Anti-Adhesion and Antibiofilm Activity of Eruca sativa Miller Extract Targeting Cell Adhesion Proteins of Food-Borne Bacteria as a Potential Mechanism: Combined In Vitro-In Silico Approach
Abstract
:1. Introduction
2. Results
2.1. Phytochemical Screening
2.2. Antibacterial Activity of E. sativa
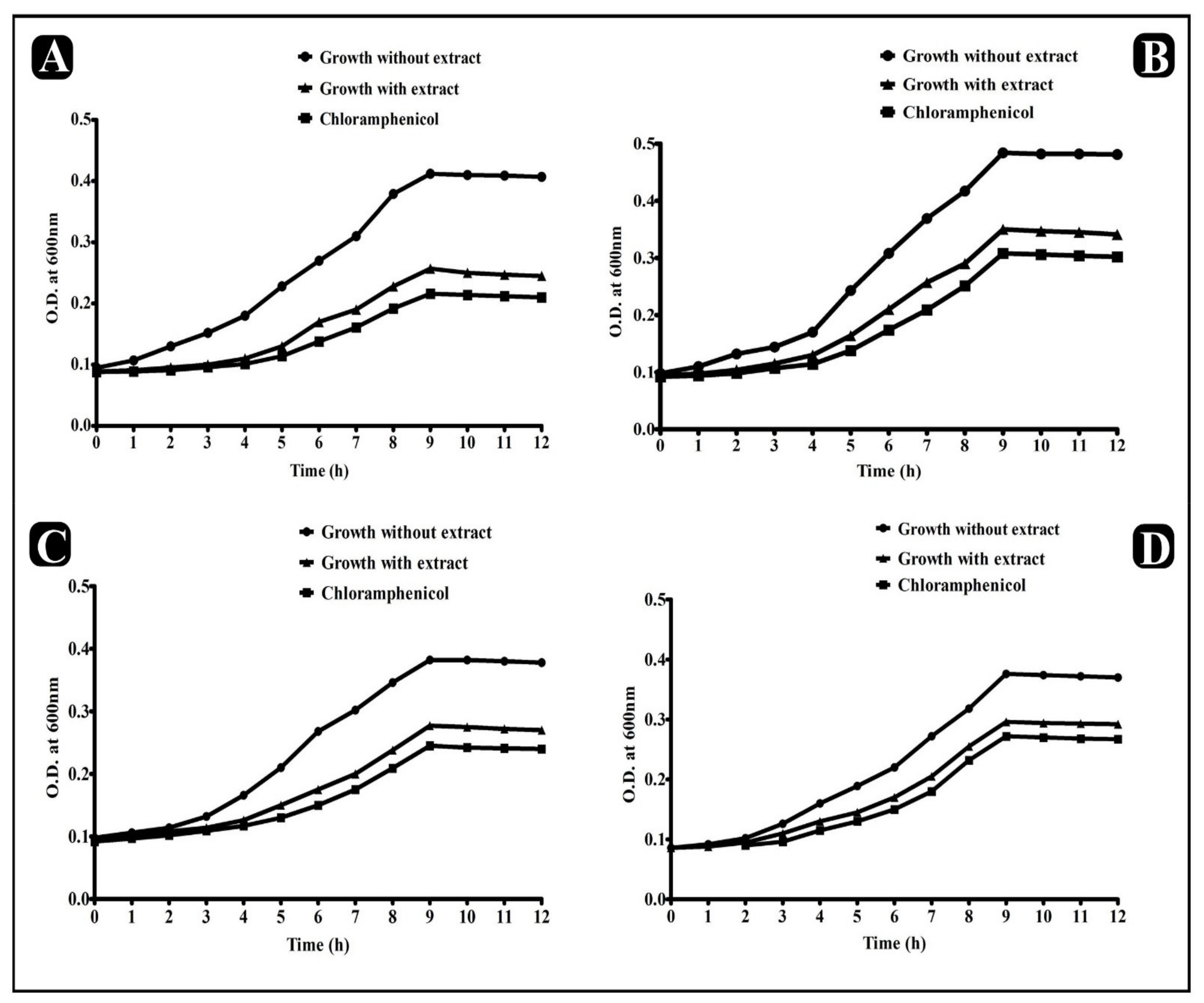
2.3. Determination of Growth Pattern of Food-Borne Bacteria
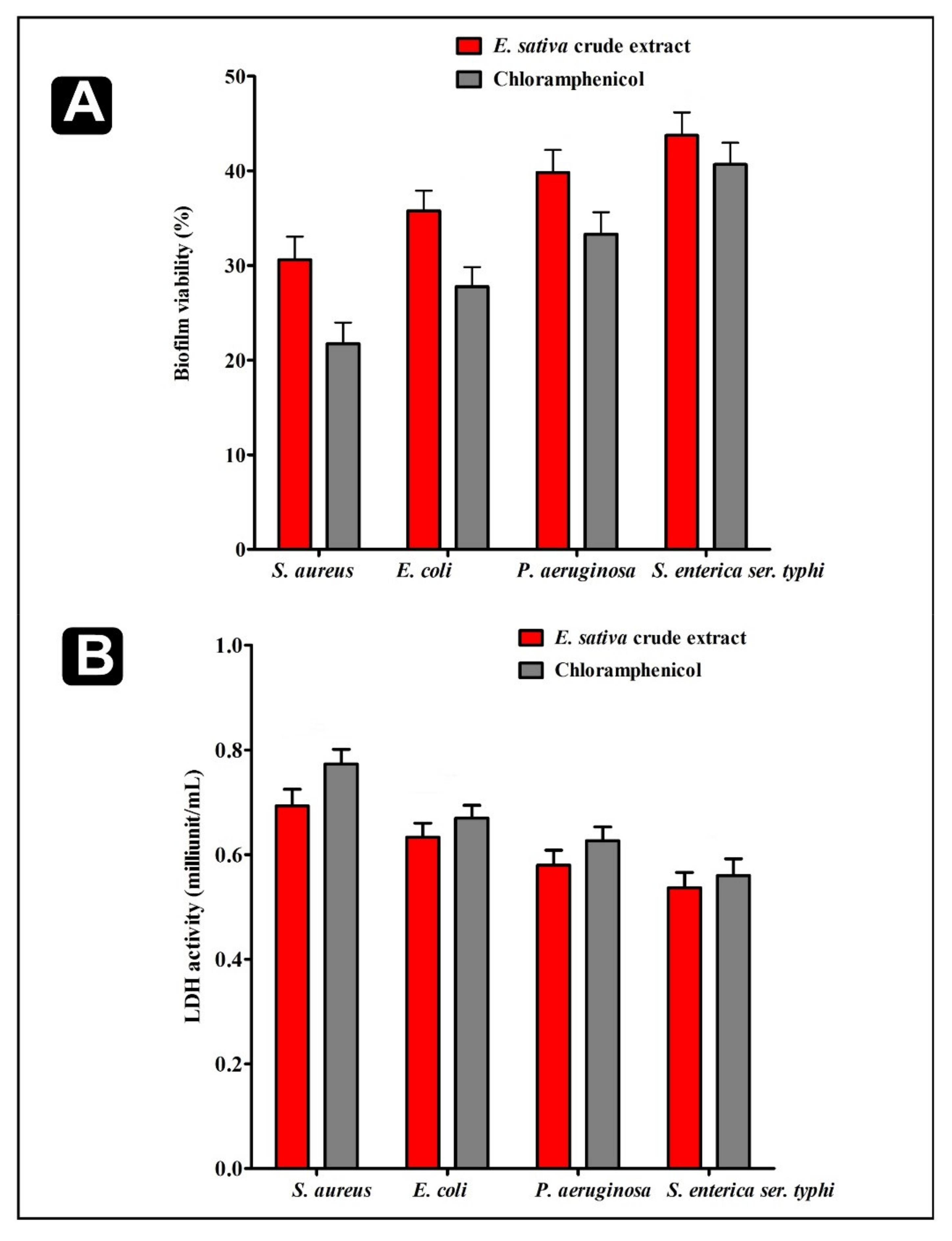
2.4. Effect of E. sativa Crude Extract on Established Biofilms and on Their Adhesion Properties
2.5. Effect of E. sativa Crude Extract on Bacterial Cells Inside Biofilms
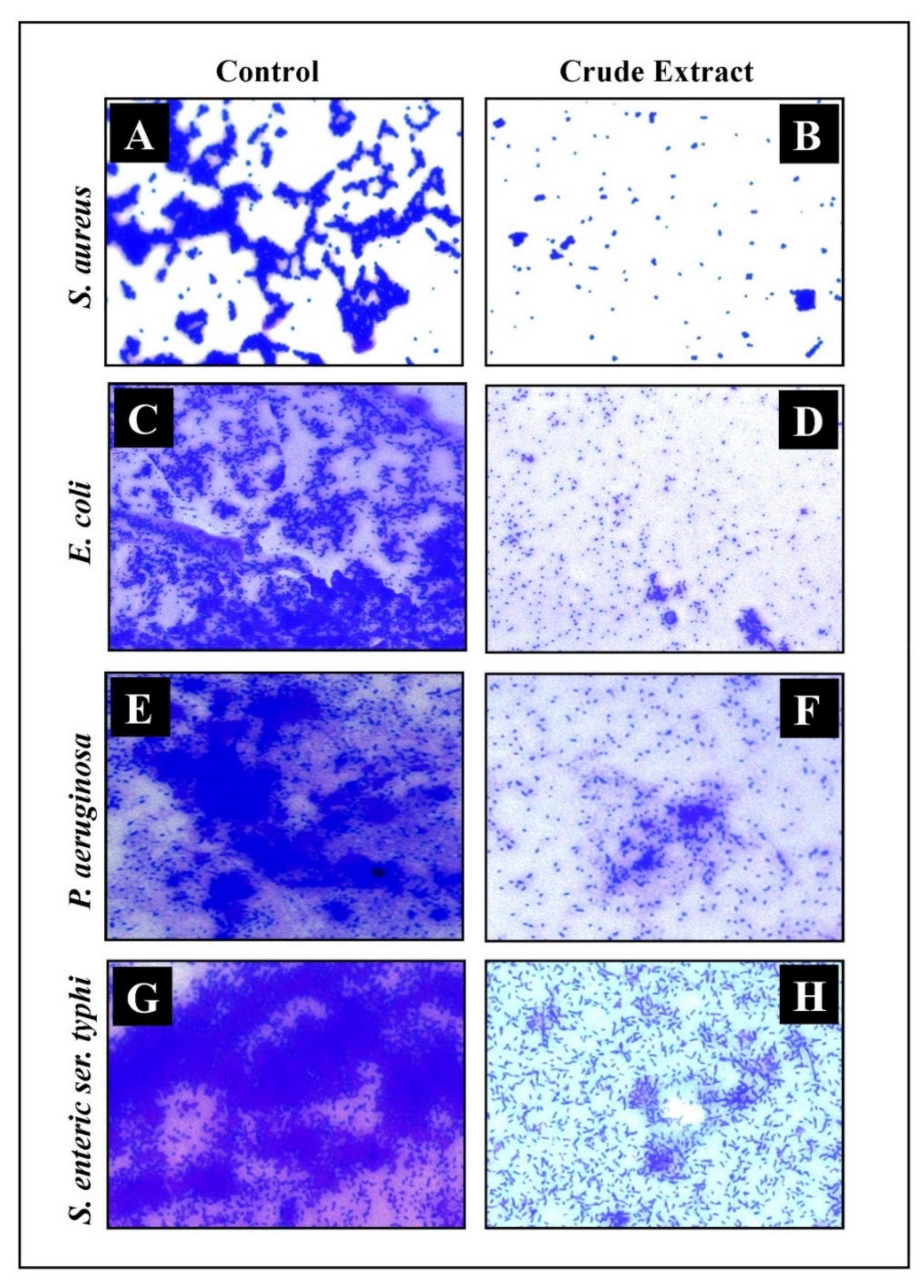
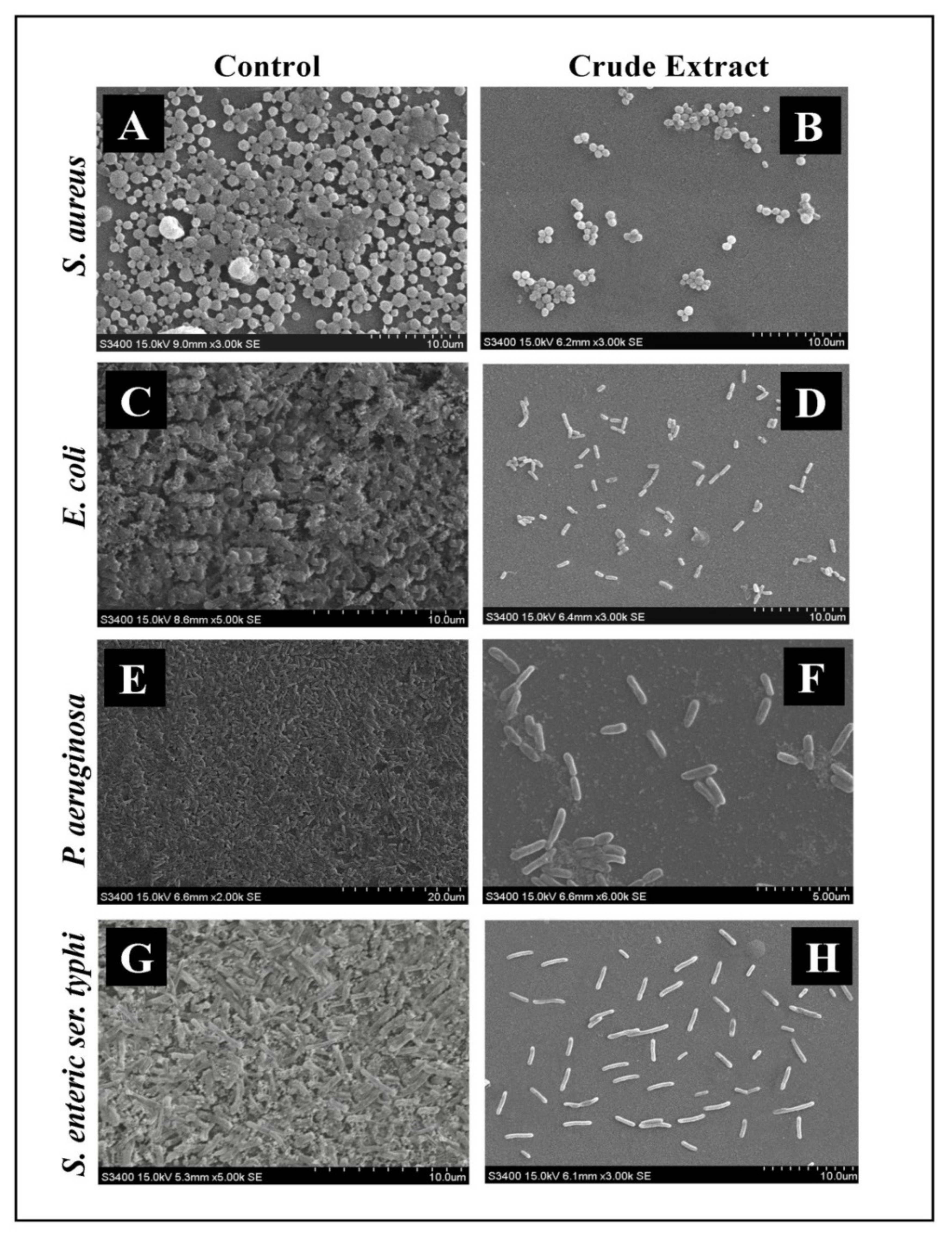
2.6. Microscopic Analysis of Disruption of Preformed Biofilm
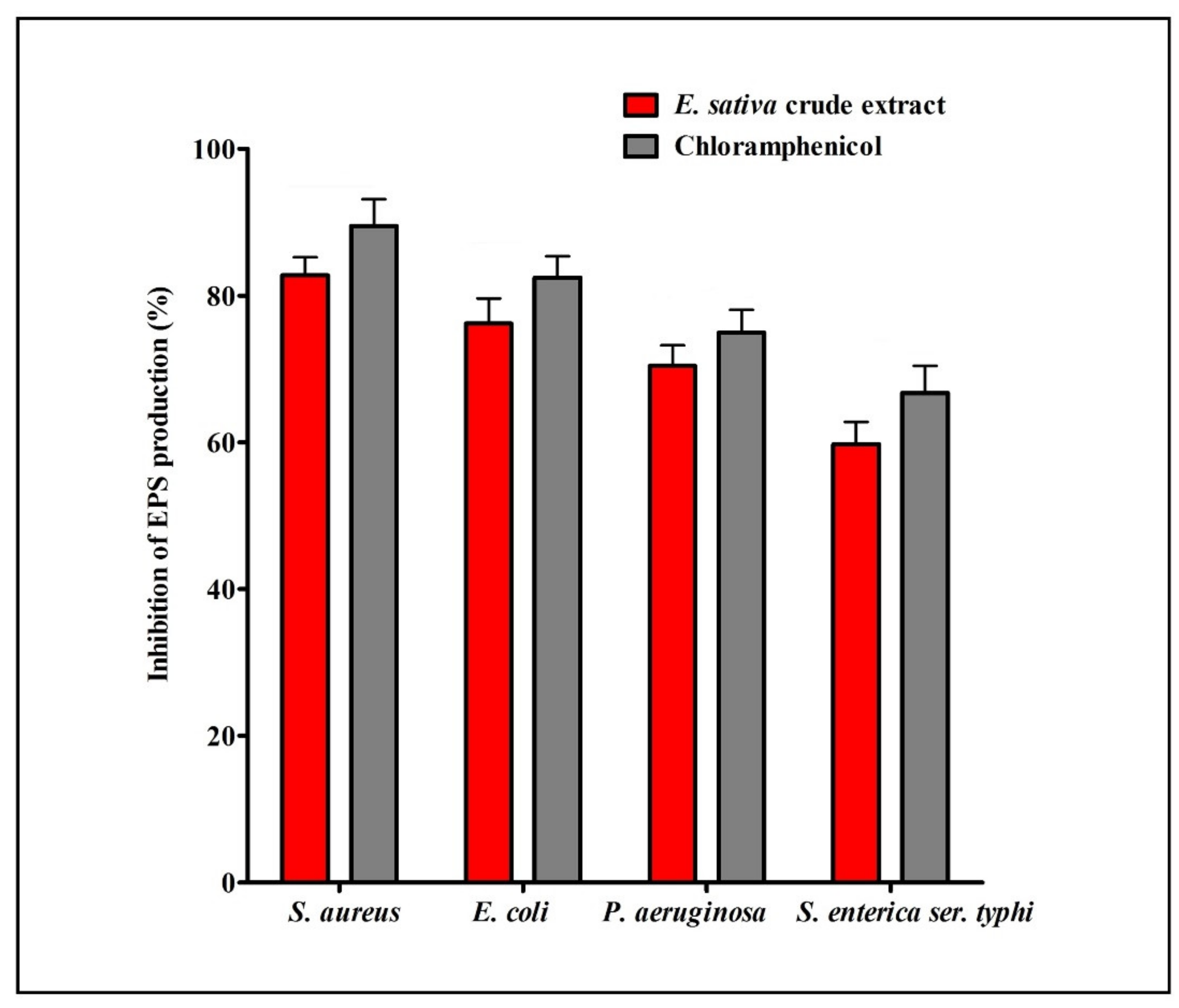
2.7. Effect on Exopolysaccharide (EPS) Production
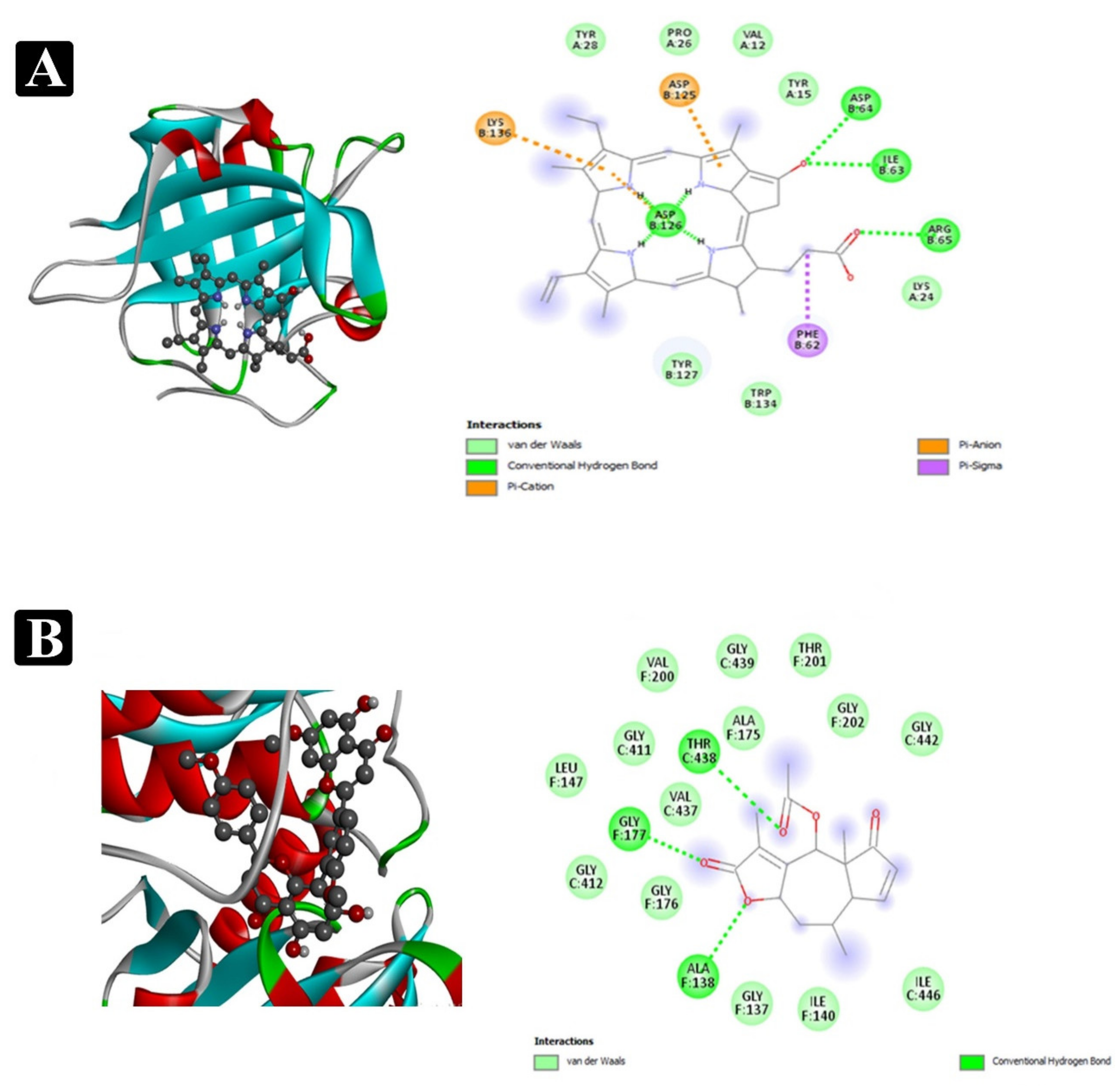
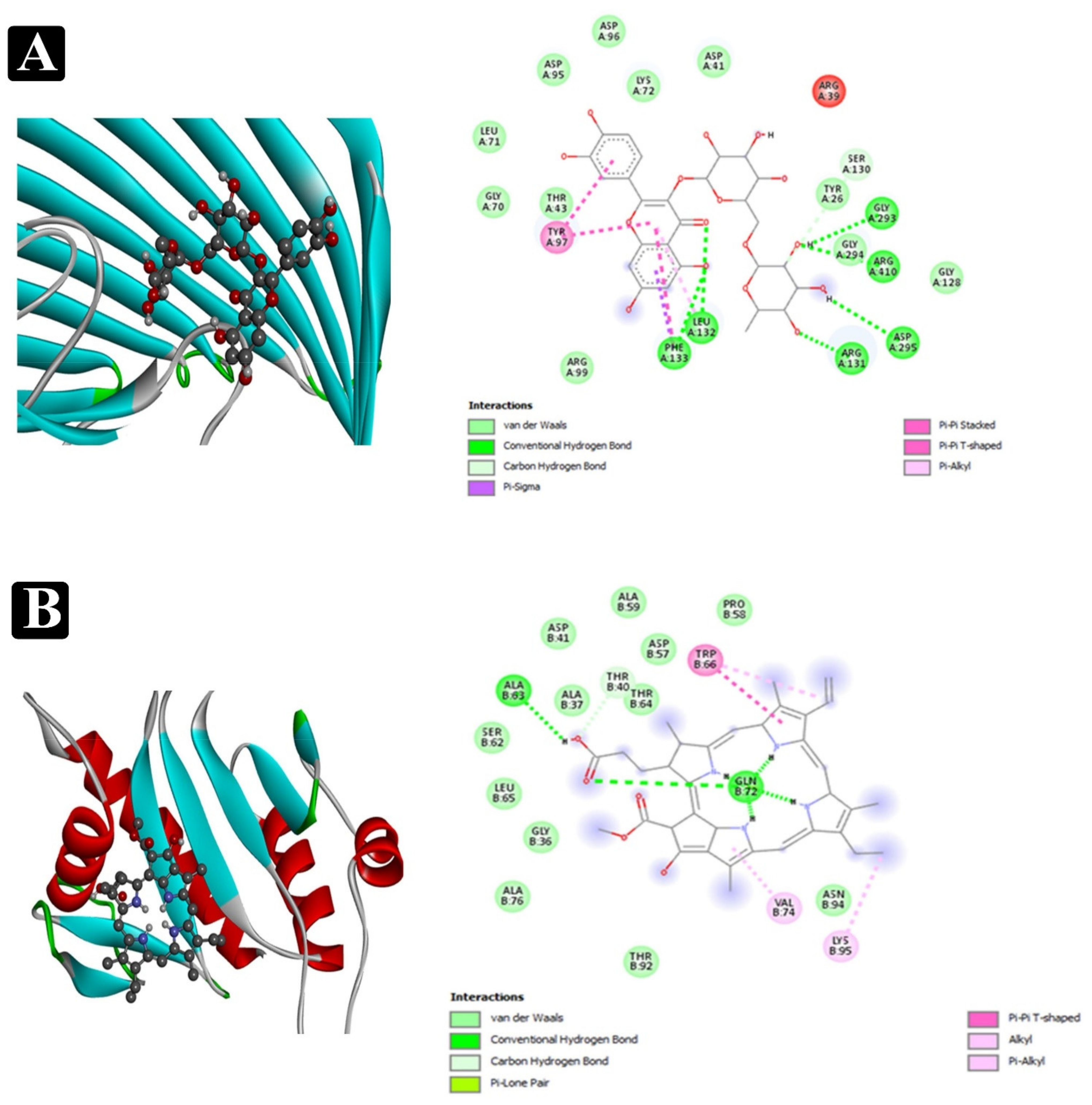
2.8. Molecular Docking Analysis
3. Discussion
4. Materials and Methods
4.1. Sample Collection, Identification and Extraction Process
4.2. Qualitative Phytochemical Analysis
4.3. Antibacterial Assay
4.3.1. Bacterial Strains
4.3.2. Agar Well Diffusion Method
4.3.3. Estimation of Minimum Inhibitory Concentration (MIC) Values
4.3.4. Estimation of Minimum Bactericidal Concentration (MBC)
4.3.5. Effect of E. sativa Crude Extract on Growth Kinetics of Bacteria
4.4. Antibiofilm Assays
4.4.1. Effect of E. sativa Crude Extract on Established Biofilms
4.4.2. Effect of E. sativa Crude Extract on the Adherence of Biofilms
4.5. Microscopic Analysis
4.5.1. Determination of Antibiofilm Activity Using Light Microscopy
4.5.2. Determination of Antibiofilm Activity via Scanning Electron Microscopy
4.5.3. Determination of Biofilm Metabolic Activity
4.5.4. Determination of Cell Damage within Biofilms
4.5.5. Determination of Extracellular Polysaccharide (EPS) Production
4.6. Molecular Docking Analysis of Phytochemicals of E. sativa with Adhesion Proteins of Food-Borne Pathogens
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Marwat, S.; Rehman, F.; Khan, A. Phytochemistry and Pharmacological Values of Rocket (Eruca sativa Miller)—A Review. Int. J. Hortic. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Sastry, E.V.D. Taramira (Eruca sativa) and its improvement A review. Agric. Rev. 2003, 24, 235–249. [Google Scholar]
- Bukhsh, E.; Malik, S.A.; Ahmad, S.S. Estimation of nutritional value and trace elements content of Carthamus oxyacantha, Eruca sativa and Plantago ovata. Pak. J. Bot. 2007, 39, 1181. [Google Scholar]
- Yehuda, H.; Khatib, S.; Sussan, I.; Musa, R.; Vaya, J.; Tamir, S. Potential skin antiinflammatory effects of 4-methylthiobutylisothiocyanate (MTBI) isolated from rocket (Eruca sativa) seeds. BioFactors 2009, 35, 295–305. [Google Scholar] [CrossRef]
