Root and Leaf Anatomy, Ion Accumulation, and Transcriptome Pattern under Salt Stress Conditions in Contrasting Genotypes of Sorghum bicolor
Abstract
:1. Introduction
2. Results
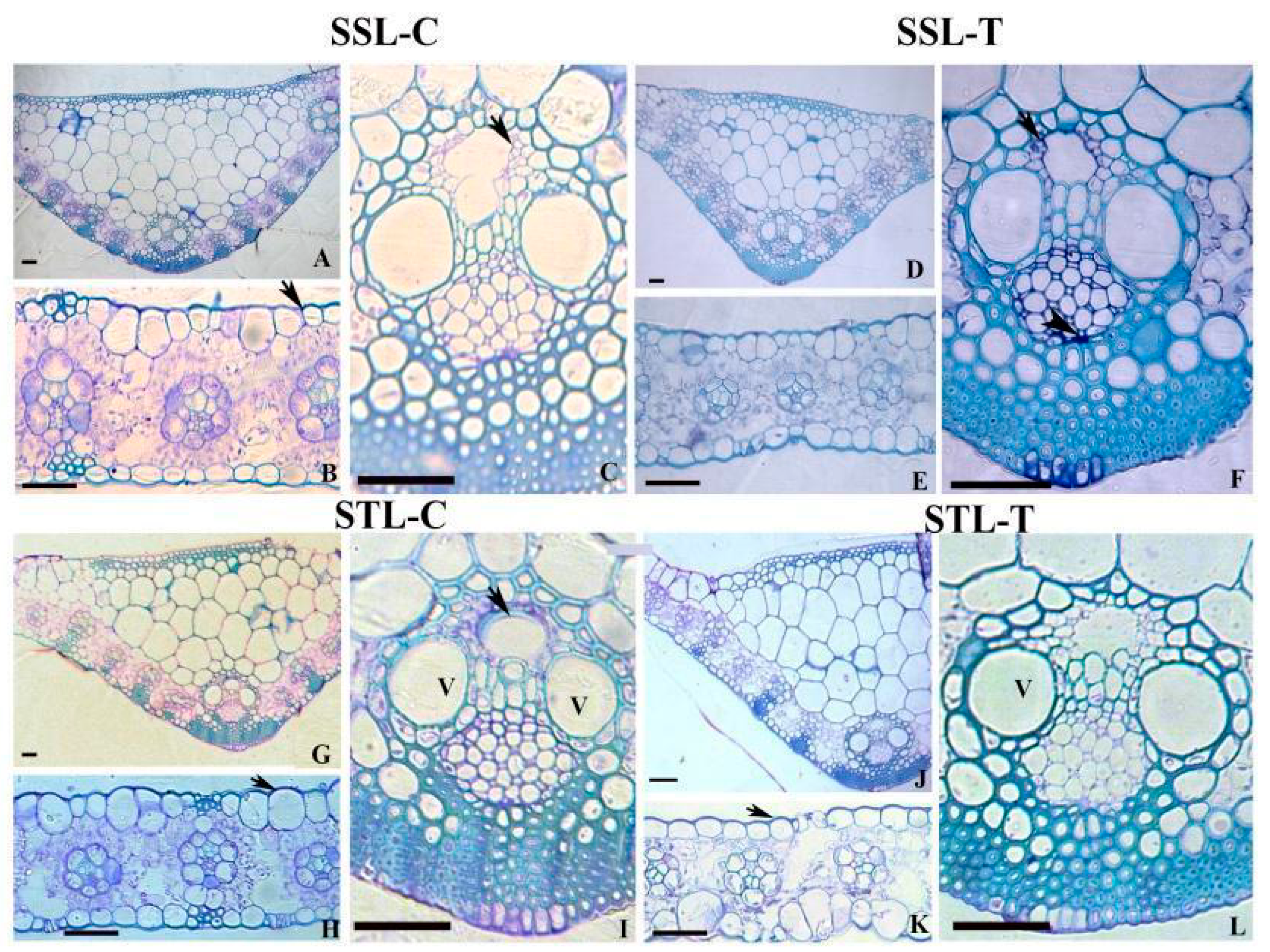
2.1. Leaf Anatomy in SS and ST Lines under Control and Stress Conditions
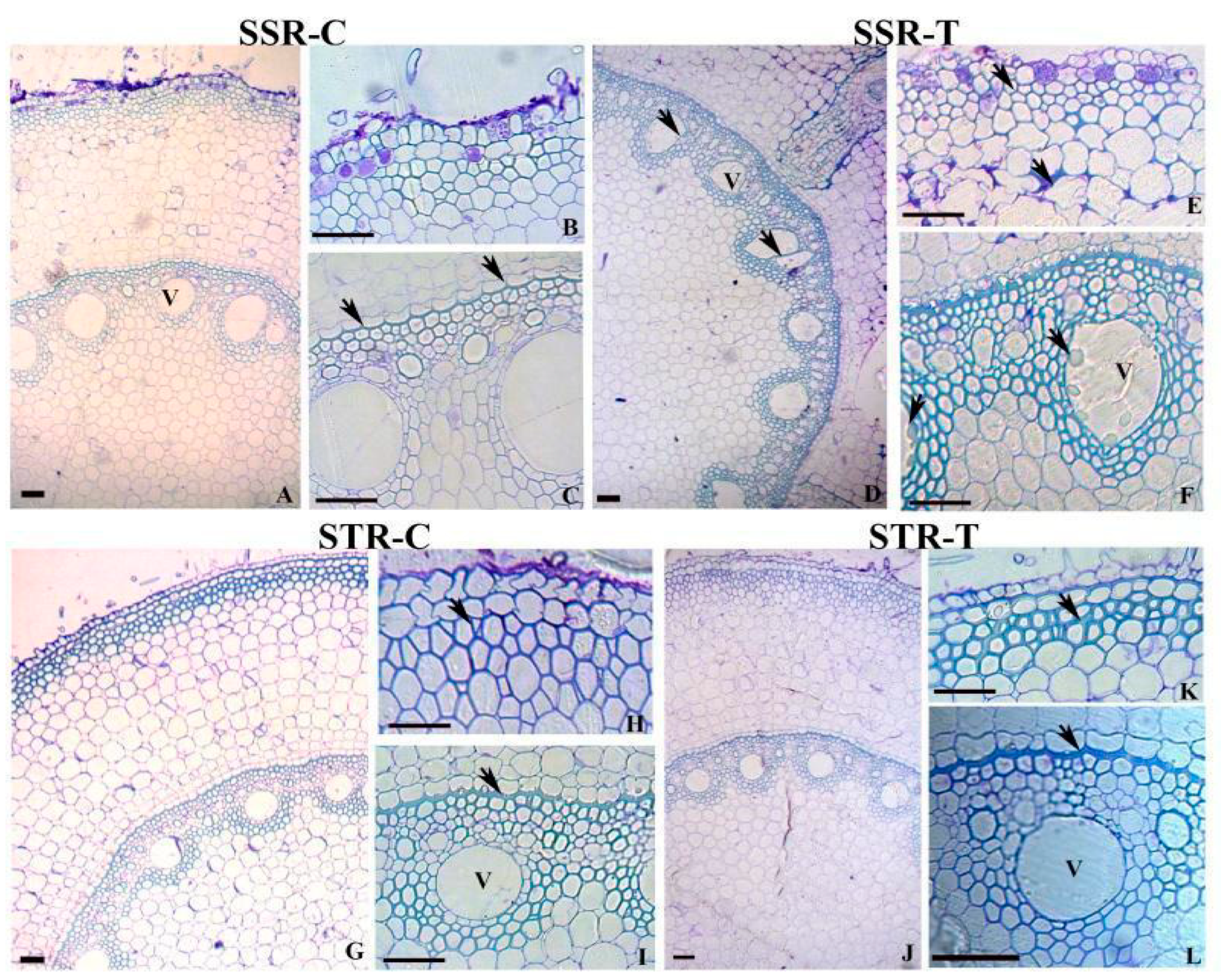
2.2. Root Anatomy in SS and ST Lines under Stress Conditions
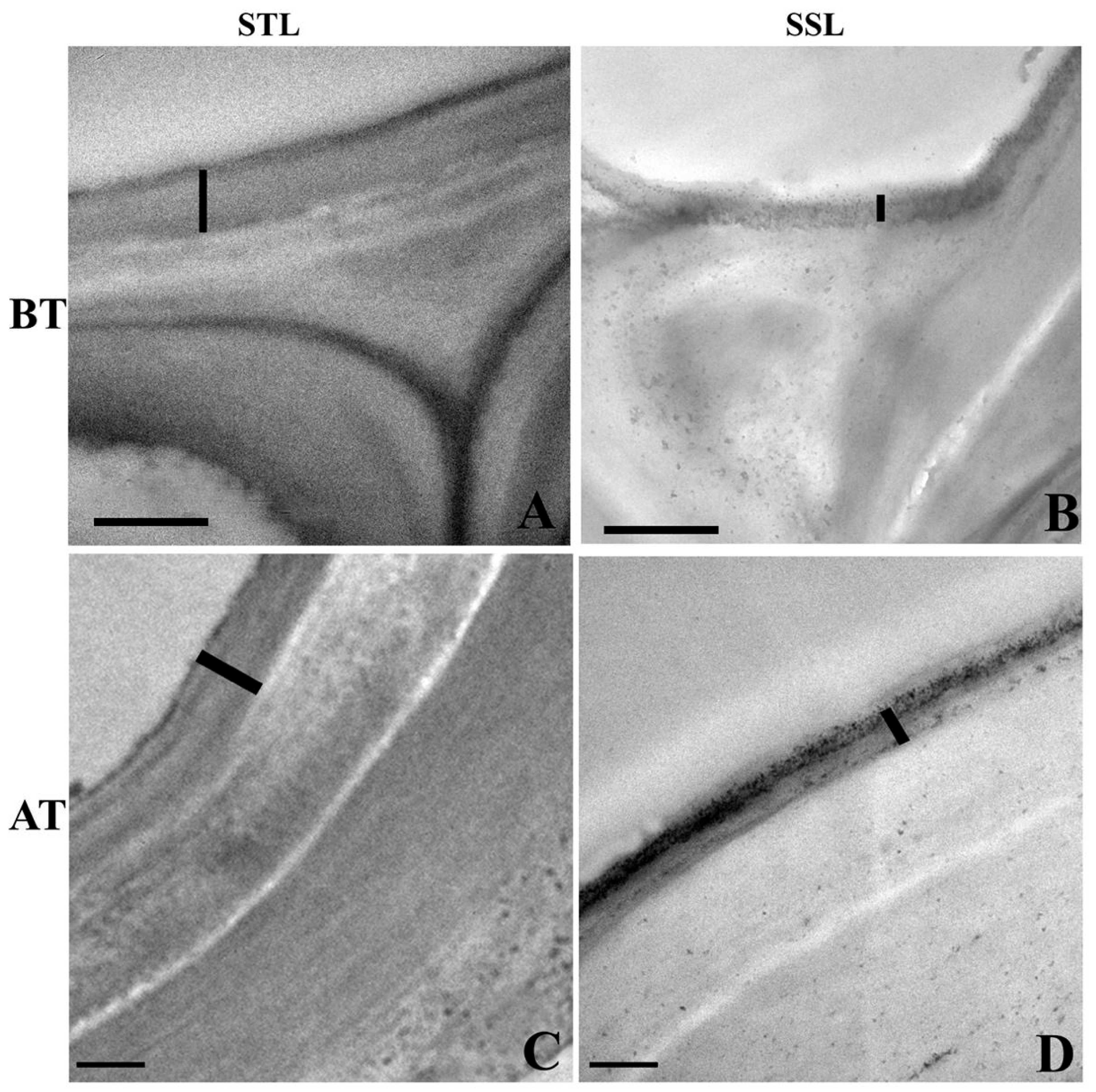
2.3. Ultra-Structural Changes in the Leaf Cuticle
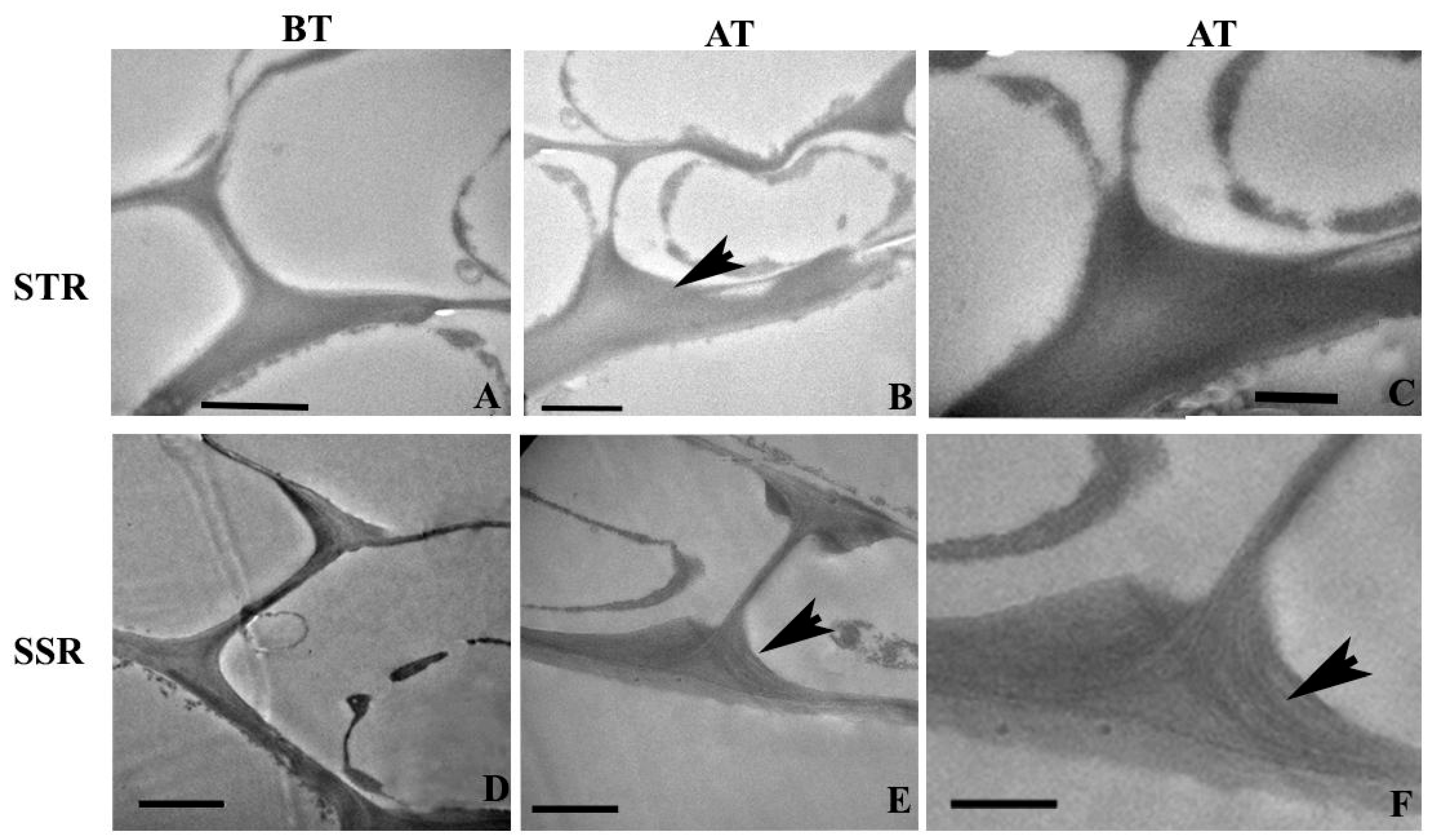
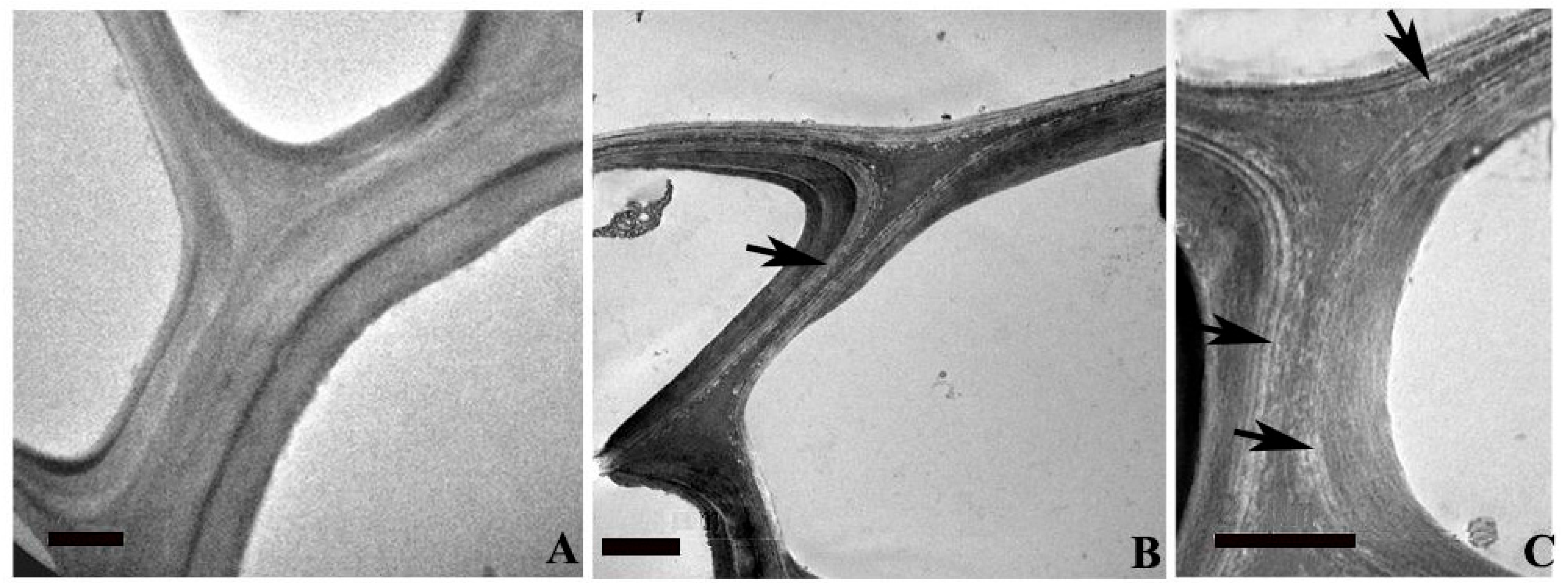
2.4. Ultra-Structural Changes and Lignin Distribution Pattern in the Root
2.5. Accumulation of Ions in Different Organs in SS, SSN, ST and STN Plants
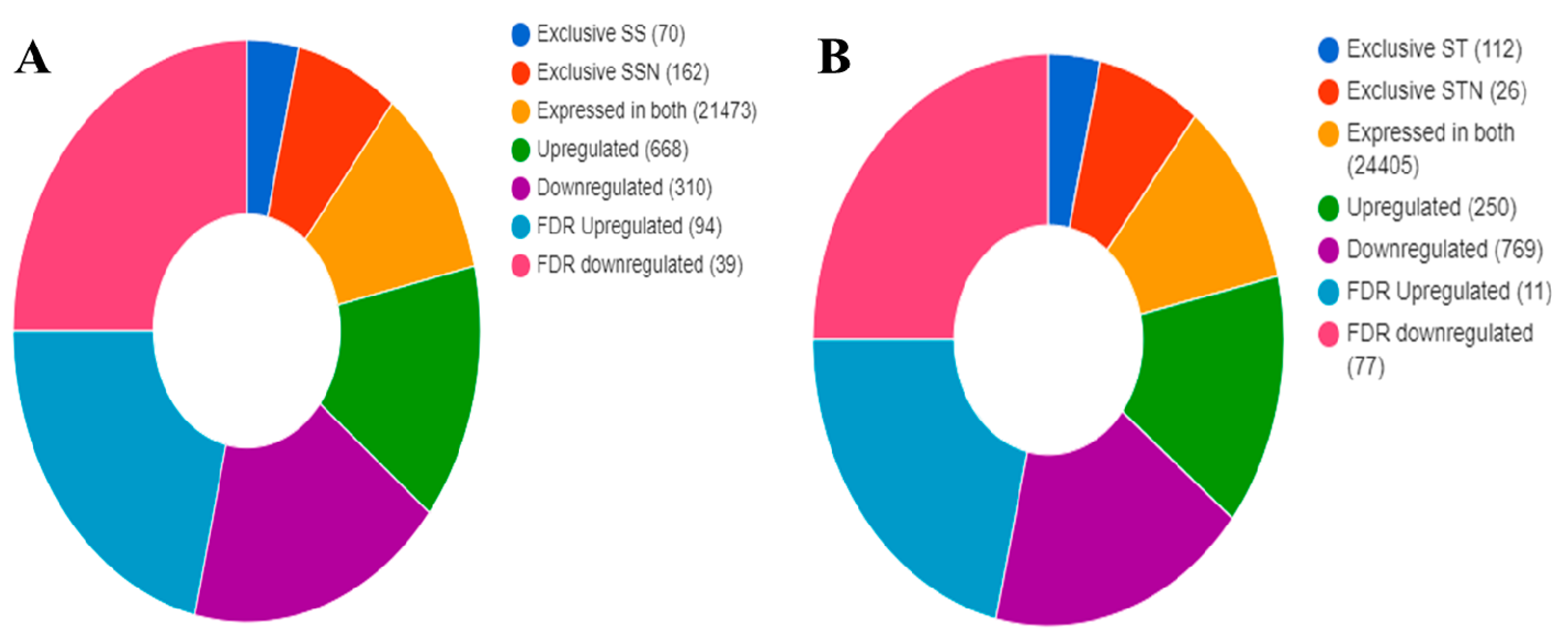
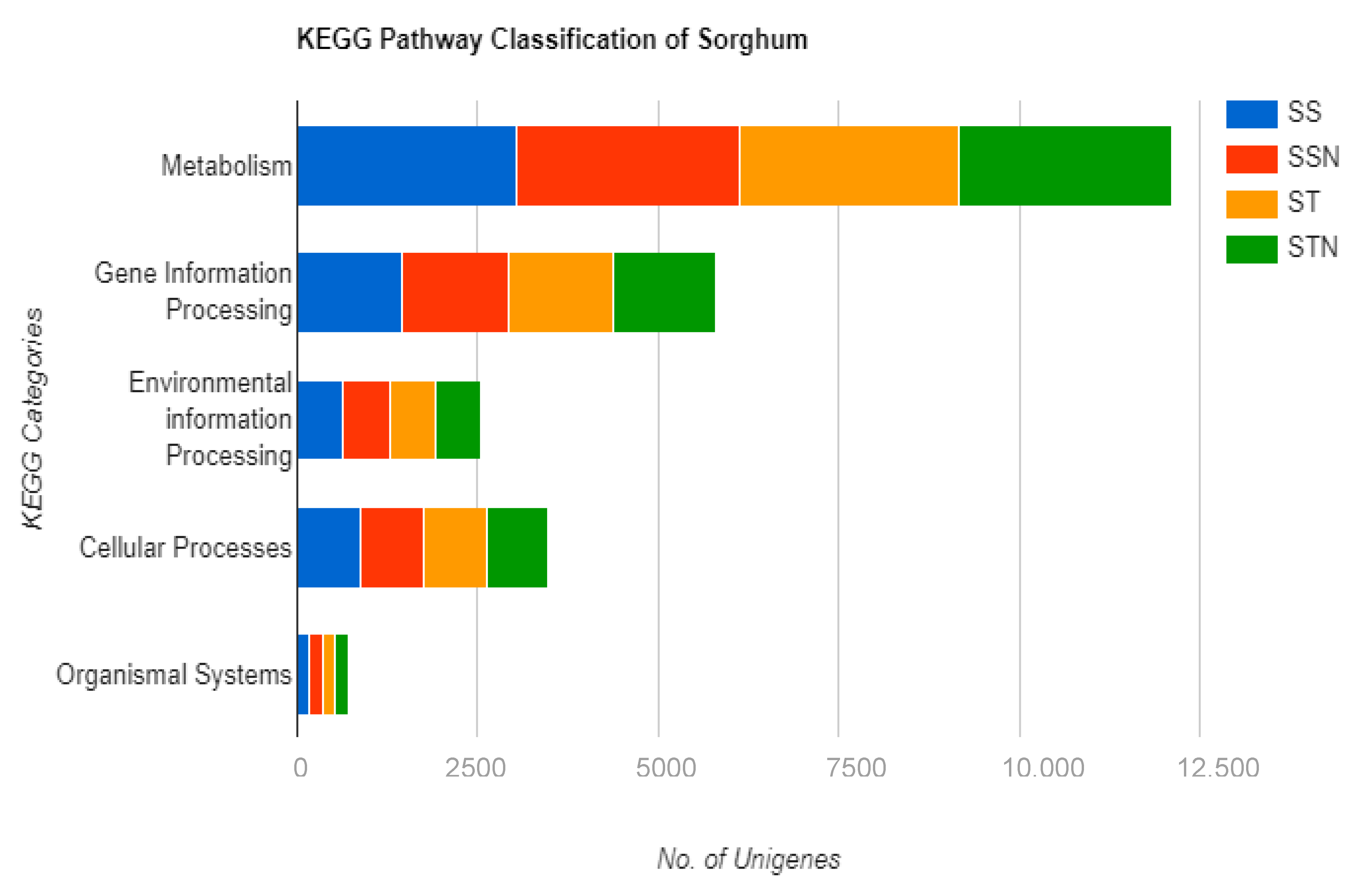
2.6. Identification of DEGs, KEGG, and GO Pathways
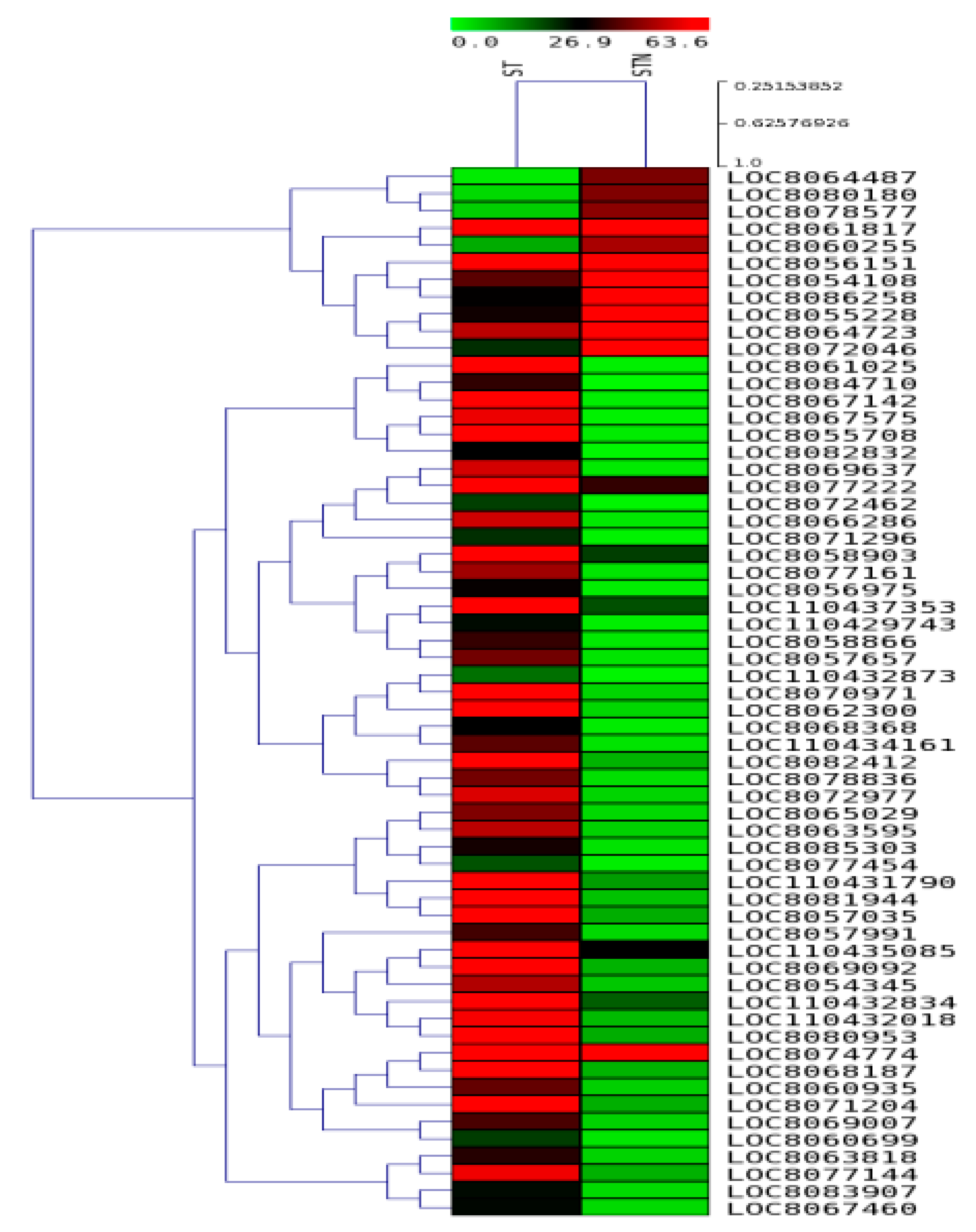
2.7. DEGs in SS and ST Genotypes under Control and Salt-Treatment
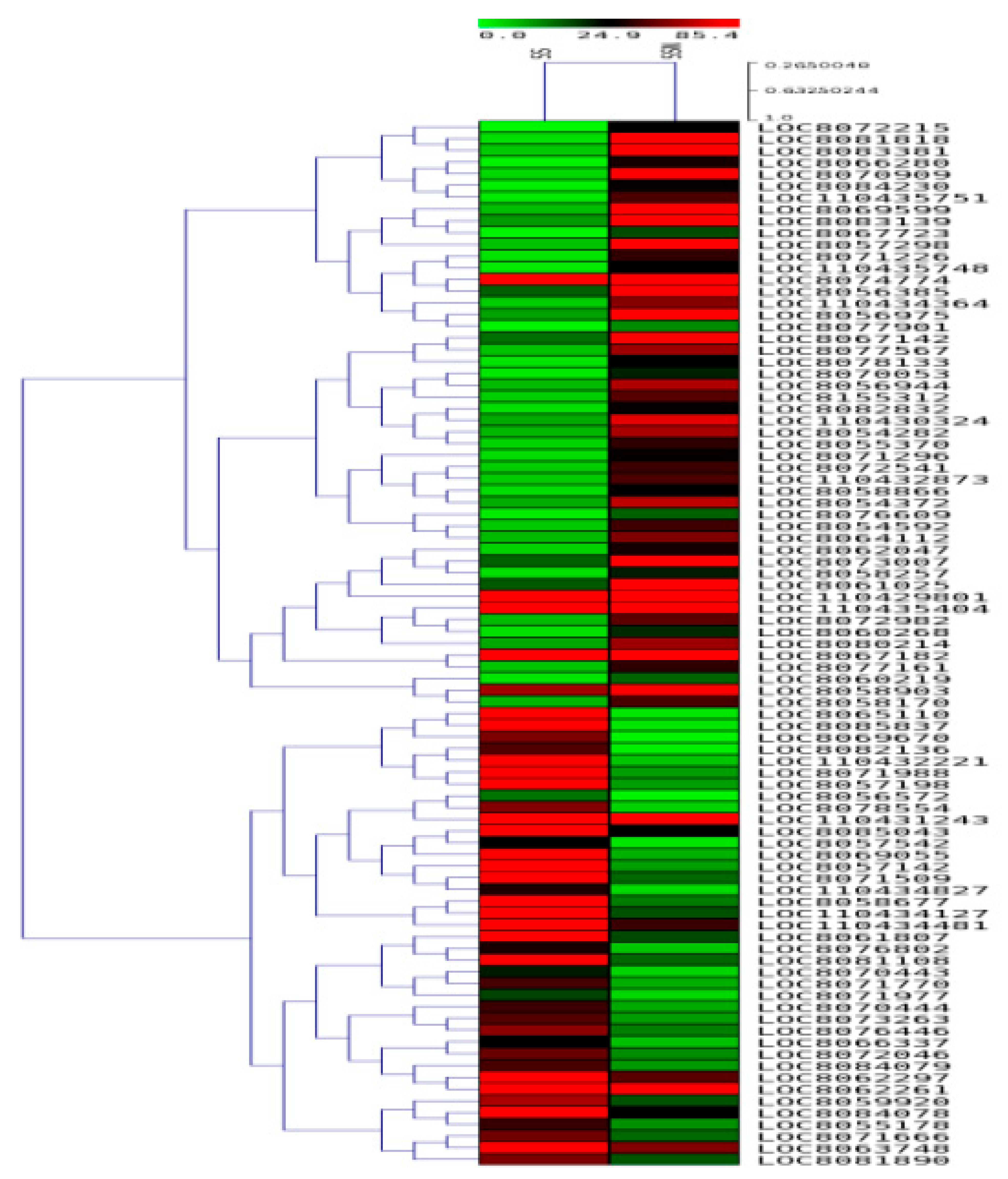
2.8. Validation of Selected DEGs by qRT-pCR
3. Discussion
3.1. Anatomical Modifications in Tolerant and Susceptible Sorghum Genotypes
3.2. Estimation of Na+ and K+ Ions in SS and ST Lines
3.3. Transcript Profiling
3.4. Validation of Genes via qRT-PCR
4. Material and Methods
4.1. Sample Preparation for Microscopy
4.2. Ion Analysis
4.3. Plant Material and RNA Isolation
4.4. Nextseq Paired End (PE) Library Preparation and RNA-Seq
4.5. Downstream Annotation and Bioinformatic Analysis
4.6. Downstream Annotation and Bioinformatic Analysis
4.7. DEG, KEGG and GO Enrichment and Pathway Analysis
4.8. GO Enrichment and Pathway Analysis
4.9. Validation of DEGs by qRT-PCR Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, J.-K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, P.K.; Shukla, P.S.; Gupta, K.; Jha, B. Bioengineering for Salinity Tolerance in Plants: State of the Art. Mol. Biotechnol. 2012, 54, 102–123. [Google Scholar] [CrossRef]
- Wani, S.H.; Kumar, V.; Khare, T.; Guddimalli, R.; Parveda, M.; Solymosi, K.; Suprasanna, P.; Kishor, P.B.K. Engineering salinity tolerance in plants: Progress and prospects. Planta 2020, 251, 76. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Ahmad, P.; Abdel Latef, A.A.; Hashem, A.; Abd Allah, E.F.; Gucel, S.; Tran, L.S. Nitric Oxide Mitigates Salt Stress by Regulating Levels of Osmolytes and Antioxidant Enzymes in Chickpea. Front. Plant Sci. 2016, 7, 347. [Google Scholar] [CrossRef] [Green Version]
- Tavakkoli, E.; Rengasamy, P.; McDonald, G.K. High concentrations of Na+ and Cl− ions in soil solution have simultaneous detrimental effects on growth of faba bean under salinity stress. J. Exp. Bot. 2010, 61, 4449–4459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhtar, N.; Hameed, M.; Nawaz, F.; Ahmad, K.S.; Hamid, A.; Segovia-Salcedo, C.; Shahnaz, M.M. Leaf anatomical and biochemical adaptations in Typha domingensis Pers. ecotypes for salinity tolerance. Bot. Sci. 2017, 95, 807–821. [Google Scholar] [CrossRef] [Green Version]
- Nassar, R.M.A.; Kamel, H.A.; Ghoniem, A.E.; Alarcón, J.J.; Sekara, A.; Ulrichs, C.; Abdelhamid, M.T. Physiological and Anatomical Mechanisms in Wheat to Cope with Salt Stress Induced by Seawater. Plants 2020, 9, 237. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; White, J.F.; Li, C. Fungal endophyte Epichloë bromicola infection regulates anatomical changes to account for salt stress tolerance in wild barley (Hordeum brevisubulatum). Plant Soil 2021, 461, 533–546. [Google Scholar] [CrossRef]
- Janz, D.; Lautner, S.; Wildhagen, H.; Behnke, K.; Schnitzler, J.; Rennenberg, H.; Fromm, J.; Polle, A. Salt stress induces the formation of a novel type of ‘pressure wood’ in two Populus species. New Phytol. 2011, 194, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Assaha, D.V.M.; Ueda, A.; Saneoka, H.; Al-Yahyai, R.; Yaish, M.W. The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front. Physiol. 2017, 8, 509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, P.S.; Jogeswar, G.; Rasineni, G.K.; Maheswari, M.; Reddy, A.R.; Varshney, R.K.; Kishor, P.K. Proline over-accumulation alleviates salt stress and protects photosynthetic and antioxidant enzyme activities in transgenic sorghum [Sorghum bicolor (L.) Moench]. Plant Physiol. Biochem. 2015, 94, 104–113. [Google Scholar] [CrossRef] [Green Version]
- Shabala, S.; Wu, H.; Bose, J. Salt stress sensing and early signalling events in plant roots: Current knowledge and hypothesis. Plant Sci. 2015, 241, 109–119. [Google Scholar] [CrossRef]
