Integrating Transcriptional, Metabolic, and Physiological Responses to Drought Stress in Ilex paraguariensis Roots
Abstract
:1. Introduction
2. Results
2.1. Root Physiological and Morphological Changes in Response to Drought
2.2. Transcriptome Sequencing, Assembly and Differential Expression Analysis
2.3. Transcriptome Functional Annotation and Metabolic Pathways Analysis
2.3.1. Hormone Signalling
2.3.2. Transcription Factors
2.3.3. Carbohydrates
2.3.4. Amino Acids and Proteins
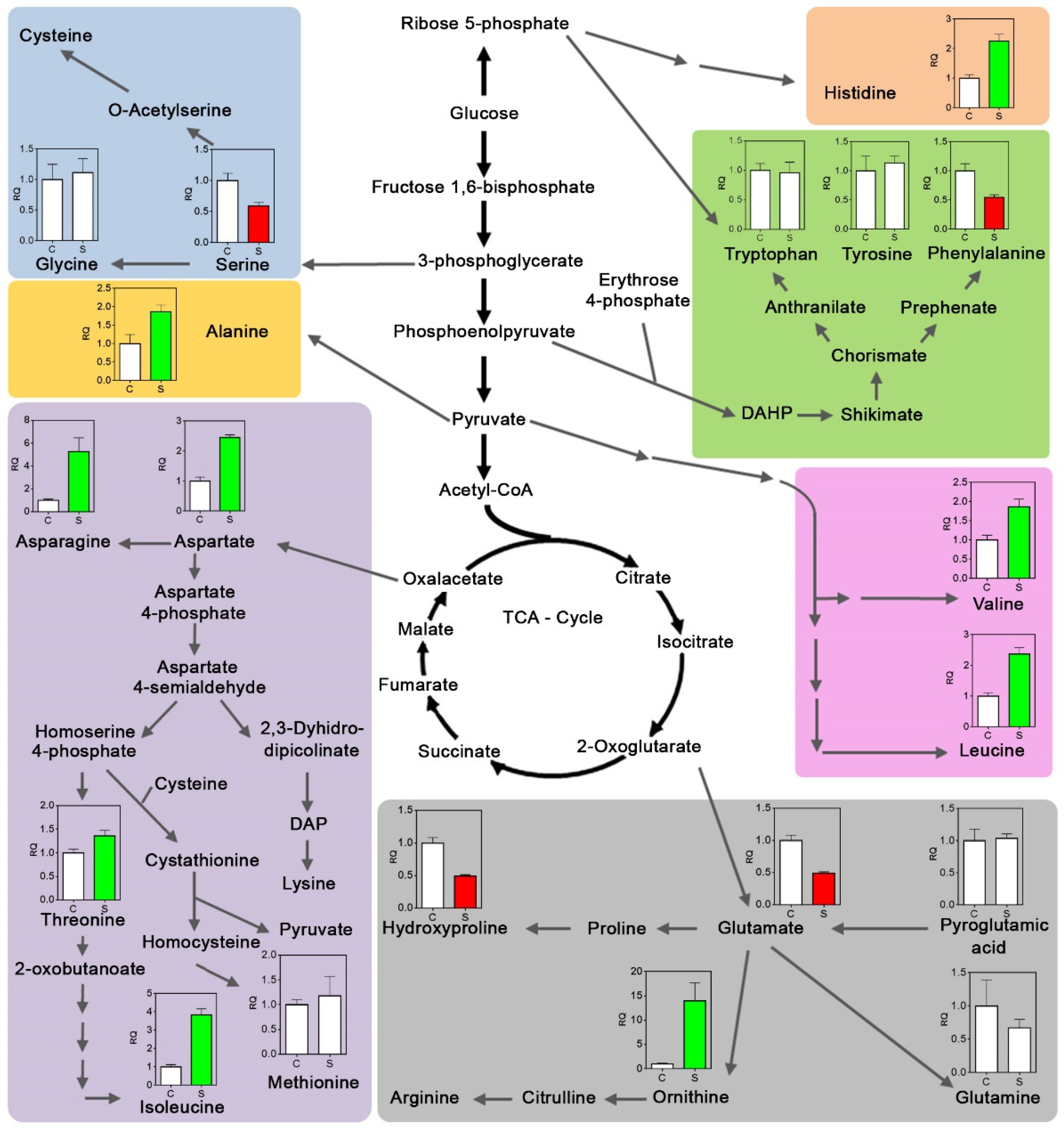
2.4. Metabolites Variation
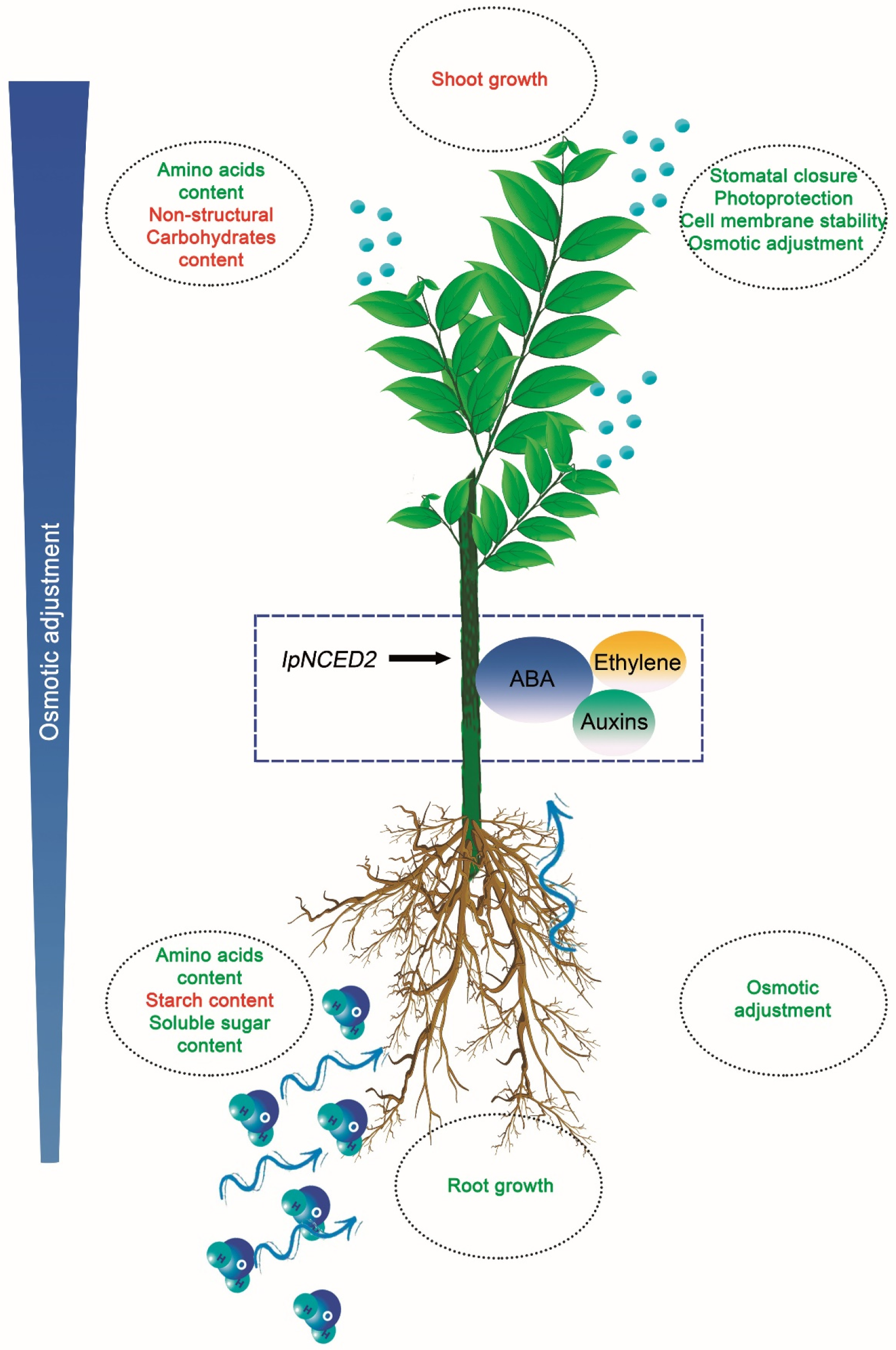
3. Discussion
3.1. Root Phenotypic Plasticity in Response to Drought
3.2. Gene Identification Related to Hydrotropic Responses
3.2.1. Hormonal Interaction and Signalling
3.2.2. Transcription Factors
3.2.3. TOR Signalling
3.3. Metabolic Responses to Dehydration
3.3.1. Non-Structural Carbohydrates
3.3.2. Organic Acids
3.3.3. Amino Acids
4. Materials and Methods
4.1. Plant Material, Drought Assay, and Sample Collection
4.2. RNA Isolation, cDNA Synthesis, and RNA-Seq Library Preparation
4.3. RNA-Seq Data Processing, de Novo Assembly, and Gene Expression Analysis
4.4. Functional Annotation of Transcripts and Metabolic Pathways Analysis
4.5. Quantitative PCR Analysis
4.6. Metabolite Profiling Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dünisch, O.; Reissmann, C.B.; Oliszeski, A. Variability of vessel characteristics in the xylem of Ilex paraguariensis (maté-tree) from south Brazil. IAWA J. 2004, 25, 449–458. [Google Scholar] [CrossRef]
- Gawron-Gzella, A.; Chanaj-Kaczmarek, J.; Cielecka-Piontek, J. Yerba Mate—A Long but Current History. Nutrients 2021, 13, 3706. [Google Scholar] [CrossRef] [PubMed]
- IPCC. Climate Change 2022: Impacts, Adaptation and Vulnerability. In Contribution of Working Group II to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Pörtner, H.-O., Roberts, D.C., Tignor, M., Poloczanska, E.S., Mintenbeck, K., Alegría, A., Craig, M., Langsdorf, S., Löschke, S., Möller, V., et al., Eds.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2022; p. 3056. [Google Scholar]
- Acevedo, R.M.; Maiale, S.J.; Pessino, S.C.; Bottini, R.; Ruiz, O.A.; Sansberro, P. A succinate dehydrogenase flavoprotein subunit-like transcript is up-regulated in Ilex paraguariensis leaves in response to water deficit and abscisic acid. Plant Physiol. Biochem. 2013, 65, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, R.M.; Avico, E.H.; González, S.; Rodrigues Salvador, A.; Rivarola, M.; Paniego, N.; Nunes-Nesi, A.; Ruiz, O.A.; Sansberro, P. Transcript and metabolic adjustments triggered by drought in Ilex paraguariensis leaves. Planta 2019, 250, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Brunner, I.; Herzog, C.; Dawes, M.A.; Arend, M.; Sperisen, C. How tree roots respond to drought. Front. Plant Sci. 2015, 6, 547. [Google Scholar] [CrossRef] [Green Version]