- Melchini, A.; Costa, C.; Traka, M.; Miceli, N.; Mithen, R.; De Pasquale, R.; Trovato, A. Erucin, a new promising cancer chemopreventive agent from rocket salads, shows anti-proliferative activity on human lung carcinoma A549 cells. Food Chem. Toxicol. 2009, 47, 1430–1436. [Google Scholar] [CrossRef]
- Saad, B.; Said, O. Greco-Arab and Islamic Herbal Medicine: Traditional System, Ethics, Safety, Efficacy, and Regulatory Issues; John Wiley & Sons: Hoboken, NJ, USA, 2011; p. 552. [Google Scholar]
- Teixeira, A.F.; de Souza, J.; Dophine, D.D.; de Souza Filho, J.D.; Saúde-Guimarães, D.A. Chemical Analysis of Eruca sativa Ethanolic Extract and Its Effects on Hyperuricaemia. Molecules 2022, 27, 1506. [Google Scholar] [CrossRef]
- Qaddoumi, S.; El-Banna, N. Antimicrobial Activity of Arugula (Eruca Sativa) Leaves on Some Pathogenic Bacteria. Int. J. Biol. 2019, 11, 10–15. [Google Scholar] [CrossRef]
- Quinlan, J.J. Foodborne Illness Incidence Rates and Food Safety Risks for Populations of Low Socioeconomic Status and Minority Race/Ethnicity: A Review of the Literature. Int. J. Environ. Res. Public Health 2013, 10, 3634–3652. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the Natural environment to infectious diseases. Nat. Rev. Genet. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, H.-C.F.J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Genet. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Adnan, M.; Alshammari, E.; Patel, M.; Ashraf, S.A.; Khan, S.; Hadi, S. Significance and potential of marine microbial natural bioactive compounds against biofilms/biofouling: Necessity for green chemistry. PeerJ 2018, 6, e5049. [Google Scholar] [CrossRef] [PubMed]
- Adnan, M.; Siddiqui, A.J.; Hamadou, W.S.; Ashraf, S.A.; Hassan, I.; Snoussi, M.; Badraoui, R.; Jamal, A.; Bardakci, F.; Awadelkareem, A.M.; et al. Functional and Structural Characterization of Pediococcus pentosaceus-Derived Biosurfactant and Its Biomedical Potential against Bacterial Adhesion, Quorum Sensing, and Biofilm Formation. Antibiotics 2021, 10, 1371. [Google Scholar] [CrossRef]
- Patel, M.; Ashraf, M.S.; Siddiqui, A.J.; Ashraf, S.A.; Sachidanandan, M.; Snoussi, M.; Adnan, M.; Hadi, S. Profiling and Role of Bioactive Molecules from Puntius sophore (Freshwater/Brackish Fish) Skin Mucus with Its Potent Antibacterial, Antiadhesion, and Antibiofilm Activities. Biomolecules 2020, 10, 920. [Google Scholar] [CrossRef] [PubMed]
- Adnan, M.; Patel, M.; Deshpande, S.; Alreshidi, M.; Siddiqui, A.J.; Reddy, M.N.; Emira, N.; De Feo, V. Effect of Adiantum philippense Extract on Biofilm Formation, Adhesion with Its Antibacterial Activities against Foodborne Pathogens, and Characterization of Bioactive Metabolites: An in vitro-in silico Approach. Front. Microbiol. 2020, 11, 823. [Google Scholar] [CrossRef] [PubMed]