- Shabala, S. Ionic and osmotic components of salt stress specifically modulate net ion fluxes from bean leaf mesophyll. Plant Cell Environ. 2000, 23, 825–837. [Google Scholar] [CrossRef]
- Pandolfi, C.; Pottosin, I.; Cuin, T.; Mancuso, S.; Shabala, S. Specificity of Polyamine Effects on NaCl-induced Ion Flux Kinetics and Salt Stress Amelioration in Plants. Plant Cell Physiol. 2010, 51, 422–434. [Google Scholar] [CrossRef]
- Kumari, P.H.; Kumar, S.A.; Ramesh, K.; Reddy, P.S.; Nagaraju, M.; Prakash, A.B.; Shah, T.; Henderson, A.; Srivastava, R.K.; Rajasheker, G.; et al. Genome-Wide Identification and Analysis of Arabidopsis Sodium Proton Antiporter (NHX) and Human Sodium Proton Exchanger (NHE) Homologs in Sorghum bicolor. Genes 2018, 9, 236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.A.; Kumari, P.H.; Nagaraju, M.; Reddy, P.S.; Dheeraj, T.D.; Mack, A.; Katam, R.; Kishor, P.B.K. Genome-wide identification and multiple abiotic stress transcript profiling of potassium transport gene homologs in Sorghum bicolor. Front. Plant Sci. 2022, 13, 3088. [Google Scholar] [CrossRef]
- Miranda, R.D.S.; Mesquita, R.O.; Costa, J.H.; Alvarez-Pizarro, J.C.; Prisco, J.T.; Gomes-Filho, E. Integrative Control Between Proton Pumps and SOS1 Antiporters in Roots is Crucial for Maintaining Low Na+ Accumulation and Salt Tolerance in Ammonium-Supplied Sorghum bicolor. Plant Cell Physiol. 2017, 58, 522–536. [Google Scholar] [CrossRef] [Green Version]
- Shahzad, B.; Yun, P.; Shabala, L.; Zhou, M.; Sellamuthu, G.; Venkataraman, G.; Chen, Z.H.; Shabala, S.; Wu, H. Unravelling the physiological basis of salinity stress tolerance in cultivated and wild rice species. Funct. Plant Biol. 2022, 49, 351–364. [Google Scholar] [CrossRef]
- Seifikalhor, M.; Aliniaeifard, S.; Shomali, A.; Azad, N.; Hassani, B.; Lastochkina, O.; Li, T. Calcium signaling and salt tolerance are diversely entwined in plants. Plant Signal. Behav. 2019, 14, 1665455. [Google Scholar] [CrossRef]
- El Mahi, H.; Pérez-Hormaeche, J.; De Luca, A.; Villalta, I.; Espartero, J.; Gámez-Arjona, F.; Fernández, J.L.; Bundó, M.; Mendoza, I.; Mieulet, D.; et al. A Critical Role of Sodium Flux via the Plasma Membrane Na+/H+ Exchanger SOS1 in the Salt Tolerance of Rice. Plant Physiol. 2019, 180, 1046–1065. [Google Scholar] [CrossRef] [Green Version]
- Kumari, S.; Chhillar, H.; Chopra, P.; Khanna, R.R.; Khan, M.I.R. Potassium: A track to develop salinity tolerant plants. Plant Physiol. Biochem. 2021, 167, 1011–1023. [Google Scholar] [CrossRef]
- Zhu, M.; Shabala, L.; Cuin, A.T.; Huang, X.; Zhou, M.; Munns, R.; Shabala, S. Naxloci affect SOS1-like Na+/H+exchanger expression and activity in wheat. J. Exp. Bot. 2015, 67, 835–844. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [Green Version]
- Guddimalli, R.; Somanaboina, A.K.; Palle, S.R.; Edupuganti, S.; Kummari, D.; Palakolanu, S.R.; Naravula, J.; Gandra, J.; Qureshi, I.A.; Marka, N.; et al. Overexpression of RNA-binding bacterial chaperones in rice leads to stay-green phenotype, improved yield and tolerance to salt and drought stresses. Physiol. Plant. 2021, 173, 1351–1368. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.; Zhang, W.-H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef] [Green Version]
- Song, Q.; Joshi, M.; Joshi, V. Transcriptomic Analysis of Short-Term Salt Stress Response in Watermelon Seedlings. Int. J. Mol. Sci. 2020, 21, 6036. [Google Scholar] [CrossRef]
- Haussmann, B.I.G.; Obilana, A.B.; Ayiecho, P.O.; Blum, A.; Schipprack, W.; Geiger, H.H. Yield and Yield Stability of Four Population Types of Grain Sorghum in a Semi-Arid Area of Kenya. Crop. Sci. 2000, 40, 319–329. [Google Scholar] [CrossRef]
- Rani, C.R.; Reema, C.; Alka, S.P.K.S. Salt tolerance of Sorghum bicolor cultivars during germination and seedling growth. Res. J. Recent. Sci. 2012, 2277, 2502. [Google Scholar]
- Wang, H.; Chen, G.; Zhang, H.; Liu, B.; Yang, Y.; Qin, L.; Chen, E.; Guan, Y. Identification of QTLs for salt tolerance at germination and seedling stage of Sorghum bicolor L. Moench. Euphytica 2013, 196, 117–127. [Google Scholar] [CrossRef]
- Bavei, V.; Shiran, B.; Arzani, A. Evaluation of salinity tolerance in sorghum (Sorghum bicolor L.) using ion accumulation, proline and peroxidase criteria. Plant Growth Regul. 2011, 64, 275–285. [Google Scholar] [CrossRef]
- Nawaz, K.; Talat, A.; Hussain, K.; Majeed, A. Induction of salt tolerance in two cultivars of sorghum (Sorghum bicolor L.) by exogenous application of proline at seedling stage. World Appl. Sci. J. 2010, 10, 93–99. [Google Scholar]