- Watt, M.; Fiorani, F.; Usadel, B.; Rascher, U.; Muller, O.; Schurr, U. Phenotyping: New windows into the plant for breeders. Ann. Rev. Plant Biol. 2020, 71, 689–712. [Google Scholar] [CrossRef]
- Usman, M.; Ho-Plágaro, T.; Frank, H.E.; Calvo-Polanco, M.; Gaillard, I.; Garcia, K.; Zimmermann, S.D. Mycorrhizal of trees to abiotic stress caused by climate change in temperate and boreal forests. Front. For. Glob. Change 2021, 4, 742392. [Google Scholar] [CrossRef]
- Janiak, A.; Kwaśniewski, M.; Szarejko, I. Gene expression regulation in roots under drought. J. Exp. Bot. 2016, 67, 1003–1014. [Google Scholar] [CrossRef] [Green Version]
- Comas, L.H.; Becker, S.R.; Cruz, V.M.; Byrne, P.F.; Dierig, D.A. Root traits contributing to plant productivity under drought. Front. Plant Sci. 2013, 5, 442. [Google Scholar] [CrossRef] [Green Version]
- Gorgues, L.; Li, X.; Maurel, C.; Martinière, A.; Nacry, P. Root osmotic sensing from local perception to systemic responses. Stress Biol. 2022, 2, 36. [Google Scholar] [CrossRef]
- Schneider, H.M.; Lynch, J.P. Should root plasticity be a crop breeding target? Front. Plant Sci. 2020, 11, 546. [Google Scholar] [CrossRef]
- Calleja-Cabrera, J.; Boter, M.; Oñate-Sánchez, L.; Pernas, M. Root growth adaptation to climate change in crops. Front. Plant Sci. 2020, 11, 544. [Google Scholar] [CrossRef]
- Galindo-Castañeda, T.; Lynch, J.P.; Six, J.; Hartmann, M. Improving soil resource uptake by plants through capitalising on synergies between root architecture and anatomy and root-associated microorganisms. Front. Plant Sci. 2022, 13, 827369. [Google Scholar] [CrossRef]
- Labdelli, A.; Adda, A.; Halis, Y.; Soualem, S. Effects of water regime on the structure of roots and stems of durum wheat (Triticum durum Desf.). J. Bot. 2014, 2014, 703834. [Google Scholar] [CrossRef]
- Motte, H.; Vanneste, S.; Beeckman, T. Molecular and environmental regulation of root development. Annu. Rev. Plant Biol. 2019, 70, 465–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Q.; Essemine, J.; Pang, X.; Chen, H.; Jin, J.; Cai, W. Abscisic acid regulates the root growth trajectory by reducing auxin transporter PIN2 protein levels in Arabidopsis thaliana. Front. Plant Sci. 2021, 12, 632676. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, D.; Pang, L.; Kobayashi, A.; Fozard, J.A.; Boudolf, V.; Bhosale, R.; Antoni, R.; Nguyen, T.; Hiratsuka, S.; Fujii, N.; et al. Root hydrotropism is controlled via a cortex-specific growth mechanism. Nat. Plants 2017, 3, 17057. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Goto, N.; Okada, K.; Takahashi, H. Hydrotropism in abscisic acid, wavy, and gravitropic mutants of Arabidopsis thaliana. Planta 2002, 216, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Shkolnik, D.M.; Krieger, G.; Nuriel, R.; Fromm, H. Hydrotropism: Root bending does not require auxin redistribution. Mol. Plant 2016, 9, 757–759. [Google Scholar] [CrossRef]
