Phylogeography of 912 Cherry Accessions Insight into Independent Origins of Fruiting Cherries and Domestication Footprints of Cultivated Chinese Cherry (Prunus pseudocerasus Lindl.)
Abstract
:1. Introduction
2. Results
2.1. Sequence Characteristics
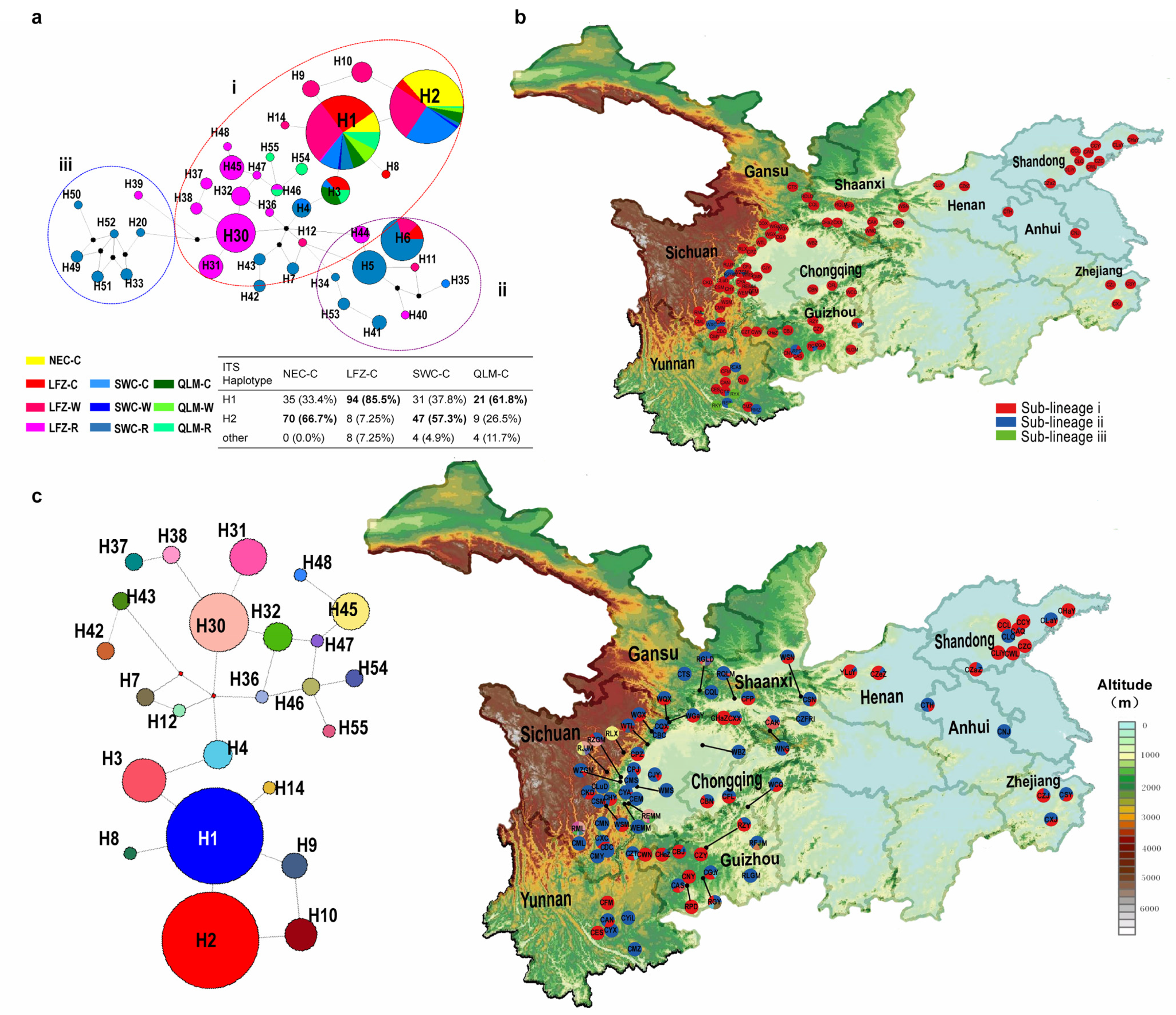
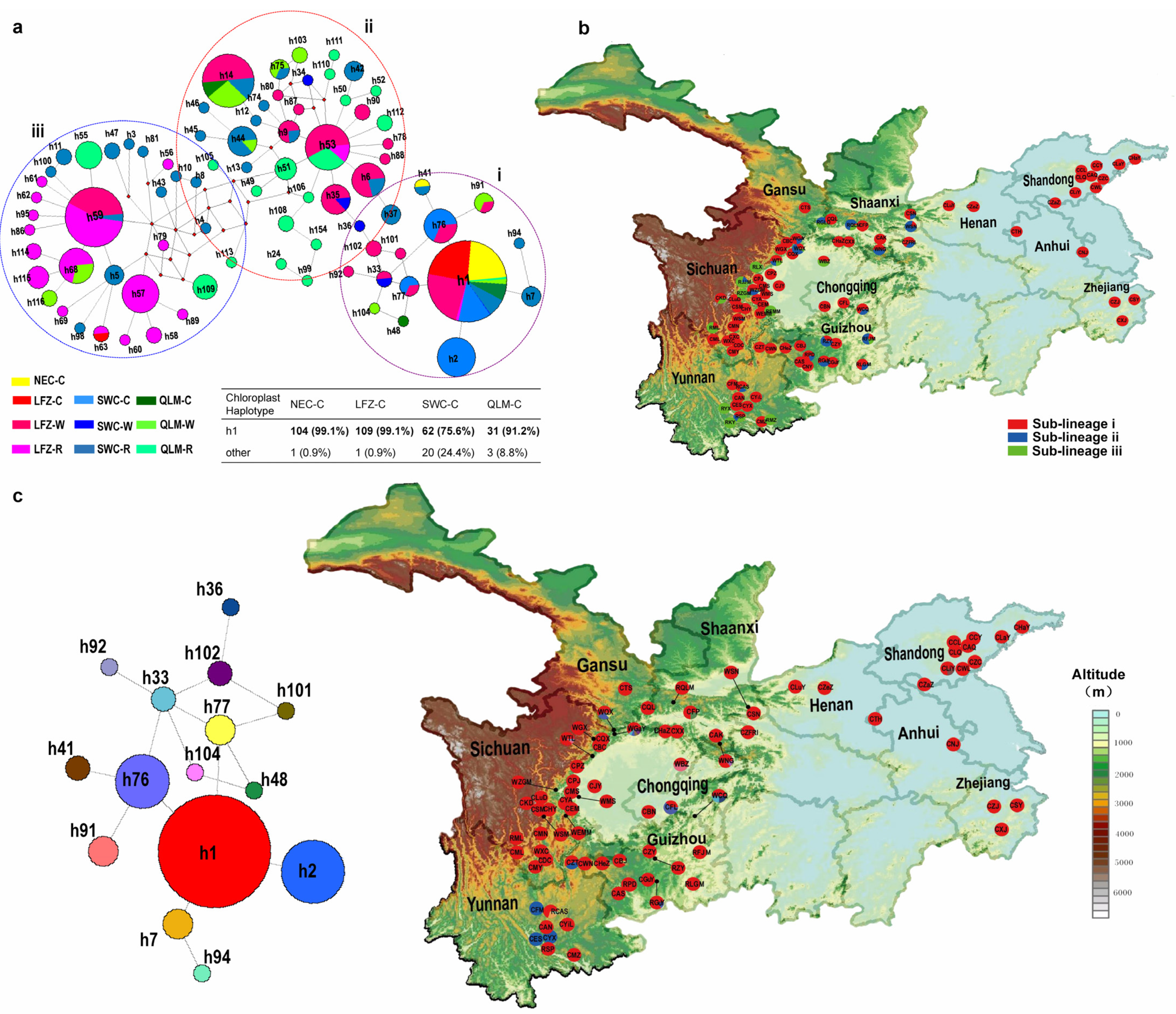
2.2. Haplotype Genealogy
2.3. Genetic Diversity and Demographic History
2.4. Phylogeographic Structure of Chinese Cherry
3. Discussion
3.1. Independent Origins of Economically Cultivated Fruiting Cherry
3.2. Geographical Origin and Domestication of Cultivated Chinese Cherry
4. Materials and Methods
4.1. Plant Materials
4.2. DNA Isolation, PCR Amplification, and Sequencing
4.3. Sequence Alignment
4.4. Haplotype Genealogy Construction
4.5. Molecular Diversity and Population Structure
4.6. Historical Demographic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yü, D.J.; Lu, L.T.; Ku, T.C.; Li, C.L.; Chen, S.X. Flora of China; Science Press: Beijing, China, 1986; Volume 38. [Google Scholar]
- Yü, D.J. Taxonomy of Fruit Trees in China; Agricultural Press: Beijing, China, 1979. [Google Scholar]
- Chen, T.; Hu, G.P.; Wang, Y.; Chen, Q.; Zhang, J.; Wang, L.; Tang, H.R.; Wang, X.R. Survey, collection and conservation of wild Cerasus Mill. germplasm resources in China. J. Plant Genet. Resour. 2020, 21, 532–541. [Google Scholar]
- Huang, X.; Wang, X.; Chen, T.; Chen, J.; Tang, H. Research progress of germplasm diversity in Chinese cherry (Cerasus pseudocerasus). J. Fruit Sci. 2013, 30, 470–479. [Google Scholar]
- Chen, T.; Huang, X.-J.; Zhang, J.; Chen, Q.; Liu, Y.; Tang, H.-R.; Pan, D.-M.; Wang, X.-R. Genetic diversity and population structure patterns in Chinese cherry (Prunus pseudocerasus Lindl) landraces. Plant Mol. Biol. Rep. 2016, 34, 440–453. [Google Scholar] [CrossRef]
- Keightley, D.N. The origins of Chinese Civilization; University of California Press: Berkeley, CA, USA, 2022. [Google Scholar]
- Pathirana, R.; Carimi, F. Management and utilization of plant genetic resources for a sustainable agriculture. Plants 2022, 11, 2038. [Google Scholar] [CrossRef] [PubMed]
- Vavilov, N.I.; Vavylov, M.I.; Dorofeev, V.F. Origin and Geography of Cultivated Plants; Cambridge University Press: Cambridge, UK, 1992. [Google Scholar]
- Yu, Y.; Fu, J.; Xu, Y.; Zhang, J.; Ren, F.; Zhao, H.; Tian, S.; Guo, W.; Tu, X.; Zhao, J.; et al. Genome re-sequencing reveals the evolutionary history of peach fruit edibility. Nat. Commun. 2018, 9, 5404. [Google Scholar] [CrossRef]
- The International Peach Genome Initiative; Verde, I.; Abbott, A.G.; Scalabrin, S.; Jung, S.; Shu, S.; Marroni, F.; Zhebentyayeva, T.; Dettori, M.T.; Grimwood, J.; et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 2013, 45, 487–494. [Google Scholar] [CrossRef]
- Cornille, A.; Gladieux, P.; Smulders, M.J.M.; Roldán-Ruiz, I.; Laurens, F.; Le Cam, B.; Nersesyan, A.; Clavel, J.; Olonova, M.; Feugey, L.; et al. New insight into the history of domesticated apple: Secondary contribution of the European wild apple to the genome of cultivated varieties. PLoS Genet. 2012, 8, e1002703. [Google Scholar] [CrossRef]