- Johnson, S.M.; Lim, F.-L.; Finkler, A.; Fromm, H.; Slabas, A.R.; Knight, M.R. Transcriptomic analysis of Sorghum bicolor responding to combined heat and drought stress. BMC Genom. 2014, 15, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Sui, N.; Yang, Z.; Liu, M.; Wang, B. Identification and transcriptomic profiling of genes involved in increasing sugar content during salt stress in sweet sorghum leaves. BMC Genom. 2015, 16, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Shang, X.; Sun, M.; Tang, S.; Khan, A.; Zhang, D.; Yan, H.; Jiang, Y.; Yu, F.; Wu, Y.; et al. Comparative transcriptome analysis of two sweet sorghum genotypes with different salt tolerance abilities to reveal the mechanism of salt tolerance. Int. J. Mol. Sci. 2022, 23, 2272. [Google Scholar] [CrossRef]
- Balasubramanian, V.; Vashisht, D.; Cletus, J.; Sakthivel, N.; Balasubramanian, V.; Vashisht, D.; Cletus, J.; Sakthivel, N. Plant β-1,3-glucanases: Their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol. Lett. 2012, 34, 1983–1990. [Google Scholar] [CrossRef]
- Razzaque, S.; Elias, S.M.; Haque, T.; Biswas, S.; Jewel, G.M.N.A.; Rahman, S.; Weng, X.; Ismail, A.M.; Walia, H.; Juenger, T.E.; et al. Gene Expression analysis associated with salt stress in a reciprocally crossed rice population. Sci. Rep. 2019, 9, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Svensson, B.; Svendsen, I.; Hoejrup, P.; Roepstorff, P.; Ludvigsen, S.; Poulsen, F.M. Primary structure of barwin: A barley seed protein closely related to the C-terminal domain of proteins encoded by wound-induced plant genes. Biochemistry 1992, 31, 8767–8770. [Google Scholar] [CrossRef]
- Jin, T.; Sun, Y.; Zhao, R.; Shan, Z.; Gai, J.; Li, Y. Overexpression of peroxidase gene GsPRX9 confers salt tolerance in soybean. Int. J. Mol. Sci. 2019, 20, 3745. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Yu, Y.; Liu, Q.; Deng, S.; Jin, X.; Yin, Y.; Guo, J.; Li, N.; Liu, Y.; Han, S.; et al. A Na2CO3-Responsive Chitinase Gene from Leymus chinensis Improve Pathogen Resistance and Saline-Alkali Stress Tolerance in Transgenic Tobacco and Maize. Front. Plant Sci. 2020, 11, 504. [Google Scholar] [CrossRef]
- de Abreu-Neto, J.B.; Turchetto-Zolet, A.C.; de Oliveira, L.F.V.; Zanettini, M.H.B.; Margis-Pinheiro, M. Heavy metal-associated isoprenylated plant protein (HIPP): Characterization of a family of proteins exclusive to plants. FEBS J. 2013, 280, 1604–1616. [Google Scholar] [CrossRef]
- Shamsunnaher; Chen, X.; Zhang, X.; Wu, X.-X.; Huang, X.; Song, W.-Y. Rice immune sensor XA21 differentially enhances plant growth and survival under distinct levels of drought. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Liu, J.; Shabala, S.; Shabala, L.; Zhou, M.; Meinke, H.; Venkataraman, G.; Chen, Z.-H.; Zeng, F.; Zhao, Q. Tissue-Specific Regulation of Na+ and K+ Transporters Explains Genotypic Differences in Salinity Stress Tolerance in Rice. Front. Plant Sci. 2019, 10, 1361. [Google Scholar] [CrossRef] [Green Version]
- Chun, H.J.; Baek, D.; Jin, B.J.; Cho, H.M.; Park, M.S.; Lee, S.H.; Lim, L.H.; Cha, Y.J.; Bae, D.-W.; Kim, S.; et al. Microtubule Dynamics Plays a Vital Role in Plant Adaptation and Tolerance to Salt Stress. Int. J. Mol. Sci. 2021, 22, 5957. [Google Scholar] [CrossRef]
- Pan, Y.; Li, J.; Jiao, L.; Li, C.; Zhu, D.; Yu, J. A Non-specific Setaria italica Lipid Transfer Protein Gene Plays a Critical Role under Abiotic Stress. Front. Plant Sci. 2016, 7, 1752. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zheng, X.; Song, Y.; Zhu, L.; Yu, Z.; Gan, L.; Zhou, S.; Liu, H.; Wen, F.; Zhu, C. NtLTP4, a lipid transfer protein that enhances salt and drought stresses tolerance in Nicotiana tabacum. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Dong, J.; Deng, F.; Wang, W.; Cheng, Y.; Song, L.; Hu, M.; Shen, J.; Xu, Q.; Shen, F. The long non-coding RNA lncRNA973 is involved in cotton response to salt stress. BMC Plant Biol. 2019, 19, 459. [Google Scholar] [CrossRef]
- He, Y.; Li, W.; Lv, J.; Jia, Y.; Wang, M.; Xia, G. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 2011, 63, 1511–1522. [Google Scholar] [CrossRef]
- Qin, Y.; Wang, M.; Tian, Y.; He, W.; Han, L.; Xia, G. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol. Biol. Rep. 2012, 39, 7183–7192. [Google Scholar] [CrossRef]