- Verma, V.; Pratibha, R.; Prakash, P.K. Plant Hormone-Mediated Regulation of Stress Responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [Green Version]
- Acevedo, R.M.; Angelina, E.L.; Guidalevic, V.; Cascales, J.; Duarte, M.J.; Peruchena, N.; Ruiz, O.A.; Sansberro, P. Ilex paraguariensis modulates the expression of 9-cis-epoxycarotenoid dioxygenase isoforms to cope with drought strain. Plant Gene 2023. under review. [Google Scholar]
- Watanabe, S.; Sato, M.; Sawada, Y.; Tanaka, M.; Matsui, A.; Kanno, Y.; Yokota, H.M.; Seki, M.; Seo, M. Arabidopsis molybdenum cofactor sulfurase ABA3 contributes to anthocyanin accumulation and oxidative stress tolerance in ABA-dependent and independent ways. Sci Rep. 2018, 81, 16592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Li, G.-J.; Bressan, R.A.; Song, C.-P.; Zhu, J.-K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Int. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Xue, L.; Batelli, G.; Lee, S.; Hou, Y.-J.; Van Oosten, M.J.; Zhang, H.; Tao, A.; Zhu, J.-K. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 2013, 110, 11205–11210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neale, D.B.; Martínez García, P.J.; De La Torre, A.R.; Montanari, S.; Wei, X.-X. Novel insights into tree biology and genome evolution as revealed through genomics. Annu. Rev. Plant Biol. 2017, 68, 457–483. [Google Scholar] [CrossRef]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Ge, H.; Wang, X.; Tang, R.; Wang, Y.; Zhao, F.; Lan, W.; Luan, S.; Yang, L. Overexpression of pyrabactin resistance-like abscisic acid receptors enhances drought, osmotic, and cold tolerance in transgenic poplars. Front. Plant Sci. 2017, 8, 1752. [Google Scholar] [CrossRef]
- Arshad, M.; Mattsson, J. A putative poplar PP2C-encoding gene negatively regulates drought and abscisic acid responses in transgenic Arabidopsis thaliana. Trees 2014, 28, 531–543. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, D.; Zhang, C.; Xia, X.; Yin, W.; Tian, Q. A putative PP2C-encoding gene negatively regulates ABA signaling in Populus euphratica. PLoS ONE 2015, 10, e0139466. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Liu, Y.; Li, M.; Lamin-Samu, A.T.; Yang, D.; Yu, X.; Izhar, M.; Jan, I.; Ali, M.; Lu, G. The Arabidopsis Small Auxin Up RNA32 protein regulates ABA—Mediated responses to drought stress. Front. Plant Sci. 2021, 12, 259. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Husain, T.; Fatima, A.; Suhel, M.; Singh, S.; Sharma, A.; Prasad, S.M.; Singh, V.P. A brief appraisal of ethylene signaling under abiotic stress in plants. Plant Signal Behav. 2020, 100, 128–142. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Laxmi, A. Transcriptional regulation of drought response: A tortuous network of transcriptional factors. Front. Plant Sci. 2015, 6, 895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, T.; Zhang, J.; Xie, M.; Yuan, G.; Tschaplinski, T.J.; Muchero, W.; Chen, J.-G. Transcriptional regulation of drought response in Arabidopsis and woody plants. Front. Plant Sci. 2021, 11, 572137. [Google Scholar] [CrossRef]
- Takahashi, F.; Kuromori, T.; Sato, H.; Shinozaki, K. Regulatory gene networks in drought stress responses and resistance in plants. In Survival Strategies in Extreme Cold and Desiccation: Adaptation Mechanisms and Their Applications; Iwaya-Inoue, M., Sakurai., M., Uemura, M., Eds.; Springer: Singapore, 2018; pp. 189–214. [Google Scholar]
- Xiao, Y.; Chu, L.; Zhang, Y.; Bian, Y.; Xiao, J.; Xu, D. HY5: A pivotal regulator of light-dependent development in higher plants. Front. Plant Sci. 2022, 12, 800989. [Google Scholar] [CrossRef]
- Lee, S.; Wang, W.; Huq, E. Spatial regulation of thermomorphogenesis by HY5 and PIF4 in Arabidopsis. Nat. Commun. 2021, 12, 3656. [Google Scholar] [CrossRef]
- Huang, X.; Ouyang, X.; Yang, P.; Lau, O.S.; Li, G.; Li, J.; Chen, H.; Deng, X.W. Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 2012, 24, 4590–4606. [Google Scholar] [CrossRef] [Green Version]
- Moriwaki, T.M.; Miyazawa, Y.; Fujii, N.; Takahashi, H. Light and abscisic acid signalling are integrated by MIZ1 gene expression and regulate hydrotropic response in roots of Arabidopsis thaliana. Plant Cell Environ. 2012, 35, 1359–1368. [Google Scholar] [CrossRef]
- Nibau, C.; Gibbs, D.J.; Coates, J.C. Branching out in new directions: The control of root architecture by lateral root formation. New Phytol. 2008, 179, 595–614. [Google Scholar] [CrossRef]
- Chen, X.; Li, C.; Wang, H.; Guo, Z. WRKY transcription factors: Evolution, binding, and action. Phytopathol. Res. 2019, 1, 13. [Google Scholar] [CrossRef] [Green Version]
- Lai, Z.; Vinod, K.; Zheng, Z.; Fan, B.; Chen, Z. Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol. 2008, 8, 68. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.; Qin, G. EXB1/WRKY71 transcription factor regulates both shoot branching and responses to abiotic stresses. Plant Signal Behav. 2016, 11, e1150404. [Google Scholar] [CrossRef]
- Rasheed, S.; Bashir, K.; Matsui, A.; Tanaka, M.; Seki, M. Transcriptomic analysis of soil-grown Arabidopsis thaliana roots and shoots in response to drought stress. Front. Plant Sci. 2016, 7, 180. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, Y.; Tan, S.; Li, Z.; Yuan, Z.; Glanc, M.; Domjan, D.; Wang, K.; Xuan, W.; Guo, Y.; et al. Root growth adaptation is mediated by PYLs ABA receptor—PP2A protein phosphatase complex. Adv. Sci. 2020, 7, 1901455. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Dong, Q.; Shao, Y.; Jiang, H.; Zhu, S.; Cheng, B.; Xiang, Y. Genome wide survey and characterisation of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012, 31, 1199–1217. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Tang, B.; Xie, Z.; Nolan, T.; Ye, H.; Song, G.-Y.; Walley, J.; Yin, Y. GSK 3-like kinase BIN 2 phosphorylates RD26 to potentiate drought signaling in Arabidopsis. Plant J. 2019, 100, 923–937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Vannozzi, A.; Wang, G.; Liang, Y.H.; Tornielli, G.B.; Zenoni, S.; Cavallini, E.; Pezzotti, M.; Cheng, Z.-M. Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hortic. Res. 2014, 1, 14016. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Ng, C.K.-Y.; Fan, L.-M. MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 2015, 114, 80–91. [Google Scholar] [CrossRef]
- Quan, R.; Hu, S.; Zhang, Z.; Zhang, H.; Huang, R. Overexpression of an ERF transcription factor TSRF1 improves rice drought tolerance. Plant Biotechnol. J. 2010, 8, 476–488. [Google Scholar] [CrossRef] [PubMed]
- Fasani, E.; DalCorso, G.; Costa, A.; Zenoni, S.; Furini, A. The Arabidopsis thaliana transcription factor MYB59 regulates calcium signalling during plant growth and stress response. Plant Mol. Biol. 2019, 99, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Du, X.-Q.; Wang, F.-L.; Li, H.; Jing, S.; Yu, M.; Li, J.; Wu, W.H.; Kudla, J.; Wang, Y. The transcription factor MYB59 regulates K+/NO3—Translocation in the Arabidopsis response to low K+ stress. Plant Cell 2019, 31, 699–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor-Teeples, M.; Lin, L.; de Lucas, M.; Turco, G.; Toal, T.W.; Gaudinier, A.; Young, N.F.; Trabucco, G.M.; Veling, M.T.; Lamothe, R.; et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 2015, 517, 571–575. [Google Scholar] [CrossRef] [Green Version]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, M. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef] [Green Version]
- Dossa, K.; Mmadi, M.A.; Zhou, R.; Liu, A.; Yang, Y.; Diouf, D.; You, J.; Zhang, X. Ectopic expression of the sesame MYB transcription factor SiMYB305 promotes root growth and modulates ABA-mediated tolerance to drought and salt stresses in Arabidopsis. AoB Plants 2020, 12, plz081. [Google Scholar] [CrossRef] [Green Version]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.J.; Campbell, M.M. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009, 149, 981–993. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Cai, W.; Liu, Y.; Li, H.; Fu, L.; Liu, Z.; Xu, L.; Liu, H.; Xu, T.; Xiong, Y. Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proc. Natl. Acad. Sci. USA 2017, 114, 2765–2770. [Google Scholar] [CrossRef] [Green Version]
