.; Ye, Y.; Ni, Z.; Xu, M.; Zhang, W.; Xu, L. An Transcriptomic analysis of flower color variation in the ornamental crabapple (Malus spp.) half-sib family through Illumina and PacBio Sequel sequencing. Plant Physiol. Biochem. 2020, 149, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Roberts, W.R.; Roalson, E.H. Comparative transcriptome analyses of flower development in four species of Achimenes (Gesneriaceae). BMC Genom. 2017, 18, 240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Pan, D.; Liang, M.; Abubakar, Y.S.; Li, J.; Lin, J.; Chen, S.; Chen, W. Regulation of anthocyanin biosynthesis in purple leaves of zijuan tea (Camellia sinensis var. kitamura). Int. J. Mol. Sci. 2017, 18, 833. [Google Scholar] [CrossRef] [Green Version]
- Broadley, M.R.; White, P.J.; Hammond, J.P.; Zelko, I.; Lux, A. Zinc in plants. New Phytol. 2007, 173, 677–702. [Google Scholar] [CrossRef]
- Luna-Vital, D.; Cortez, R.; Ongkowijoyo, P.; Gonzalez de Mejia, E. Protection of color and chemical degradation of anthocyanin from purple corn (Zea mays L.) by zinc ions and alginate through chemical interaction in a beverage model. Food Res. Int. 2018, 105, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, S.A.; Lee, J.; Guerinot, M.L.; An, G. Zinc deficiency-inducible OsZIP8 encodes a plasma membrane-localized zinc transporter in rice. Mol. Cells 2010, 29, 551–558. [Google Scholar] [CrossRef]
- Sasaki, A.; Yamaji, N.; Mitani-Ueno, N.; Kashino, M.; Ma, J.F. A node-localized transporter OsZIP3 is responsible for the preferential distribution of Zn to developing tissues in rice. Plant J. 2015, 84, 374–384. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.S.; Yang, G.M. Development of new hybrid cultivar ‘Senock’ in Zoysiagrass. Kor. Turfgrass Sci. 2004, 18, 201–209. [Google Scholar]
- Emmons, R.; Rossi, F. Turfgrass Science and Mnagement, 5th ed.; Cengage Learning: Stamford, CT, USA, 2015; pp. 40–41. [Google Scholar]
- Choi, J.-S.; Yang, G.-M. Development of new cultivar ’Millock’ in zoysiagrass. Asian J. Turfgrass Sci. 2006, 20, 1–10. [Google Scholar]
- Meng, X.; Yin, B.; Feng, H.L.; Zhang, S.; Liang, X.Q.; Meng, Q.W. Overexpression of R2R3-MYB gene leads to accumulation of anthocyanin and enhanced resistance to chilling and oxidative stress. Biol. Plant. 2014, 58, 121–130. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Tian, X.; Li, S.; Fu, Y.; Xu, J.; Wang, G. The AabHLH35 Transcription Factor Identified from Anthurium andraeanum is Involved in Cold and Drought Tolerance. Plants 2019, 8, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karcher, D.E.; Richardson, M.D. Batch analysis of digital images to evaluate turfgrass characteristics. Crop Sci. 2005, 45, 1536–1539. [Google Scholar] [CrossRef]
- Karcher, D.E.; Richardson, M.D. Quantifying turfgrass color using digital image analysis. Crop Sci. 2003, 43, 943–951. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.D.; Fernandez-Pozo, N.; Drake-Stowe, K.; Humphry, M.; Evans, A.D.; Bombarely, A.; Allen, F.; Hurst, R.; White, B.; Kernodle, S.P.; et al. A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics 2017, 18, 1–14. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omi. A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Ling, Q.L.; Feng, Y.X.; Lu, C.J.; Lin, Y.J.; Yu, X.Z. Genetic variation and gene expression of anthocyanin synthesis and transport related enzymes in Oryza sativa against thiocyanate. Plant Physiol. Biochem. 2021, 160, 18–26. [Google Scholar] [CrossRef]




| Genotype | 2nd Leaf | 3rd Leaf | ||
|---|---|---|---|---|
| Length (cm) | Width (mm) | Length (cm) | Width (mm) | |
| LY-3 | 10.19 ± 1.78 | 4.02 ± 0.55 | 11.93 ± 2.59 | 4.05 ± 0.58 |
| YN-9 | 12.47 ± 2.28 ** | 3.92 ± 0.41 | 14.75 ± 2.59 ** | 3.93 ± 0.48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, H.-X.; Jiang, M.; Yang, J.-F.; Wu, Z.-H.; Ma, L.-L.; Wang, C.-C.; Liang, C.; Ning, X.-Y.; Ge, L.-F.; Chen, S. A Survey of Enhanced Cold Tolerance and Low-Temperature-Induced Anthocyanin Accumulation in a Novel Zoysia japonica Biotype. Plants 2022, 11, 429. https://doi.org/10.3390/plants11030429
Jin H-X, Jiang M, Yang J-F, Wu Z-H, Ma L-L, Wang C-C, Liang C, Ning X-Y, Ge L-F, Chen S. A Survey of Enhanced Cold Tolerance and Low-Temperature-Induced Anthocyanin Accumulation in a Novel Zoysia japonica Biotype. Plants. 2022; 11(3):429. https://doi.org/10.3390/plants11030429
Chicago/Turabian StyleJin, Hai-Xiang, Ming Jiang, Jian-Feng Yang, Zhi-Hao Wu, Long-Long Ma, Cong-Cong Wang, Chen Liang, Xin-Yi Ning, Liang-Fa Ge, and Shu Chen. 2022. "A Survey of Enhanced Cold Tolerance and Low-Temperature-Induced Anthocyanin Accumulation in a Novel Zoysia japonica Biotype" Plants 11, no. 3: 429. https://doi.org/10.3390/plants11030429