- Yang, L.; Ma, C.; Wang, L.; Chen, S.; Li, H. Salt stress induced proteome and transcriptome changes in sugar beet monosomic addition line M14. J. Plant Physiol. 2012, 169, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, G.; Xia, C.; Jia, J.; Liu, X.; Kong, X. Overexpression of a wheat MYB transcription factor gene, TaMYB56-B, enhances tolerances to freezing and salt stresses in transgenic Arabidopsis. Gene 2012, 505, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple Functions of MYB Transcription Factors in Abiotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.; Li, C.; Chen, F.; Zhao, S.; Xia, G. A Bowman-Birk type protease inhibitor is involved in the tolerance to salt stress in wheat. Plant Cell Environ. 2008, 31, 1128–1137. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Seifert, G.J. Fasciclin like arabinogalactan protein 4 and respiratory burst oxidase homolog D and F independently modulate abscisic acid signaling. Plant Signal. Behav. 2015, 10, 2. [Google Scholar] [CrossRef] [Green Version]
- Mito, T.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M.; Matsui, K. Generation of chimeric repressors that confer salt tolerance in Arabidopsis and rice. Plant Biotechnol. J. 2010, 9, 736–746. [Google Scholar] [CrossRef]
- Su, P.; Yan, J.; Li, W.; Wang, L.; Zhao, J.; Ma, X.; Li, A.; Wang, H.; Kong, L. A member of wheat class III peroxidase gene family, TaPRX-2A, enhanced the tolerance of salt stress. BMC Plant Biol. 2020, 20, 1–15. [Google Scholar] [CrossRef]
- Hluska, T.; Šebela, M.; Lenobel, R.; Frébort, I.; Galuszka, P. Purification of Maize Nucleotide Pyrophosphatase/Phosphodiesterase Casts Doubt on the Existence of Zeatin Cis–Trans Isomerase in Plants. Front. Plant Sci. 2017, 8, 1473. [Google Scholar] [CrossRef] [Green Version]
- Kosma, D.K.; Jenks, M.A. Eco-physiological and molecular-genetic determinants of plant cuticle function in drought and salt stress tolerance. In Advances in Molecular Breeding toward Drought and Salt Tolerant Crops; Springer: Dordrecht, The Netherlands, 2007; pp. 91–120. [Google Scholar]
- Samuels, L.; Kunst, L.; Jetter, R. Sealing Plant Surfaces: Cuticular Wax Formation by Epidermal Cells. Annu. Rev. Plant Biol. 2008, 59, 683–707. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Cai, X.; Wu, X.; Karahara, I.; Schreiber, L.; Lin, J. Casparian strip development and its potential function in salt tolerance. Plant Signal. Behav. 2011, 6, 1499–1502. [Google Scholar] [CrossRef]
- Goyal, S.S.; Sharma, S.K.; Rains, D.W. Crop Production in Saline Environments: Global and Integrative Perspectives; Haworth Press: New York, NY, USA, 2003. [Google Scholar]
- Liu, C.; Yu, H.; Rao, X.; Li, L.; Dixon, R.A. Abscisic acid regulates secondary cell-wall formation and lignin deposition in Arabidopsis thaliana through phosphorylation of NST1. Proc. Natl. Acad. Sci. USA 2021, 118, e2010911118. [Google Scholar] [CrossRef]
- Augstein, F.; Carlsbecker, A. Salinity induces discontinuous protoxylem via a DELLA-dependent mechanism promoting salt tolerance in Arabidopsis seedlings. New Phytol. 2022, 236, 195–209. [Google Scholar] [CrossRef]
- Junghans, U.; Polle, A.; Düchting, P.; Weiler, E.; Kuhlman, B.; Gruber, F.; Teichmann, T. Adaptation to high salinity in poplar involves changes in xylem anatomy and auxin physiology. Plant Cell Environ. 2006, 29, 1519–1531. [Google Scholar] [CrossRef]
- Endler, A.; Kesten, C.; Schneider, R.; Zhang, Y.; Ivakov, A.; Froehlich, A.; Funke, N.; Persson, S. A mechanism for sustained cellulose synthesis during salt stress. Cell 2015, 162, 1353–1364. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Zayed, O.; Zeng, F.; Liu, C.; Zhang, L.; Zhu, P.; Hsu, C.C.; Tuncil, Y.E.; Tao, W.A.; Carpita, N.C.; et al. Arabinose biosynthesis is critical for salt stress tolerance in Arabidopsis. New Phytol. 2019, 224, 274–290. [Google Scholar] [CrossRef]
- Novaković, L.; Guo, T.; Bacic, A.; Sampathkumar, A.; Johnson, K.L. Hitting the Wall—Sensing and Signaling Pathways Involved in Plant Cell Wall Remodeling in Response to Abiotic Stress. Plants 2018, 7, 89. [Google Scholar] [CrossRef] [Green Version]
- Vaahtera, L.; Schulz, J.; Hamann, T. Cell wall integrity maintenance during plant development and interaction with the environment. Nat. Plants 2019, 5, 924–932. [Google Scholar] [CrossRef]
- Rui, Y.; Dinneny, J.R. A wall with integrity: Surveillance and maintenance of the plant cell wall under stress. New Phytol. 2020, 225, 1428–1439. [Google Scholar] [CrossRef] [Green Version]
- Feng, W.; Kita, D.; Peaucelle, A.; Cartwright, H.N.; Doan, V.; Duan, Q.; Liu, M.C.; Maman, J.; Steinhorst, L.; Schmitz-Thom, I.; et al. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr. Biol. 2018, 28, 666–675. [Google Scholar] [CrossRef] [Green Version]