- Colagiorgi, A.; Bruini, I.; Di Ciccio, P.A.; Zanardi, E.; Ghidini, S.; Ianieri, A. Listeria monocytogenes Biofilms in the Wonderland of Food Industry. Pathogens 2017, 6, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Silveira, S.M.; Luciano, F.B.; Fronza, N.; Cunha, A., Jr.; Scheuermann, G.N.; Werneck Vieira, C.R. Chemical composition and antibacterial activity of Laurus nobilis essential oil towards foodborne pathogens and its application in fresh Tuscan sausage stored at 7 °C. LWT-Food Sci. Technol. 2014, 59, 86–93. [Google Scholar] [CrossRef]
- Molina, R.D.I.; Campos-Silva, R.; Díaz, M.A.; Macedo, A.J.; Blázquez, M.A.; Alberto, M.R.; Arena, M.E. Laurel extracts inhibit Quorum sensing, virulence factors and biofilm of foodborne pathogens. LWT 2020, 134, 109899. [Google Scholar] [CrossRef]
- Srey, S.; Jahid, I.K.; Ha, S.-D. Biofilm formation in food industries: A food safety concern. Food Control 2013, 31, 572–585. [Google Scholar] [CrossRef]
- Jessen, B.; Lammert, L. Biofilm and disinfection in meat processing plants. Int. Biodeterior. Biodegrad. 2003, 51, 265–269. [Google Scholar] [CrossRef]
- Al-Shabib, N.A.; Husain, F.M.; Ahmad, I.; Khan, M.S.; Khan, R.A.; Khan, J.M. Rutin inhibits mono and multi-species biofilm formation by foodborne drug resistant Escherichia coli and Staphylococcus aureus. Food Control 2017, 79, 325–332. [Google Scholar] [CrossRef]
- Charlebois, A.; Jacques, M.; Boulianne, M.; Archambault, M. Tolerance of Clostridium perfringens biofilms to disinfectants commonly used in the food industry. Food Microbiol. 2017, 62, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.D.; Plewa, M.J.; Wagner, E.D.; Schoeny, R.; DeMarini, D. Occurrence, genotoxicity, and carcinogenicity of regulated and emerging disinfection by-products in drinking water: A review and roadmap for research. Mutat. Res. Mutat. Res. 2007, 636, 178–242. [Google Scholar] [CrossRef] [PubMed]
- Nazzaro, F.; Fratianni, F.; Coppola, R. Quorum Sensing and Phytochemicals. Int. J. Mol. Sci. 2013, 14, 12607–12619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alreshidi, M.; Noumi, E.; Bouslama, L.; Ceylan, O.; Veettil, V.N.; Adnan, M.; Danciu, C.; Elkahoui, S.; Badraoui, R.; Al-Motair, K.A.; et al. Phytochemical Screening, Antibacterial, Antifungal, Antiviral, Cytotoxic, and Anti-Quorum-Sensing Properties of Teucrium polium L. Aerial Parts Methanolic Extract. Plants 2020, 9, 1418. [Google Scholar] [CrossRef]
- Reddy, M.N.; Adnan, M.; Alreshidi, M.M.; Saeed, M.; Patel, M. Evaluation of Anticancer, Antibacterial and Antioxidant Properties of a Medicinally Treasured Fern Tectaria coadunata with its Phytoconstituents Analysis by HR-LCMS. Anti-Cancer Agents Med. Chem. 2020, 20, 1845–1856. [Google Scholar] [CrossRef]
- Mseddi, K.; Alimi, F.; Noumi, E.; Veettil, V.N.; Deshpande, S.; Adnan, M.; Hamdi, A.; Elkahoui, S.; Alghamdi, A.; Kadri, A.; et al. Thymus musilii Velen. as a promising source of potent bioactive compounds with its pharmacological properties: In vitro and in silico analysis. Arab. J. Chem. 2020, 13, 6782–6801. [Google Scholar] [CrossRef]