- Gross, B.L.; Henk, A.D.; Richards, C.M.; Fazio, G.; Volk, G.M. Genetic diversity in Malus× domestica (Rosaceae) through time in response to domestication. Am. J. Bot. 2014, 101, 1770–1779. [Google Scholar] [CrossRef]
- Yi, X.-G.; Yu, X.-Q.; Chen, J.; Zhang, M.; Liu, S.-W.; Zhu, H.; Li, M.; Duan, Y.-F.; Chen, L.; Wu, L.; et al. The genome of Chinese flowering cherry (Cerasus serrulata) provides new insights into Cerasus species. Hortic. Res. 2020, 7, 165. [Google Scholar] [CrossRef]
- Janick, J. The origins of fruits, fruit growing, and fruit breeding. In Plant Breeding Reviews; Wiley & Sons Oxford: Oxford, UK, 2005; Volume 25, pp. 255–320. [Google Scholar]
- Meyer, R.S.; Purugganan, M.D. Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 2013, 14, 840–852. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Y.; Xu, J.; Korban, S.S.; Fei, Z.; Tao, S.; Ming, R.; Tai, S.; Khan, A.M.; Postman, J.D.; et al. Diversification and independent domestication of Asian and European pears. Genome Biol. 2018, 19, 77. [Google Scholar] [CrossRef]
- Groppi, A.; Liu, S.; Cornille, A.; Decroocq, S.; Bui, Q.T.; Tricon, D.; Cruaud, C.; Arribat, S.; Belser, C.; Marande, W. Population genomics of apricots unravels domestication history and adaptive events. Nat. Commun. 2021, 12, 3956. [Google Scholar] [CrossRef] [PubMed]
- Chin, S.W.; Shaw, J.; Haberle, R.; Wen, J.; Potter, D. Diversification of almonds, peaches, plums and cherries—Molecular systematics and biogeographic history of Prunus (Rosaceae). Mol. Phylogenet. Evol. 2014, 76, 34–48. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Matsumoto, A.; Yoshimura, K.; Katsuki, T.; Iwamoto, K.; Kawahara, T.; Mukai, Y.; Tsuda, Y.; Ishio, S.; Nakamura, K.; et al. Origins of Japanese flowering cherry (Prunus subgenus Cerasus) cultivars revealed using nuclear SSR markers. Tree Genet. Genomes 2014, 10, 477–487. [Google Scholar] [CrossRef]
- Shirasawa, K.; Isuzugawa, K.; Ikenaga, M.; Saito, Y.; Yamamoto, T.; Hirakawa, H.; Isobe, S. The genome sequence of sweet cherry (Prunus avium) for use in genomics-assisted breeding. DNA Res. 2017, 24, 499–508. [Google Scholar] [CrossRef]
- Baek, S.; Choi, K.; Kim, G.-B.; Yu, H.-J.; Cho, A.; Jang, H.; Kim, C.; Kim, H.-J.; Chang, K.S.; Kim, J.-H.; et al. Draft genome sequence of wild Prunus yedoensis reveals massive inter-specific hybridization between sympatric flowering cherries. Genome Biol. 2018, 19, 127. [Google Scholar] [CrossRef]
- Pinosio, S.; Marroni, F.; Zuccolo, A.; Vitulo, N.; Morgante, M. A draft genome of sweet cherry (Prunus avium L.) reveals genome-wide and local effects of domestication. Plant J. 2020, 103, 1420–1432. [Google Scholar] [CrossRef]
- Yan, J.; Li, J.; Bai, W.; Yu, L.; Nie, D.; Xiong, Y.; Li, B.; Wu, S. Genotyping and genetic diversity analysis of 47 flowering cherry (Cerasus) germplasms. Int. J. Hortic. 2020, 1–10. [Google Scholar] [CrossRef]