- Guo, M.; Liu, J.-H.; Ma, X.; Luo, D.-X.; Gong, Z.-H.; Lu, M.-H. The plant heat stress transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef] [Green Version]
- ElSayed, A.I.; Rafudeen, M.S.; Golldack, D. Physiological aspects of raffinose family oligosaccharides in plants: Protection against abiotic stress. Plant Biol. 2014, 16, 1–8. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF Transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binder, B.M. Ethylene signaling in plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heyman, J.; Cools, T.; Vandenbussche, F.; Heyndrickx, K.S.; Van Leene, J.; Vercauteren, I.; Vanderauwera, S.; Vandepoele, K.; De Jaeger, G.; Van Der Straeten, D.; et al. ERF115 controls root quiescent center cell division and stem cell replenishment. Science 2013, 342, 860–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Duan, Y.; Yin, J.; Ye, S.; Zhu, J.; Zhang, F.; Lu, W.; Fan, D.; Luo, K. Genome-wide identification and characterisation of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J. Exp. Bot. 2014, 65, 6629–6644. [Google Scholar] [CrossRef]
- Ye, H.; Liu, S.; Tang, B.; Chen, J.; Xie, Z.; Nolan, T.M.; Jiang, H.; Guo, H.; Lin, H.Y.; Li, L.; et al. RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun. 2017, 8, 14573. [Google Scholar] [CrossRef] [Green Version]
- Jensen, M.K.; Lindemose, S.; De Masi, F.; Reimer, J.J.; Nielsen, M.; Perera, V.; Workman, C.T.; Turck, F.; Grant, M.R.; Mundy, J.; et al. ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio. 2013, 3, 321–327. [Google Scholar] [CrossRef] [Green Version]
- Shim, J.S.; Oh, N.; Chung, P.J.; Kim, Y.S.; Choi, Y.D.; Kim, J.K. Overexpression of OsNAC14 improves drought tolerance in rice. Front. Plant Sci. 2018, 9, 310. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Zhang, X.; Duan, H.; Lian, C.; Liu, C.; Yin, W.; Xia, X. Three stress-responsive NAC transcription factors from Populus euphratica differentially regulate salt and drought tolerance in transgenic plants. Physiol. Plant. 2018, 162, 73–97. [Google Scholar] [CrossRef] [Green Version]
- Fang, L.; Su, L.; Sun, X.; Li, X.; Sun, M.; Karungo, S.K.; Fang, S.; Chu, J.; Li, S.; Xin, H. Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synthesis. J. Exp. Bot. 2016, 67, 2829–2845. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Wang, Z.; Xu, X.; Zhang, H.; Li, C. Genome-wide analysis of C2H2 zinc-finger family transcription factors and their responses to abiotic stresses in Poplar (Populus trichocarpa). PLoS ONE 2015, 10, e0134753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Yang, Z.; Zhang, Y.; Guo, J.; Liu, L.; Wang, C.; Wang, B.; Han, G. The roles of HD-ZIP proteins in plant abiotic stress tolerance. Front. Plant Sci. 2022, 13, 1027071. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-M.; An, J.; Han, H.J.; Kim, S.H.; Lim, C.O.; Yun, D.J.; Chung, W.S. ZAT11, a zinc finger transcription factor, is a negative regulator of nickel ion tolerance in Arabidopsis. Plant Cell Rep. 2014, 33, 2015–2021. [Google Scholar] [CrossRef]