- Surti, M.; Patel, M.; Adnan, M.; Moin, A.; Ashraf, S.A.; Siddiqui, A.J.; Snoussi, M.; Deshpande, S.; Reddy, M.N. Ilimaquinone (marine sponge metabolite) as a novel inhibitor of SARS-CoV-2 key target proteins in comparison with suggested COVID-19 drugs: Designing, docking and molecular dynamics simulation study. RSC Adv. 2020, 10, 37707–37720. [Google Scholar] [CrossRef]
- Adnan, M.; Siddiqui, A.J.; Hamadou, W.S.; Patel, M.; Ashraf, S.A.; Jamal, A.; Awadelkareem, A.M.; Sachidanandan, M.; Snoussi, M.; De Feo, V. Phytochemistry, Bioactivities, Pharmacokinetics and Toxicity Prediction of Selaginella repanda with Its Anticancer Potential against Human Lung, Breast and Colorectal Carcinoma Cell Lines. Molecules 2021, 26, 768. [Google Scholar] [CrossRef]
- Gopu, V.; Meena, C.K.; Murali, A.; Shetty, P.H. Petunidin as a competitive inhibitor of acylated homoserine lactones in Klebsiella pneumoniae. RSC Adv. 2016, 6, 2592–2601. [Google Scholar] [CrossRef]
- Koubaa, M.; Driss, D.; Bouaziz, F.; Ghorbel, R.E.; Chaabouni, S.E. Antioxidant and antimicrobial activities of solvent extract obtained from rocket (Eruca sativa L.) flowers. Free Radic. Antioxid. 2015, 5, 29–34. [Google Scholar] [CrossRef] [Green Version]
- De Cássia Orlandi Sardi, J.; Freires, I.A.; Lazarini, J.G.; Infante, J.; de Alencar, S.M.; Rosalen, P.L. Unexplored endemic fruit species from Brazil: Antibiofilm properties, insights into mode of action, and systemic toxicity of four Eugenia spp. Microb. Pathog. 2017, 105, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Khoobchandani, M.; Ojeswi, B.; Ganesh, N.; Srivastava, M.; Gabbanini, S.; Matera, R.; Iori, R.; Valgimigli, L. Antimicrobial properties and analytical profile of traditional Eruca sativa seed oil: Comparison with various aerial and root plant extracts. Food Chem. 2010, 120, 217–224. [Google Scholar] [CrossRef]
- Rizwana, H.; Alwahibi, M.; Khan, F.; Soliman, D. Chemical composition and antimicrobial activity of Eruca sativa seeds against pathogenic bacteria and fungi. J. Anim. Plant Sci. 2016, 26, 1859–1871. [Google Scholar]
- Foerster, S.; Unemo, M.; Hathaway, L.J.; Low, N.; Althaus, C.L. Time-kill curve analysis and pharmacodynamic modelling for in vitro evaluation of antimicrobials against Neisseria gonorrhoeae. BMC Microbiol. 2016, 16, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anantharaman, A.; Rizvi, M.S.; Sahal, D. Synergy with rifampin and kanamycin enhances potency, kill kinetics, and selectivity of de novo-designed antimicrobial peptides. Antimicrob. Agents Chemother. 2010, 54, 1693–1699. [Google Scholar] [CrossRef] [Green Version]
- Rutherford, S.T.; Bassler, B.L. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef]
- Sandasi, M.; Leonard, C.; Viljoen, A. The in vitro antibiofilm activity of selected culinary herbs and medicinal plants against Listeria monocytogenes. Lett. Appl. Microbiol. 2010, 50, 30–35. [Google Scholar] [CrossRef]