- Hussain, H.; Nisar, M. Assessment of plant genetic variations using molecular markers: A review. J. Appl. Biol. Biotechnol. 2020, 8, 99–109. [Google Scholar]
- Chen, T.; Chen, Q.; Luo, Y.; Huang, Z.-L.; Zhang, J.; Tang, H.-R.; Pan, D.-M.; Wang, X.R. Phylogeography of Chinese cherry (Prunus pseudocerasus Lindl.) inferred from chloroplast and nuclear DNA: Insights into evolutionary patterns and demographic history. Plant Biol. 2015, 17, 787–797. [Google Scholar] [CrossRef]
- Chen, T.; Wang, X.R.; Tang, H.R.; Chen, Q.; Huang, X.J.; Chen, J. Genetic diversity and population structure of Chinese cherry revealed by chloroplast DNA trnQ-rps16 intergenic spacers variation. Genet. Resour. Crop Ev. 2013, 60, 1859–1871. [Google Scholar] [CrossRef]
- Shi, S.; Li, J.; Sun, J.; Yu, J.; Zhou, S. Phylogeny and classification of Prunus sensu lato (Rosaceae). J. Integr. Plant Biol. 2013, 55, 1069–1079. [Google Scholar] [CrossRef]
- Batnini, M.A.; Bourguiba, H.; Trifi-Farah, N.; Krichen, L. Analysis on relationship and taxonomic status of some Species in Subg. Cerasus Koehne with chloroplast DNA atpB-rbcL fragment. Sci. Hortic. 2019, 245, 99–106. [Google Scholar]
- Bailey, C.D.; Carr, T.G.; Harris, S.A.; Hughes, C.E. Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Mol. Phylogenetics Evol. 2003, 29, 435–455. [Google Scholar] [CrossRef] [PubMed]
- Osuna-Mascaro, C.; Rubio De Casas, R.; Berbel, M.; Gómez, J.M.; Perfectti, F. Lack of ITS sequence homogenization in congeneric plant species with different ploidy levels. bioRxiv 2022, 29, 493735. [Google Scholar]
- Zheng, X.; Cai, D.; Yao, L.; Teng, Y. Non-concerted ITS evolution, early origin and phylogenetic utility of ITS pseudogenes in Pyrus. Mol. Phylogenetics Evol. 2008, 48, 892–903. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Zhang, J.; Wang, S.; Yang, L.; Luo, Y.; Gao, S.; Zhang, M.; Wu, S.; Hu, S.; Sun, H.; et al. The apricot (Prunus armeniaca L.) genome elucidates Rosaceae evolution and beta-carotenoid synthesis. Hortic. Res. 2019, 6, 128. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, W.; Zhu, D.; Hong, P.; Zhang, S.; Xiao, S.; Tan, Y.; Chen, X.; Xu, L.; Zong, X.; et al. Chromosome-scale genome assembly of sweet cherry (Prunus avium L.) cv. Tieton obtained using long-read and Hi-C sequencing. Hortic. Res. 2020, 7, 122. [Google Scholar] [CrossRef]
- Wang, P.; Yi, S.; Mu, X.; Zhang, J.; Du, J. Chromosome-level genome assembly of Cerasus humilis using PacBio and Hi-C technologies. Front. Genet. 2020, 11, 956. [Google Scholar] [CrossRef]
- Turkoglu, Z.; Bilgener, S.; Ercisli, S.; Yildirim, N. Simple sequence repeat (SSR) analysis for assessment of genetic variability in wild cherry germplasm. J. Appl. Bot. Food Qual. 2013, 85, 229–234. [Google Scholar]
- Favre, A.; Päckert, M.; Pauls, S.U.; Jhnig, S.C.; Uhl, D.; Michalak, I.; Muellner-Riehl, A.N. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. 2015, 90, 236–253. [Google Scholar] [CrossRef]