- Skirycza, A.; Camila, C.; Fernie, A.R. Regulation of plant primary metabolism—How results from novel technologies are extending our understanding from classical targeted approaches. Crit. Rev. Plant Sci. 2022, 41, 32–51. [Google Scholar] [CrossRef]
- McCready, K.; Spencer, V.; Kim, M. The Importance of TOR kinase in plant development. Front. Plant Sci. 2020, 11, 16. [Google Scholar] [CrossRef] [Green Version]
- Brunkard, J.O.; Xu, M.; Regina Scarpin, M.; Chatterjee, S.; Shemyakina, E.A.; Goodman, H.M.; Zambryski, P. TOR dynamically regulates plant cell–cell transport. Proc. Natl. Acad. Sci. USA 2020, 117, 5049–5058. [Google Scholar] [CrossRef]
- Rodriguez, M.; Parola, R.; Andreola, S.; Pereyra, C.; Martínez-Noël, G. TOR and SnRK1 signaling pathways in plant response to abiotic stresses: Do they always act according to the “yin-yang” model? Plant Sci. 2019, 288, 110–220. [Google Scholar] [CrossRef] [PubMed]
- Henriques, R.; Calderan-Rodrigues, M.J.; Crespo, J.L.; Baena-González, E.; Caldana, C. Growing of the TOR world. J. Exp. Bot. 2022, 73, 6987–6992. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Li, C.; Cao, X.-Y.; Sun, X.-D.; Bao, Q.-X.; Mu, X.-R.; Liu, C.-Y.; Loake, G.J.; Chen, H.-H.; Meng, L.-S. Long-distance transport of sucrose in source leaves promotes sink root growth by the EIN3-SUC2 module. PLoS Genet. 2022, 18, e1010424. [Google Scholar] [CrossRef]
- Zulfiqar, F.; Chen, J.; Finnegan, P.M.; Younis, A.; Hamed, K. Application of trehalose and salicylic acid mitigates drought stress in sweet basil and improves plant growth. Plants 2021, 10, 1078. [Google Scholar] [CrossRef]
- Iturriaga, G.; Suárez, R.; Nova-Franco, B. Trehalose metabolism: From osmoprotection to signaling. Int. J. Mol. Sci. 2009, 10, 3793–3810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitchner, F.; Lunn, J.E. The role of trehalose-6-phosphate (Tre6P) in plant metabolism and development. Ann. Rev. Plant Biol. 2021, 72, 737–760. [Google Scholar] [CrossRef]
- Velázquez, M.S.; Fabisik, J.C.; Abarca, C.L.; Allegrucci, N.; Cabello, M.N. Colonization dynamics of arbuscular mycorrhizal fungi (AMF) in Ilex paraguariensis crops: Seasonality and influence of management practices. J. King Saud Univ.-Sci. 2020, 32, 183–188. [Google Scholar] [CrossRef]
- Panchal, P.; Miller, A.J.; Giri, J. Organic acids: Versatile stress-response roles in plants. J. Exp. Bot. 2021, 72, 4038–4052. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Xu, Y.; Wang, J.; Singer, S.D.; Chen, G. The Role of triacylglycerol in plant stress response. Plants 2020, 9, 472. [Google Scholar] [CrossRef] [Green Version]