- Razak, F.A.; Rahim, Z.H.A. The anti-adherence effect of Piper betle and Psidium guajava extracts on the adhesion of early settlers in dental plaque to saliva-coated glass surfaces. J. Oral Sci. 2003, 45, 201–206. [Google Scholar] [CrossRef] [Green Version]
- Namasivayam, S.K.R.; Roy, E.A. Anti biofilm effect of medicinal plant extracts against clinical isolate of biofilm of Escherichia coli. Int. J. Pharm. Pharm. Sci. 2013, 5, 486–489. [Google Scholar]
- Miyazawa, M.; Maehara, T.; Kurose, K. Composition of the essential oil from the leaves of Eruca sativa. Flavour Fragr. J. 2002, 17, 187–190. [Google Scholar] [CrossRef]
- Gulfraz, M.; Sadiq, A.; Tariq, H.B.; Imran, M.; Qureshi, R.; Zeenat, A. Phytochemical analysis and antibacterial activity of Eruca sativa seed. Pak. J. Bot. 2011, 43, 1351–1359. [Google Scholar]
- Hussein, Z.F. Study the Effect of Eruca Sativa Leaves Extract on Male Fertility in Albino Mice. Al-Nahrain J. Sci. 2013, 16, 143–146. [Google Scholar] [CrossRef]
- Orhan, I.E.; Kartal, M.; Sekeroglu, N.; Esiyok, D.; Şener, B.; Ugur, A.; Süntar, I.; Aslan, S. Variations in fatty acid compositions of the seed oil of Eruca sativa Mill. caused by different sowing periods and nitrogen forms. Pharmacogn. Mag. 2010, 6, 305–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jirovetz, L.; Smith, D.; Buchbauer, G. Aroma Compound Analysis of Eruca sativa (Brassicaceae) SPME Headspace Leaf Samples Using GC, GC−MS, and Olfactometry. J. Agric. Food Chem. 2002, 50, 4643–4646. [Google Scholar] [CrossRef]
- Abdul-Jalil, T.Z. Phytochemicals screening by GC/MS and determination of some flavonol in cultivated Iraqi Eruca sativa dried leaves extract and its biological activity as antioxidant. Int. J. Pharmacogn. Phytochem. Res. 2016, 8, 1722–1730. [Google Scholar]
- Villatoro-Pulido, M.; Priego-Capote, F.; Álvarez-Sánchez, B.; Saha, S.; Philo, M.; Obregón-Cano, S.; De Haro-Bailón, A.; Font, R.; Del Río-Celestino, M. An approach to the phytochemical profiling of rocket [Eruca sativa (Mill.) Thell]. J. Sci. Food Agric. 2013, 93, 3809–3819. [Google Scholar] [CrossRef] [PubMed]
- Michael, H.; Shafik, R.; Rasmy, G. Studies on the chemical constituents of fresh leaf of Eruca sativa extract and its biological activity as anticancer agent in vitro. J. Med. Plants Res. 2011, 5, 1184–1191. [Google Scholar]
- Hussain, S.A. Phytochemical study of vsome medicinal compounds present in Hedera helix L. plant cultivated in Iraq. Master’s Thesis, University of Baghdad, Baghdad, Iraq, 2014. [Google Scholar]
- Blažević, I.; Mastelić, J. Free and bound volatiles of rocket (Eruca sativa Mill.). Flavour Fragr. J. 2008, 23, 278–285. [Google Scholar]
- Kitchen, D.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef]