- Xu, H.; Su, T.; Zhou, Z.K. Leaf and infructescence fossils of Alnus (Betulaceae) from the late Eocene of the southeastern Qinghai-Tibetan Plateau: Alnus from the Eocene of the Qinghai-Tibetan Plateau. J. Syst. Evol. 2018, 57, 105–113. [Google Scholar] [CrossRef]
- Maddison, W.P. Gene trees in species trees. Syst. Biol. 1997, 46, 523–536. [Google Scholar] [CrossRef]
- Wendel, J.F.; Doyle, J.J. Phylogenetic incongruence. In Window into Geno Ehistory and Molecular Evolution; Kluwer Academic Publishers: Norwell, MA, USA, 1998. [Google Scholar]
- Zou, X.H.; Ge, S. Conflicting gene trees and phylogenomics. J. Syst. Evol. 2008, 46, 795–807. [Google Scholar]
- Van Do, T.; Xu, B.; Gao, X.-F. Molecular phylogeny and character evolution of Flemingia (Leguminosae, Papilionoideae, Phaseoleae, Cajaninae) inferred from three cpDNA and nrITS sequence data. Plant Syst. Evol. 2021, 307, 30. [Google Scholar] [CrossRef]
- Li, Y.; Zhai, S.-N.; Qiu, Y.-X.; Guo, Y.-P.; Ge, X.-J.; Comes, H.P. Glacial survival east and west of the ‘Mekong–Salween Divide’ in the Himalaya–Hengduan Mountains region as revealed by AFLPs and cpDNA sequence variation in Sinopodophyllum hexandrum (Berberidaceae). Mol. Phylogenetics Evol. 2011, 59, 412–424. [Google Scholar] [CrossRef] [PubMed]
- López-Pujol, J.; Zhang, F.-M.; Sun, H.-Q.; Ying, T.-S.; Ge, S. Centres of plant endemism in China: Places for survival or for speciation? J. Biogeogr. 2011, 38, 1267–1280. [Google Scholar] [CrossRef]
- Jian, H.-Y.; Tang, K.-X.; Sun, H. Phylogeography of Rosa soulieana (Rosaceae) in the Hengduan Mountains: Refugia and ‘melting’ pots in the Quaternary climate oscillations. Plant Syst. Evol. 2015, 301, 1819–1830. [Google Scholar] [CrossRef]
- Meng, L.; Chen, G.; Li, Z.; Yang, Y.; Wang, Z.; Wang, L. Refugial isolation and range expansions drive the genetic structure of Oxyria sinensis (Polygonaceae) in the Himalaya-Hengduan Mountains. Sci. Rep. 2015, 5, 10396. [Google Scholar] [CrossRef]
- Meng, H.-H.; Zhou, S.-S.; Jiang, X.-L.; Gugger, P.F.; Li, L.; Tan, Y.-H.; Li, J. Are mountaintops climate refugia for plants under global warming? A lesson from high-mountain oaks in tropical rainforest. Alp. Bot. 2019, 129, 175–183. [Google Scholar] [CrossRef]
- He, L.; Wagner, N.D.; Hörandl, E. Restriction-site associated DNA sequencing data reveal a radiation of willow species (Salix L., Salicaceae) in the Hengduan Mountains and adjacent areas. J. Syst. Evol. 2020, 59, 44–57. [Google Scholar] [CrossRef]
- Howe, C.J. Chloroplast Genome; John Wiley & Sons, Ltd.: Chichester, UK, 2016. [Google Scholar]
- Tang, L.; Tang, J.-M.; Tan, S.; Li, J.; Ma, X.; Zhou, Z.-Q. ITS sequence variation and concerted evolution in the natural hybrid species Malus toringoides. Nord. J. Bot. 2015, 33, 109–119. [Google Scholar] [CrossRef]
- Zarrei, M.; Stefanović, S.; Dickinson, T.A. Reticulate evolution in North American black-fruited hawthorns (Crataegus section Douglasia; Rosaceae): Evidence from nuclear ITS2 and plastid sequences. Ann. Bot. 2014, 114, 253–269. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, T.; Wang, Y.; Chen, Q.; Sun, B.; Luo, Y.; Zhang, Y.; Tang, H.; Wang, X. Genetic diversity and domestication footprints of Chinese cherry [Cerasus pseudocerasus (Lindl.) G. Don] as revealed by nuclear microsatellites. Front. Plant Sci. 2018, 9, 238. [Google Scholar] [CrossRef]