- Singh, D.; Singh, C.K.; Kumari, S.; Singh Tomar, R.S.; Karwa, S.; Singh, R.; Singh, R.B.; Sarkar, S.K.; Pal, M. Discerning morpho-anatomical, physiological and molecular multiformity in cultivated and wild genotypes of lentil with reconciliation to salinity stress. PLoS ONE 2017, 12, e0177465. [Google Scholar] [CrossRef]
- Tu, Y.; Jiang, A.; Gan, L.; Hossain, M.; Zhang, J.; Peng, B.; Xiong, Y.; Song, Z.; Cai, D.; Xu, W.; et al. Genome duplication improves rice root resistance to salt stress. Rice 2014, 7, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adem, G.D.; Roy, S.J.; Zhou, M.; Bowman, J.P.; Shabala, S. Evaluating contribution of ionic, osmotic and oxidative stress components towards salinity tolerance in barley. BMC Plant Biol. 2014, 14, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bose, J.; Shabala, L.; Pottosin, I.; Zeng, F.; Velarde-Buendía, A.M.; Massart, A.; Poschenrieder, C.; Hariadi, Y.; Shabala, S. Kinetics of xylem loading, membrane potential maintenance, and sensitivity of K+-permeable channels to reactive oxygen species: Physiological traits that differentiate salinity tolerance between pea and barley. Plant Cell Environ. 2014, 37, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, K.; Bose, J.; Shabala, L.; Shabala, S. Difference in root K+ retention ability and reduced sensitivity of K+-permeable channels to reactive oxygen species confer differential salt tolerance in three Brassica species. J. Exp. Bot. 2016, 67, 4611–4625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, Z.-H.; Gao, J.-P.; Li, L.-G.; Cai, X.-L.; Huang, W.; Chao, D.-Y.; Zhu, M.-Z.; Wang, Z.-Y.; Luan, S.; Lin, H.-X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef]
- Cotsaftis, O.; Plett, D.; Johnson, A.A.; Walia, H.; Wilson, C.; Ismail, A.M.; Close, T.J.; Tester, M.; Baumann, U. Root-specific transcript profiling of contrasting rice genotypes in response to salinity stress. Mol. Plant 2011, 4, 25–41. [Google Scholar] [CrossRef] [Green Version]
- Henderson, S.W.; Wege, S.; Qiu, J.; Blackmore, D.H.; Walker, A.R.; Tyerman, S.D.; Walker, R.R.; Gilliham, M. Grapevine and Arabidopsis cation-chloride cotransporters localize to the golgi and trans-golgi network and indirectly influence long-distance ion transport and plant salt tolerance. Plant Physiol. 2015, 169, 2215–2229. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Zhou, M.; Shabala, L.; Shabala, S. Physiological and molecular mechanisms mediating xylem Na+ loading in barley in the context of salinity stress tolerance. Plant Cell Environ. 2017, 40, 1009–1020. [Google Scholar] [CrossRef]
- Wu, B.; Munkhtuya, Y.; Li, J.; Hu, Y.; Zhang, Q.; Zhang, Z. Comparative transcriptional profiling and physiological responses of two contrasting oat genotypes under salt stress. Sci. Rep. 2018, 8, 16248. [Google Scholar] [CrossRef] [Green Version]
- Shabala, S.; Demidchik, V.; Shabala, L.; Cuin, T.A.; Smith, S.J.; Miller, A.J.; Davies, J.M.; Newman, I.A. Extracellular Ca2+ ameliorates NaCl-induced K+ loss from Arabidopsis root and leaf cells by controlling plasma membrane K+-permeable channels. Plant Physiol. 2006, 141, 1653–1665. [Google Scholar] [CrossRef] [Green Version]
- Lang, T.; Sun, H.; Li, N.; Lu, Y.; Shen, Z.; Jing, X.; Xiang, M.; Shen, X.; Chen, S. Multiple signaling networks of extracellular ATP, hydrogen peroxide, calcium, and nitric oxide in the mediation of root ion fluxes in secretor and non-secretor mangroves under salt stress. Aquat. Bot. 2014, 119, 33–43. [Google Scholar] [CrossRef]
- Wiens, C.P.; Shabala, L.; Zhang, J.; Pottosin, I.I.; Bose, J.; Zhu, M.; Fuglsang, A.T.; Velarde, A.; Massart, A.; Hill, C.B.; et al. Cell-type specific H+-ATPase activity enables root K+ retention and mediates acclimatation to salinity. Plant Physiol. 2016, 172, 2445–2458. [Google Scholar] [CrossRef] [Green Version]
- Zhao, N.; Wang, S.; Ma, X.; Zhu, H.; Sa, G.; Sun, J.; Li, N.; Zhao, C.; Zhao, R.; Chen, S. Extracellular ATP mediates cellular K+/Na+ homeostasis in two contrasting poplar species under NaCl stress. Trees 2016, 30, 825–837. [Google Scholar] [CrossRef]
- Almodares, A.; Hadi, M.R.; Kholdebarin, B.; Samedani, B.; Kharazian, Z.A. The response of sweet sorghum cultivars to salt stress and accumulation of Na+, Cl− and K+ ions in relation to salinity. J. Environ. Biol. 2014, 35, 733–739. [Google Scholar]