- Zong, Y.; Bice, T.W.; Ton-That, H.; Schneewind, O.; Narayana, S.V.L. Crystal structures of Staphylococcus aureus sortase A and its substrate complex. J. Biol. Chem. 2004, 279, 31383–31389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.; Staahl, B.T.; Alla, R.K.; Doudna, J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 2014, 3, e04766. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam, P.K.; Kuruva, J.; Hari, S.; Annamalai, D.S.K.; Baskaran, K. HPTLC Finger print analysis and Phytochemical Investigation of Morinda tinctoria Roxb. Leaf extracts by HPLC and GS MS. Int. J. Pharm. Pharm. Sci. 2014, 7, 360–366. [Google Scholar]
- Ashraf, S.A.; Al-Shammari, E.; Hussain, T.; Tajuddin, S.; Panda, B.P. In-vitro antimicrobial activity and identification of bioactive components using GC–MS of commercially available essential oils in Saudi Arabia. J. Food Sci. Technol. 2017, 54, 3948–3958. [Google Scholar] [CrossRef]
- Ashraf, S.A.; Khan, M.A.; Awadelkareem, A.M.; Tajuddin, S.; Ahmad, F.; Hussain, T. GC-MS Analysis of Commercially Available Allium sativum and Trigonella foenum-graecum Essential Oils and their Antimicrobial Activities. J. Pure Appl. Microbiol. 2019, 13, 2545–2552. [Google Scholar] [CrossRef] [Green Version]
- CLSI, Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2014.
- Lemos, A.S.O.; Campos, L.M.; Melo, L.; Guedes, M.C.M.R.; Oliveira, L.G.; Silva, T.P.; Melo, R.; Rocha, V.N.; Aguiar, J.; Apolonio, A.C.; et al. Antibacterial and Antibiofilm Activities of Psychorubrin, a Pyranonaphthoquinone Isolated from Mitracarpus frigidus (Rubiaceae). Front. Microbiol. 2018, 9, 724. [Google Scholar] [CrossRef] [Green Version]
- Plyuta, V.; Zaitseva, J.; Lobakova, E.; Zagoskina, N.; Kuznetsov, A.; Khmel, I. Effect of plant phenolic compounds on biofilm formation by Pseudomonas aeruginosa. Apmis 2013, 121, 1073–1081. [Google Scholar] [CrossRef]
- Musthafa, K.S.; Ravi, A.V.; Annapoorani, A.; Packiavathy, S.V.; Pandian, S.K. Evaluation of Anti-Quorum-Sensing Activity of Edible Plants and Fruits through Inhibition of the N-Acyl-Homoserine Lactone System in Chromobacterium violaceum and Pseudomonas aeruginosa. Chemotherapy 2010, 56, 333–339. [Google Scholar] [CrossRef]
- Ramage, G.; Walle, K.V.; Wickes, B.; Lopez-Ribot, J.L. Standardized Method for In Vitro Antifungal Susceptibility Testing of Candida albicans Biofilms. Antimicrob. Agents Chemother. 2001, 45, 2475–2479. [Google Scholar] [CrossRef] [Green Version]
- Nett, J.E.; Cain, M.; Crawford, K.; Andes, D. Optimizing a Candida Biofilm Microtiter Plate Model for Measurement of Antifungal Susceptibility by Tetrazolium Salt Assay. J. Clin. Microbiol. 2011, 49, 1426–1433. [Google Scholar] [CrossRef] [Green Version]
- Borucki, M.K.; Peppin, J.D.; White, D.; Loge, F.; Call, D.R. Variation in biofilm formation among strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2003, 69, 7336–7342. [Google Scholar] [PubMed] [Green Version]