- Chen, T.; Li, L.; Zhang, J.; Huang, Z.L.; Zhang, H.W.; Liu, Y.; Chen, Q.; Tang, H.R.; Wang, X.R. Investigation, collection and preliminary evaluation of genetic resources of Chinese cherry [Cerasus pseudocerasus (Lindl.) G.Don]. J. Fruit Sci. 2016, 33, 917–933. [Google Scholar]
- Liu, Y.; Chen, T.; Zhang, J.; Wang, J.; Wang, X.R. Genetic diversity analysis of Chinese cherry landraces (Prunus pseudocerasus) based on phenotypic traits. Acta Hortic. Sinica 2016, 43, 2119–2132. [Google Scholar]
- Meyer, R.S.; Duval, A.E.; Jensen, H.R. Patterns and processes in crop domestication: An historical review and quantitative analysis of 203 global food crops. New Phytol. 2012, 196, 29–48. [Google Scholar] [CrossRef]
- Cao, K.; Zheng, Z.J.; Wang, L.R.; Liu, X.; Zhu, G.R.; Fang, W.C.; Cheng, S.F.; Zeng, P.; Chen, C.W.; Wang, X.W.; et al. Comparative population genomics reveals the domestication history of the peach, Prunus persica, and human influences on perennial fruit crops. Genome Biol. 2014, 15, 415. [Google Scholar] [CrossRef]
- Cornille, A.; Giraud, T.; Smulders, M.J.M.; Roldán-Ruiz, I.; Gladieux, P. The domestication and evolutionary ecology of apples. Trends Genet. 2014, 30, 57–65. [Google Scholar] [CrossRef]
- Duan, N.; Bai, Y.; Sun, H.; Wang, N.; Ma, Y.; Li, M.; Wang, X.; Jiao, C.; Legall, N.; Mao, L.; et al. Genome re-sequencing reveals the history of apple and supports a two-stage model for fruit enlargement. Nat. Commun. 2017, 8, 249. [Google Scholar] [CrossRef]
- Liu, C.J.; Jin, G.Y.; Kong, Z.C. Archaeobotany-Research on Seeds and Fruits; Science Press: Beijing, China, 2008. [Google Scholar]
- Jiang, H.; Yang, J.; Liang, T.; Zhang, Z.; Wang, S.; Qi, X.; Sheng, P. Palaeoethnobotanical analysis of plant remains discovered in the graveyard of the Haihun Marquis, Nanchang, China. Veg. Hist. Archaeobotany 2021, 30, 119–135. [Google Scholar] [CrossRef]
- Feliner, G.N.; Rosselló, J.A. Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Mol. Phylogenet. Evol. 2007, 44, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 2.3, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 7, 1870. [Google Scholar]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Farris, J.S.; Källersjö, M.; Kluge, A.G.; Bult, C. Constructing a Significance Test for Incongruence. Syst. Biol. 1995, 44, 570–572. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b10; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Borchsenius, F. FastGap 1.2; University of Aarhus: Aarhus, Denmark, 2009. [Google Scholar]
- Librado, P.; Rozas, J. Dnasp v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Forster, P.; Bandelt, H.; Röhl, A. Fluxus Technology Ltda. Network 4.2.0.1. 2004. Available online: www.fluxus-engineering.com (accessed on 17 December 2022).