- Calone, R.; Sanoubar, R.; Lambertini, C.; Speranza, M.; Vittori Antisari, L.; Vianello, G.; Barbanti, L. Salt tolerance and Na allocation in Sorghum bicolor under variable soil and water salinity. Plants 2020, 9, 561. [Google Scholar] [CrossRef]
- Pasini, L.; Bergonti, M.; Fracasso, A.; Marocco, A.; Amaducci, S. Microarray analysis of differentially expressed mRNAs and miRNAs in young leaves of sorghum under dry-down conditions. J. Plant Physiol. 2014, 171, 537–548. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Jagendorf, A.; Zhu, J.K. Understanding and Improving Salt Tolerance in Plants. Crop Sci. 2005, 45, 437–448. [Google Scholar] [CrossRef] [Green Version]
- Fracasso, A.; Trindade, L.M.; Amaducci, S. Drought stress tolerance strategies revealed by RNA-Seq in two sorghum genotypes with contrasting WUE. BMC Plant Biol. 2016, 16, 115. [Google Scholar] [CrossRef]
- Devnarain, N.; Crampton, B.G.; Olivier, N.; Van der Westhuyzen, C.; Becker, J.V.; O’Kennedy, M.M. Transcriptomic analysis of a Sorghum bicolor landrace identifies a role for beta-alanine betaine biosynthesis in drought tolerance. S. Afr. J. Bot. 2019, 127, 244–255. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Ullah, F.; Ben-Hur, A.; Reddy, A.S.N. Transcriptome Analysis of Drought-Resistant and Drought-Sensitive Sorghum (Sorghum bicolor) Genotypes in Response to PEG-Induced Drought Stress. Int. J. Mol. Sci. 2020, 21, 772. [Google Scholar] [CrossRef] [Green Version]
- Azzouz-Olden, F.; Hunt, A.G.; Dinkins, R. Transcriptome analysis of drought-tolerant sorghum genotype SC56 in response to water stress reveals an oxidative stress defense strategy. Mol. Biol. Rep. 2020, 47, 3291–3303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Lu, F.; Wang, Y.; Zhang, Z.; Wang, J.; Zhang, K.; Wu, H.; Zou, J.; Duan, Y.; Ke, F.; et al. Combined transcriptomic and physiological metabolomic analyses elucidate key biological pathways in the response of two sorghum genotypes to salinity stress. Front. Plant Sci. 2022, 13, 880373. [Google Scholar] [CrossRef] [PubMed]
- Leah, R.; Tommerup, H.; Svendsen, I.B.; Mundy, J. Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J. Biol. Chem. 1991, 266, 1564–1573. [Google Scholar] [CrossRef] [PubMed]
- Mostek, A.; Börner, A.; Weidner, S. Comparative proteomic analysis of β-aminobutyric acid-mediated alleviation of salt stress in barley. Plant Physiol. Biochem. 2016, 99, 150–161. [Google Scholar] [CrossRef]
- Colin, L.; Ruhnow, F.; Zhu, J.K.; Zhao, C.; Zhao, Y.; Persson, S. The cell biology of primary cell walls during salt stress. Plant Cell 2022, 35, 201–217. [Google Scholar] [CrossRef]
- Lou, D.; Wang, H.; Yu, D. The sucrose non-fermenting-1-related protein kinases SAPK1 and SAPK2 function collaboratively as positive regulators of salt stress tolerance in rice. BMC Plant Biol. 2018, 18, 203. [Google Scholar] [CrossRef] [Green Version]
- García-Garrido, J.M.; Menossi, M.; Puigdoménech, P.; Martínez-Izquierdo, J.A.; Delseny, M. Characterization of a gene encoding an abscisic acid-inducible type-2 lipid transfer protein from rice. FEBS Lett. 1998, 428, 193–199. [Google Scholar] [CrossRef] [Green Version]
- Jang, C.S.; Lee, H.J.; Chang, S.J.; Seo, Y.W. Expression and promoter analysis of the TaLTP1 gene induced by drought and salt stress in wheat (Triticum aestivum L.). Plant Sci. 2004, 167, 995–1001. [Google Scholar] [CrossRef]
- Wang, H.W.; Kwon, H.J.; Yim, W.C.; Lim, S.D.; Moon, J.C.; Lee, B.M.; Seo, Y.W.; Kim, W.; Jang, C.S. Expressional diversity of wheat nsLTP genes: Evidence of subfunctionalization via cis-regulatory divergence. Genetica 2010, 138, 843–852. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Y.; Zhang, Y.; Li, C.; Gong, S.; Yan, S.; Li, G.; Hu, G.; Ren, H.; Yang, J.; et al. Comparative transcriptome analysis of salt-sensitive and salt-tolerant maize reveals potential mechanisms to enhance salt resistance. Genes Genom. 2019, 41, 781–801. [Google Scholar] [CrossRef]
- Fernandes, J.; Morrow, D.J.; Casati, P.; Walbot, V. Distinctive transcriptome responses to adverse environmental conditions in Zea mays L. Plant Biotechnol. J. 2008, 6, 782–798. [Google Scholar] [CrossRef]
- Caruso, C.; Bertini, L.; Tucci, M.; Caporale, C.; Leonardi, L.; Saccardo, F.; Bressan, R.A.; Veronese, P.; Buonocore, V. Isolation and characterisation of wheat cDNA clones encoding PR4 proteins. DNA Seq. 1999, 10, 301–307. [Google Scholar] [CrossRef]