- Yip, C.K.; Finlay, B.B.; Strynadka, N.C.J. Structural characterization of a type III secretion system filament protein in complex with its chaperone. Nat. Struct. Mol. Biol. 2004, 12, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Balakrishna, A.M.; Saxena, A.M.; Mok, H.Y.; Swaminathan, K. Structural basis of typhoid: Salmonella typhi type IVb pilin (PilS) and cystic fibrosis transmembrane conductance regulator interaction. Proteins 2009, 77, 253–261. [Google Scholar] [CrossRef]
- Kühnel, K.; Diezmann, D. Crystal Structure of the Autochaperone Region from the Shigella flexneri Autotransporter IcsA. J. Bacteriol. 2011, 193, 2042–2045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eren, E.; Vijayaraghavan, J.; Liu, J.; Cheneke, B.R.; Touw, D.S.; Lepore, B.W.; Indic, M.; Movileanu, L.; Berg, B.V.D. Substrate Specificity within a Family of Outer Membrane Carboxylate Channels. PLoS Biol. 2012, 10, e1001242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G. Open Babel: An open chemical toolbox. J. Cheminforma 2011, 3, 33. [Google Scholar] [CrossRef] [Green Version]



| Phytochemicals | Crude Extract |
|---|---|
| Alkaloids | Positive |
| Tannins | Positive |
| Flavonoids | Positive |
| Phenolics | Positive |
| Terpenoids | Positive |
| Glycosides | Positive |
| Saponins | Positive |
| Carbohydrates | Positive |
| Bacterial Strain | E. sativa Crude Extract (µg/mL) | Chloramphenicol (µg/mL) | ||
|---|---|---|---|---|
| MIC | MBC | MIC | MBC | |
| S. aureus | 125 | 250 | 7.812 | 15.625 |
| E. coli | 250 | 500 | 15.65 | 31.25 |
| P. aeruginosa | 250 | 500 | 62.5 | 125 |
| S. enterica ser. typhi | 500 | 1000 | 125 | 250 |
| Compound Number | Name | 1T2P | 1X0U | 3SY7 | 3FHU |
|---|---|---|---|---|---|
| 1 | (+/-)-3-[(2-methyl-3-furyl)thio]-2-butanone | −4.3 | −5.1 | −4.9 | −4.3 |
| 2 | (10Z,14E,16E)-10,14,16-Octadecatrien-12-ynoic acid | −4.9 | −5.8 | −6.1 | −5.5 |
| 3 | (6beta,8betaOH)-6,8-Dihydroxy-7(11)-eremophilen-12,8-olide | −7.2 | −7.6 | −7.7 | −6.4 |
| 4 | (S)-2-(Hydroxymethyl)glutarate | −4.6 | −5.3 | −4.7 | −4.3 |
| 5 | (±)-Rollipyrrole | −6.2 | −6.3 | −7.3 | −5.5 |
| 6 | 1,4-Dimethoxyglucobrassicin | −6.6 | −8.0 | −7.4 | −6.8 |
| 7 | 1-Methoxy-1H-indole-3-carboxaldehyde | −5.2 | −6.4 | −5.8 | −5.1 |
| 8 | 16-Hydroxy hexadecanoic acid | −4.6 | −4.8 | −5.2 | −3.6 |
| 9 | 2-Deoxy-scyllo-inosose | −5.1 | −5.5 | −5.2 | −4.8 |
| 10 | 3,4’,5,6,8-Pentamethoxyflavone | −7.0 | −8.1 | −7.0 | −6.2 |
| 11 | 4-(3-Hydroxy-7-phenyl-6-heptenyl)-1,2-benzenediol | −7.7 | −8.2 | −7.1 | −5.1 |
| 12 | 4-Amino-2-methyl-1-naphthol | −6.5 | −7.2 | −6.2 | −5.6 |
| 13 | 9Z-Octadecenedioic acid | −5.7 | −5.7 | −5.9 | −4.6 |
| 14 | Afzelechin | −7.5 | −8.5 | −7.4 | −6.3 |
| 15 | Artomunoxanthentrione epoxide | −8.3 | −10.3 | −9.1 | −7.4 |
| 16 | Corchorifatty acid F | −6.0 | −5.8 | −6.4 | −5.2 |
| 17 | Evoxine | −6.0 | −7.8 | −6.7 | −5.6 |
| 18 | Fraxidin | −6.5 | −6.8 | −6.4 | −5.8 |
| 19 | Glucoraphanin | −5.9 | −6.5 | −6.8 | −5.9 |