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef]
- Akaike, H.T. A New Look at the Statistical Model Identification; Springer: New York, NY, USA, 1974. [Google Scholar]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Donnelly, P. Case–Control studies of association in structured or admixed populations. Theor. Popul. Biol. 2001, 60, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Harpending, H.C. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum. Biol. 1994, 66, 591–600. [Google Scholar] [PubMed]
- Slatkin, M.; Hudson, R.R. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 1991, 129, 555–562. [Google Scholar] [CrossRef] [PubMed]




| Groups and Lineages | Gene Diversity (h) | Nucleotide Diversity (π) | Tajima’s D (p-Values) | Fu’s Fs (p-Values) | SSD (PSSD) | Raggedness Index (PRag) |
|---|---|---|---|---|---|---|
| ITS sequences | ||||||
| Groups | ||||||
| CC | 0.5378 ± 0.0135 | 0.0062 ± 0.0051 | −1.3853 (0.0620) | −1.1937 (0.3310) | 0.0266 (0.0000) | 0.2003 (0.0000) |
| WC | 0.5057 ± 0.0311 | 0.0076 ± 0.0059 | −1.2024 (0.1070) | −1.6268 (0.2690) | 0.0086 (0.0300) | 0.1315 (0.0300) |
| RC | 0.8730 ± 0.0185 | 0.0447 ± 0.0240 | −0.9768 (0.1730) | −0.7611 (0.4900) | 0.0127 (0.1600) | 0.0213 (0.2200) |
| EC | 0.7381 ± 0.0402 | 0.0230 ± 0.0136 | −1.5179 (0.0320) | −2.1721 (0.2660) | 0.0040 (0.0060) | 0.0273 (0.0000) |
| MC | 0.8916 ± 0.0137 | 0.0837 ± 0.0428 | 0.4441 (0.7490) | 5.4135 (0.9290) | 0.0164 (0.4100) | 0.0201 (0.2900) |
| Lineages | ||||||
| I (CC + WC + RC) | 0.6465 ± 0.0151 | 0.0189 ± 0.0115 | −1.3076 (0.0730) | −14.5974 (0.0060) | 0.0353 (0.5600) | 0.0901 (0.6700) |
| II (EC) | 0.7141 ± 0.0426 | 0.0120 ± 0.0081 | −1.2283 (0.0850) | −14.7058 (0.0030) | 0.0020 (0.7400) | 0.0305 (0.9200) |
| III (RC) | 0.9333 ± 0.0620 | 0.0413 ± 0.0248 | −0.7810 (0.2030) | −0.7369 (0.3280) | 0.0207 (0.4100) | 0.0384 (0.8300) |
| IV (EC) | 0.6667 ± 0.2041 | 0.0055 ± 0.0062 | 1.6330 (0.9650) | 0.5400 (0.5040) | 0.0898 (0.3100) | 0.5556 (0.3400) |
| V (MC) | 0.8916 ± 0.0137 | 0.0837 ± 0.0428 | 0.4441 (0.7280) | 5.4135 (0.9210) | 0.0164 (0.3800) | 0.0201 (0.1900) |
| Chloroplast sequences | ||||||
| Groups | ||||||
| CC | 0.1440 ± 0.0262 | 0.0021 ± 0.0024 | −2.1891 (0.0000) | −6.1829 (0.0020) | 0.0003 (0.3500) | 0.5633 (0.7100) |
| WC | 0.6528 ± 0.0375 | 0.0275 ± 0.0153 | 0.4693 (0.7110) | −5.8760 (0.0720) | 0.0854 (0.1200) | 0.1539 (0.0700) |
| RC | 0.9279 ± 0.0135 | 0.0436 ± 0.0230 | −1.1777 (0.0910) | 0.6622 (0.6440) | 0.0123 (0.0100) | 0.0167 (0.0900) |
| EC | 0.6771 ± 0.0444 | 0.0391 ± 0.0209 | −0.1564 (0.5040) | 0.6627 (0.6380) | 0.0617 (0.3400) | 0.1037 (0.3000) |
| MC | 0.8559 ± 0.0216 | 0.0435 ± 0.0231 | 0.0161 (0.5700) | 0.6622 (0.6460) | 0.0308 (0.1000) | 0.0371 (0.1600) |
| Lineages | ||||||
| I (CC + WC + RC) | 0.4217 ± 0.0258 | 0.0167 ± 0.0100 | −1.2293 (0.0770) | −26.1216 (0.0000) | 0.0523 (0.0900) | 0.2725 (0.4000) |
| II (CC + WC + RC) | 0.8023 ± 0.0426 | 0.0112 ± 0.0074 | −1.8758 (0.0080) | −10.5326 (0.0000) | 0.0005 (0.8800) | 0.0449 (0.5900) |
| III (EC) | 0.8603 ± 0.0554 | 0.0253 ± 0.0150 | −0.4424 (0.3410) | −0.6875 (0.3740) | 0.0414 (0.0700) | 0.0628 (0.6300) |