- Bravo, J.M.; Campo, S.; Murillo, I.; Coca, M.; San Segundo, B. Fungus-and wound-induced accumulation of mRNA containing a class II chitinase of the pathogenesis-related protein 4 (PR-4) family of maize. Plant Mol. Biol. 2003, 52, 745–759. [Google Scholar] [CrossRef]
- Zhu, T.; Song, F.; Zheng, Z. Molecular characterization of the rice pathogenesis-related protein, OsPR-4b, and its antifungal activity against Rhizoctonia solani. J. Phytopathol. 2006, 154, 378–384. [Google Scholar] [CrossRef]
- Saxena, S.C.; Salvi, P.; Kamble, N.U.; Joshi, P.K.; Majee, M.; Arora, S. Ectopic overexpression of cytosolic ascorbate peroxidase gene (Apx1) improves salinity stress tolerance in Brassica juncea by strengthening antioxidative defense mechanism. Acta Physiol. Plant 2020, 42, 45. [Google Scholar] [CrossRef]
- Asaoka, M.; Segami, S.; Ferjani, A.; Maeshima, M. Contribution of PPi-hydrolyzing function of vacuolar H+-pyrophosphatase in vegetative growth of Arabidopsis: Evidenced by expression of uncoupling mutated enzymes. Front Plant Sci. 2016, 7, 415. [Google Scholar] [CrossRef] [Green Version]
- Anjaneyulu, E.; Reddy, P.S.; Sunita, M.S.; Kishor, P.B.; Meriga, B. Salt tolerance and activity of antioxidative enzymes of transgenic finger millet overexpressing a vacuolar H+-pyrophosphatase gene (SbVPPase) from Sorghum bicolor. J. Plant Physiol. 2014, 171, 789–798. [Google Scholar] [CrossRef]
- Kumar, T.; Khan, M.R.; Abbas, Z.; Ali, G.M. Genetic improvement of sugarcane for drought and salinity stress tolerance using Arabidopsis vacuolar pyrophosphatase (AVP1) gene. Mol. Biotechnol. 2014, 56, 199–209. [Google Scholar] [CrossRef]
- Lv, S.; Jiang, P.; Wang, D.; Li, Y. H+-pyrophosphatase from Salicornia europaea enhances tolerance to low phosphate under salinity in Arabidopsis. Plant Signal. Behav. 2016, 11, 1. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Zhou, L.; Li, Y.; Zhang, D.; Gao, Y. Plant NIGT1/HRS1/HHO transcription factors: Key regulators with multiple roles in plant growth, development, and stress responses. Int. J. Mol. Sci. 2021, 22, 8685. [Google Scholar] [CrossRef]
- Pramod, S.; Rajput, K.S.; Rao, K.S. Immnolocalization of β (1-4)-D-galactan, xyloglucan ad xylan in the reaction xylem of Leucaena leucocephala (Lam) de Wit. Plant Physiol. Biochem. 2019, 142, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Berlyn, G.P.; Miksche, J.P.; Sass, J.E. Botanical Microtechnique and Cytochemistry; Iowa State University Press: Ames, IA, USA, 1976. [Google Scholar]
- Reynolds, E.S. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 1963, 17, 208–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donaldson, L.A. Lignin distribution during latewood formation in Pinus radiata D. Don. IAWA J. 1992, 13, 381–387. [Google Scholar] [CrossRef]
- Edwards, I.K.; Kalra, Y.P.; Radford, F.G. Chloride determination and levels in the soil-plant environment. Environ. Pollut. Ser. B Chem. Phys. 1981, 2, 109–117. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]


| Biological Process | Cellular Components | Molecular Function | ||
|---|---|---|---|---|
| SS vs. SSN | Downregulated | 83 | 81 | 118 |
| Exclusive SS | 6 | 6 | 7 | |
| Exclusive SSN | 29 | 36 | 35 | |
| Expressed in both | 6396 | 7340 | 7858 | |
| False discovery rate (FDR) downregulated | 14 | 13 | 16 | |
| FDR upregulated | 38 | 35 | 47 | |
| Downregulated | 239 | 245 | 296 | |
| ST vs. STN | Downregulated | 299 | 321 | 343 |
| Exclusive ST | 29 | 32 | 36 | |
| Exclusive STN | 2 | 2 | 2 | |
| Expressed in both | 6213 | 7136 | 7632 | |
| FDR downregulated | 32 | 32 | 39 | |
| FDR upregulated | 4 | 6 | 10 | |
| Downregulated | 89 | 96 | 120 | |
| S. No. | Name of Gene Selected for qRT-PCR | SSN/STN Up- and Down-Regulated/Function | References |
|---|---|---|---|
| 1 | Glucan endo-1,3-beta glucosidase 3-like Upregulated) | It stimulates and initiates the plant defence mechanism. A glucan endo-1,3-beta-glucosidase precursor has been detected in rice under salt stress. It plays a role during abiotic stress. | [37,38] |
| 2 | BARWIN (Upregulated) | BARWIN has similarity with proteins encoded by the wound-induced genes of barley (win) and others. It is involved in a common defence mechanism in plants. | [39] |
| 3 | Peroxidase 42 (Upregulated) | Peroxidases play prominent roles in antioxidant response through removing hydrogen peroxide from the system. Its overexpression results in salt stress tolerance. | [40] |
| 4 | Chitinase 2 (Upregulated) | Overexpression of a chitinase 2 gene (LcCHI2) in maize and tobacco improved salt and alkaline stress tolerance through reducing Na+ accumulation and malon-dialdehyde content. | [41] |