| 20 | Indoleacrylic acid | −6.2 | −7.1 | −6.3 | −6.3 |
| 21 | Lactucin | −6.9 | −8.0 | −7.5 | −6.9 |
| 22 | Linifolin A | −7.7 | −8.0 | −7.2 | −6.2 |
| 23 | Methyl N-methylanthranilate | −5.3 | −5.9 | −5.2 | −4.9 |
| 24 | N-(6-Oxo-6H-dibenzo[b,d]pyran-3-yl)maleamic acid | −7.4 | −9.1 | −8.6 | −6.9 |
| 25 | N-trans-Feruloyl-4-O-methyldopamine | −7.4 | −7.9 | −7.1 | −6.6 |
| 26 | N6-cis-p-Coumaroylserotonin | −7.8 | −8.5 | −7.7 | −6.8 |
| 27 | Nopaline | −5.5 | −6.6 | −6.0 | −5.2 |
| 28 | Oleamide | −5.1 | −5.2 | −5.4 | −4.0 |
| 29 | Palmitic amide | −4.6 | −4.9 | −4.9 | −4.0 |
| 30 | Petasitenine | −7.4 | −9.0 | −8.4 | −7.1 |
| 31 | Pheophorbide a | −8.1 | −9.1 | −8.5 | −8.8 |
| 32 | Pubesenolide | −6.0 | −7.6 | −7.7 | −6.1 |
| 33 | Pyrafoline D | −8.8 | −9.4 | −8.7 | −7.7 |
| 34 | Pyropheophorbide a | −9.4 | −10.0 | −8.7 | −8.0 |
| 35 | Rutin | −8.5 | −9.8 | −9.8 | −6.8 |
| 36 | Sciadopitysin | −9.0 | −10.8 | −9.1 | −8.4 |
| 37 | Serinyl-Hydroxyprolinen | −5.7 | −7.0 | −5.9 | −5.5 |
| 38 | Terminaline | −7.4 | −8.0 | −7.9 | −6.7 |
| 39 | Thalidasine | −8.1 | −9.1 | −8.8 | −7.0 |
| 40 | Trilobolide | −7.2 | −8.4 | −7.7 | −6.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Awadelkareem, A.M.; Al-Shammari, E.; Elkhalifa, A.O.; Adnan, M.; Siddiqui, A.J.; Mahmood, D.; Azad, Z.R.A.A.; Patel, M.; Mehmood, K.; Danciu, C.; et al. Anti-Adhesion and Antibiofilm Activity of Eruca sativa Miller Extract Targeting Cell Adhesion Proteins of Food-Borne Bacteria as a Potential Mechanism: Combined In Vitro-In Silico Approach. Plants 2022, 11, 610. https://doi.org/10.3390/plants11050610
Awadelkareem AM, Al-Shammari E, Elkhalifa AO, Adnan M, Siddiqui AJ, Mahmood D, Azad ZRAA, Patel M, Mehmood K, Danciu C, et al. Anti-Adhesion and Antibiofilm Activity of Eruca sativa Miller Extract Targeting Cell Adhesion Proteins of Food-Borne Bacteria as a Potential Mechanism: Combined In Vitro-In Silico Approach. Plants. 2022; 11(5):610. https://doi.org/10.3390/plants11050610
Chicago/Turabian StyleAwadelkareem, Amir Mahgoub, Eyad Al-Shammari, AbdElmoneim O. Elkhalifa, Mohd Adnan, Arif Jamal Siddiqui, Danish Mahmood, Z. R. Azaz Ahmad Azad, Mitesh Patel, Khalid Mehmood, Corina Danciu, and et al. 2022. "Anti-Adhesion and Antibiofilm Activity of Eruca sativa Miller Extract Targeting Cell Adhesion Proteins of Food-Borne Bacteria as a Potential Mechanism: Combined In Vitro-In Silico Approach" Plants 11, no. 5: 610. https://doi.org/10.3390/plants11050610
APA StyleAwadelkareem, A. M., Al-Shammari, E., Elkhalifa, A. O., Adnan, M., Siddiqui, A. J., Mahmood, D., Azad, Z. R. A. A., Patel, M., Mehmood, K., Danciu, C., & Ashraf, S. A. (2022). Anti-Adhesion and Antibiofilm Activity of Eruca sativa Miller Extract Targeting Cell Adhesion Proteins of Food-Borne Bacteria as a Potential Mechanism: Combined In Vitro-In Silico Approach. Plants, 11(5), 610. https://doi.org/10.3390/plants11050610