| IV (EC) | 0.5404 ± 0.0521 | 0.0144 ± 0.0090 | −0.2552 (0.4640) | 1.4514 (0.7710) | 0.3687 (0.0000) | 0.1874 (1.0000) |
| V (RC) | 0.8629 ± 0.0502 | 0.0360 ± 0.0199 | −0.8824 (0.1980) | −2.7766 (0.1400) | 0.0422 (0.0200) | 0.0710 (0.0300) |
| VI (EC) | 0.6667 ± 0.2041 | 0.0046 ± 0.0052 | 1.6330 (0.9610) | 0.5400 (0.4960) | 0.0898 (0.3200) | 0.5556 (0.3100) |
| VII (MC) | 0.8576 ± 0.0210 | 0.0432 ± 0.0230 | 0.0019 (−0.5700) | 0.6605 (0.6660) | 0.0305 (0.1400) | 0.0365 (0.2700) |
| Source of Variation | Among Groups (%) ITS/cpDNA | Among Populations within Groups (%) ITS/cpDNA | Within Populations (%) ITS/cpDNA | FCT ITS/cpDNA | FSC ITS/cpDNA | FST ITS/cpDNA |
|---|---|---|---|---|---|---|
| Five groups | 71.97/57.92 | 18.63/16.04 | 9.4/26.04 | 0.7196/0.5792 | 0.6647/0.3811 | 0.9060/0.7396 |
| Lineages | 84.02/75.06 | 10.75/10.75 | 5.24/14.18 | 0.8402/0.7506 | 0.6723/0.4312 | 0.9476/0.8582 |
| Sub-lineages | 79.98/82.35 | 11.40/7.30 | 8.62/10.35 | 0.7998/0.8235 | 0.5692/0.4137 | 0.9138/0.8965 |
| Three groups (CC, WC, RC) | 23.98/30.05 | 20.61/11.78 | 55.41/58.17 | 0.2398/0.3005 | 0.2712/0.1684 | 0.4460/0.4183 |
| CC (NEC-C, LFZ-C, YGP-C, QLM-C) | 21.53/3.60 | 22.07/31.05 | 56.4/65.35 | 0.2153/0.0360 | 0.2813/0.3221 | 0.4360/0.3465 |
| WC (LFZ-W, YGP-W, QLM-W) | 26.73/2.69 | 31.02/19.29 | 42.25/78.01 | 0.2674/0.0269 | 0.4234/0.1983 | 0.5775/0.2199 |
| RC (LFZ-R, YGP-R, QLM-R) | 18.41/18.39 | 40.67/19.94 | 40.92/61.67 | 0.1841/0.1839 | 0.4985/0.2443 | 0.5908/0.3833 |
| Groups and Lineages | Gene Diversity (h) | Nucleotide Diversity (π) | Tajima’s D (p-Values) | Fu’s Fs (p-Values) | SSD (PSSD) | Raggedness Index (PRag) |
|---|---|---|---|---|---|---|
| ITS sequences | ||||||
| Geographic subgroups of CC, WC and RC | ||||||
| NEC-C | 0.4487 ± 0.0313 | 0.0037 ± 0.0037 | 1.5274 (0.9460) | 2.0922 (0.7810) | 0.0128 (0.0300) | 0.2119 (0.0100) |
| LFZ-C | 0.2647 ± 0.0541 | 0.0054 ± 0.0047 | −1.6581 (0.0210) | −0.6743 (0.3650) | 0.0044 (0.3800) | 0.3250 (0.5500) |
| YGP-C | 0.5342 ± 0.0317 | 0.0065 ± 0.0053 | −1.4573 (0.0480) | −0.3691 (0.4330) | 0.0191 (0.0300) | 0.1744 (0.0000) |
| QLM-C | 0.5508 ± 0.0701 | 0.0051 ± 0.0046 | 0.5080 (0.7810) | 0.5710 (0.5820) | 0.0224 (0.0400) | 0.1840 (0.0700) |
| LFZ-W | 0.5395 ± 0.0321 | 0.0086 ± 0.0064 | −1.1186 (0.1330) | −1.3437 (0.2970) | 0.0104 (0.0300) | 0.1286 (0.0300) |
| YGP-W | 0.5333 ± 0.1721 | 0.0044 ± 0.0048 | 0.8506 (0.8870) | 0.6254 (0.4680) | 0.0303 (0.3500) | 0.2889 (0.5100) |
| QLM-W | 0.2540 ± 0.0953 | 0.0021 ± 0.0027 | −0.0187 (0.3410) | 0.4480 (0.3650) | 0.2754 (0.1900) | 0.3066 (0.1200) |
| LFZ-R | 0.8434 ± 0.0338 | 0.0249 ± 0.0146 | −1.3240 (0.0750) | −2.6787 (0.1540) | 0.0037 (0.8400) | 0.0141 (0.9600) |
| YGP-R | 0.8536 ± 0.0228 | 0.0540 ± 0.0286 | 0.2359 (0.6810) | 0.3402 (0.5990) | 0.0419 (0.0500) | 0.0554 (0.0400) |
| QLM-R | 0.3524 ± 0.1009 | 0.0109 ± 0.0077 | −0.9248 (0.1990) | −0.5961 (0.3790) | 0.0331 (0.2200) | 0.3015 (0.5200) |
| Sub-lineages | ||||||
| i | 0.5892 ± 0.0156 | 0.0117 ± 0.0079 | −1.2786 (0.067) | −12.4953 (0.0030) | 0.0198 (0.0000) | 0.1181 (0.0000) |
| ii | 0.7051 ± 0.0476 | 0.0201 ± 0.0123 | −0.9626 (0.178) | −0.3677 (0.4960) | 0.0342 (0.2500) | 0.0832 (0.5000) |