| 5 | Heavy metal-associated isoprenylated plant protein 16 (HIPP16) (Upregulated) | In addition to involvement in heavy metal homeostasis and detoxification, they are associated with abiotic stresses such as drought, cold, and plant–pathogen interactions. | [42] |
| 6 | Receptor kinase-like protein XA 21 (Upregulated) | XA 21 is an immune sensor protein, which promotes survival during dehydration stress. Its overexpression has enhanced lignin deposition and cellulose in the xylem vessels and surrounding cells. | [43] |
| 7 | K+ transporter (Upregulated) | Acquisition of K+ during salt stress is crucial. Its tissue-specific regulation of Na+ and K+ transporters explains genotypic differences in salt stress tolerance. | [44] |
| 8 | Tubulin and alpha 2-tubulin (Upregulated) | Microtubules and their dynamics play key roles in the adaptation of plants and their tolerance to salt stress. | [45] |
| 9 | Lipid transfer protein (LTP) (Downregulated) | LTPs are regulated by upstream transcription factors or kinase orphosphatases. They are critical for imparting salt/abiotic stress tolerance. However, it is downregulated in the SSN line but not in STN. | [46,47] |
| 10 | Micro RNA (miRf11471-akr) (Downregulated) | They are involved in gene regulation at the post-transcriptional level and associated with salt/abiotic stress tolerance. It is downregulated in SSN but not in STN. | [48] |
| 11 | MYB 61 (v-Myb avian myeloblastosis viral oncogene homolog transcription factor) (Upregulated) | Several MYBs have been shown to be associated with salinity stress in diverse plant systems. | [49,50,51,52,53] |
| 12 | Bowman-Birk type trypsin inhibitor (Upregulated) | It is involved in salt stress tolerance in wheat and other plants via limiting the transport of Na+ from the root to the shoot. | [54] |
| 13 | Heavy metal-associated isoprenylated plant protein (HIPP) (Upregulated) | Besides involvement in heavy metal homeostasis and detoxification, they are associated with abiotic stresses such as drought, cold, and plant–pathogen interactions. | [42] |
| 14 | Glucan endo-1,3-beta glucosidase (Upregulated) | It stimulates and initiates the plant defence mechanism. It plays a role in cell division, flower formation, and a vital role during abiotic stress. | [37,38] |
| 15 | Tubulin beta-4 chain (Upregulated) | Microtubules and their dynamics play key roles in the adaptation of plants and tolerance to salt stress. | [45] |
| 16 | Fasciclin-like arabinogalactan protein 12 (Downregulated) | This gene is involved in developing xylem and cell wall formation during salt stress. Additionally, it controls ROS levels. It is downregulated in STN, but not in SSN. | [11,55] |
| 17 | HHO5 (HRS1 homolog, (transcription factor) (Upregulated) | NIGT1/HRS1/HHO transcription factors are associated with salt stress tolerance, though the precise function is obscure. | [56] |
| 18 | Peroxidase 2 (Upregulated) | It removes ROS-like hydrogen peroxide generated during salt stress. | [57] |
| 19 | Pyrophospha-tase/phospho-diesterase (Upregulated) | It improves salt stress tolerance. | [58] |
| 20 | Micro RNA (ncRNA) (un-characterized) (miR8709c) (Upregulated) | It is suspected to be associated with cis-trans inter-conversion of cytokinin zeatin. Its overexpression led to decreased FAD and enhanced FMN and riboflavin contents, but its precise function is not known. | [48] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karumanchi, A.R.; Sivan, P.; Kummari, D.; Rajasheker, G.; Kumar, S.A.; Reddy, P.S.; Suravajhala, P.; Podha, S.; Kishor, P.B.K. Root and Leaf Anatomy, Ion Accumulation, and Transcriptome Pattern under Salt Stress Conditions in Contrasting Genotypes of Sorghum bicolor. Plants 2023, 12, 2400. https://doi.org/10.3390/plants12132400
Karumanchi AR, Sivan P, Kummari D, Rajasheker G, Kumar SA, Reddy PS, Suravajhala P, Podha S, Kishor PBK. Root and Leaf Anatomy, Ion Accumulation, and Transcriptome Pattern under Salt Stress Conditions in Contrasting Genotypes of Sorghum bicolor. Plants. 2023; 12(13):2400. https://doi.org/10.3390/plants12132400
Chicago/Turabian StyleKarumanchi, Appa Rao, Pramod Sivan, Divya Kummari, G. Rajasheker, S. Anil Kumar, Palakolanu Sudhakar Reddy, Prashanth Suravajhala, Sudhakar Podha, and P. B. Kavi Kishor. 2023. "Root and Leaf Anatomy, Ion Accumulation, and Transcriptome Pattern under Salt Stress Conditions in Contrasting Genotypes of Sorghum bicolor" Plants 12, no. 13: 2400. https://doi.org/10.3390/plants12132400