| iii | 0.9333 ± 0.0620 | 0.0413 ± 0.0248 | −0.7810 (0.235) | −0.7369 (0.3010) | 0.0207 (0.4100) | 0.0383(0.8300) |
| Chloroplast sequences | ||||||
| Geographic subgroups of CC, WC and RC | ||||||
| NEC-C | 0.0190 ± 0.0186 | 0.0003 ± 0.0008 | −1.3716 (0.0280) | −1.5192 (0.0590) | 0.0005 (0.0500) | 0.9603 (0.9200) |
| LFZ-C | 0.1823 ± 0.0178 | 0.0013 ± 0.0018 | −2.2798 (0.0020) | 0.4346 (0.3970) | 0.0005 (0.0400) | 0.9646 (0.9300) |
| YGP-C | 0.4059 ± 0.0615 | 0.0030 ± 0.0030 | −0.5075 (0.3220) | −0.8229 (0.3060) | 0.0063 (0.1000) | 0.1660 (0.3000) |
| QLM-C | 0.1693 ± 0.0841 | 0.0078 ± 0.0058 | −1.4709 (0.0680) | 2.0832 (0.8570) | 0.0273 (0.1200) | 0.7206 (0.6300) |
| LFZ-W | 0.5920 ± 0.0437 | 0.0248 ± 0.0140 | 0.3762 (0.7170) | −2.1202 (0.2860) | 0.1113 (0.1300) | 0.2040 (0.1300) |
| YGP-W | 0.9333 ± 0.1217 | 0.0276 ± 0.0185 | 1.7737 (0.9780) | −0.7460 (0.2200) | 0.0323 (0.5900) | 0.1289 (0.5200) |
| QLM-W | 0.8466 ± 0.0454 | 0.0390 ± 0.0215 | 1.9323 (0.9840) | 1.4395 (0.7800) | 0.0641 (0.0100) | 0.1753 (0.0300) |
| LFZ-R | 0.8790 ± 0.0254 | 0.0277 ± 0.0155 | −1.1694 (0.1210) | −4.1397 (0.0770) | 0.0127 (0.5400) | 0.0299 (0.4800) |
| YGP-R | 0.8176 ± 0.0433 | 0.0427 ± 0.0227 | −0.6262 (0.3020) | −7.1104 (0.0350) | 0.0488 (0.0000) | 0.0551 (0.0000) |
| QLM-R | 0.9302 ± 0.0213 | 0.0365 ± 0.0200 | 0.3232 (0.6920) | −3.8279 (0.0710) | 0.0051 (0.8300) | 0.0087 (0.9700) |
| Sub-lineages | ||||||
| i | 0.1590 ± 0.0223 | 0.0017 ± 0.0022 | −1.8953 (0.002) | −21.5495 (0.0000) | 0.3400 (0.0014) | 0.5549 (0.6400) |
| ii | 0.9168 ± 0.0152 | 0.0206 ± 0.0120 | −1.2586 (0.095) | −3.0942 (0.0800) | 0.0083 (0.1700) | 0.0309 (0.2700) |
| iii | 0.8965 ± 0.0218 | 0.0256 ± 0.0145 | −1.7462 (0.014) | −1.8477 (0.2300) | 0.0054 (0.7900) | 0.0135 (0.8900) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.; Chen, Q.; Zhang, J.; Wang, Y.; Wang, H.; Zhang, Y.; Luo, Y.; Tang, H.; Wang, X. Phylogeography of 912 Cherry Accessions Insight into Independent Origins of Fruiting Cherries and Domestication Footprints of Cultivated Chinese Cherry (Prunus pseudocerasus Lindl.). Plants 2023, 12, 2258. https://doi.org/10.3390/plants12122258
Chen T, Chen Q, Zhang J, Wang Y, Wang H, Zhang Y, Luo Y, Tang H, Wang X. Phylogeography of 912 Cherry Accessions Insight into Independent Origins of Fruiting Cherries and Domestication Footprints of Cultivated Chinese Cherry (Prunus pseudocerasus Lindl.). Plants. 2023; 12(12):2258. https://doi.org/10.3390/plants12122258
Chicago/Turabian StyleChen, Tao, Qing Chen, Jing Zhang, Yan Wang, Hao Wang, Yong Zhang, Ya Luo, Haoru Tang, and Xiaorong Wang. 2023. "Phylogeography of 912 Cherry Accessions Insight into Independent Origins of Fruiting Cherries and Domestication Footprints of Cultivated Chinese Cherry (Prunus pseudocerasus Lindl.)" Plants 12, no. 12: 2258. https://doi.org/10.3390/plants12122258
APA StyleChen, T., Chen, Q., Zhang, J., Wang, Y., Wang, H., Zhang, Y., Luo, Y., Tang, H., & Wang, X. (2023). Phylogeography of 912 Cherry Accessions Insight into Independent Origins of Fruiting Cherries and Domestication Footprints of Cultivated Chinese Cherry (Prunus pseudocerasus Lindl.). Plants, 12(12), 2258. https://doi.org/10.3390/plants12122258