- Finkemeier, I.; Sweetlove, L.J. The role of malate in plant homeostasis. Biol. Rep. 2009, 1, 47. [Google Scholar] [CrossRef]
- Fábregas, N.; Fernie, A.R. The metabolic response to drought. J. Exp. Bot. 2019, 70, 1077–1085. [Google Scholar] [CrossRef] [Green Version]
- Pires, M.V.; Pereira Júnior, A.A.; Medeiros, D.B.; Daloso, D.M.; Pham, P.A.; Barros, K.A.; Engqvist, M.K.; Florian, A.; Krahnert, I.; Maurino, V.G.; et al. The influence of alternative pathways of respiration that utilise branched-chain amino acids following water shortage in Arabidopsis. Plant Cell Environ. 2016, 39, 1304–1319. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Wu, Y.; Sheen, J. TOR Signaling in plants: Conservation and innovation. Development 2018, 145, dev160887. [Google Scholar] [CrossRef] [Green Version]
- Espasandin, F.D.; Maiale, S.J.; Calzadilla, P.; Ruiz, O.A.; Sansberro, P. Transcriptional regulation of 9-cis-epoxycarotenoid dioxygenase (NCED) gene by putrescine accumulation positively modulates ABA synthesis and drought tolerance in Lotus tenuis plants. Plant Physiol. Biochem. 2014, 76, 29–35. [Google Scholar] [CrossRef]
- Callister, A.N.; Arndt, S.K.; Adams, M.A. Comparison of four methods for measuring osmotic potential of tree leaves. Physiol. Plant. 2006, 3, 383–392. [Google Scholar] [CrossRef]
- Cseresnyés, I.; Szitár, K.; Rajkai, K.; Füzy, A.; Mikó, P.; Kovács, R.; Takács, T. Application of electrical capacitance method for prediction of plant root mass and activity in field-grown crops. Front. Plant Sci. 2018, 9, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avico, E.H.; Acevedo, R.M.; Calzadilla, P.I.; Ruiz, O.A.; Sansberro, P.A. Isolation of high-quality RNA from soil-grown Ilex paraguariensis roots suitable for next-generation sequencing and gene expression analyses. Plant Omics J. 2017, 10, 205–209. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 7, 2957–2963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 7, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualisation and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Lohse, M.; Nagel, A.; Herter, T.; May, P.; Schroda, M.; Zrenner, R.; Tohge, T.; Fernie, A.R.; Stitt, M.; Usadel, B. Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 2014, 37, 1250–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016, 45, 1040–1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasmussen, R. Quantification on the LightCycler. In Rapid Cycle Real-Time PCR, 1st ed.; Meuer, S., Wittwer, C., Nakagawara, K.I., Eds.; Springer: Berlin, Heidelberg, Germany, 2001; pp. 21–34. [Google Scholar] [CrossRef]
- Acevedo, R.M.; Avico, E.H.; Ruiz, O.A.; Sansberro, P.A. Assessment of reference genes for quantitative real-time PCR normalisation in Ilex paraguariensis leaves during drought acclimatisation. Biol. Plantarum. 2018, 62, 89–96. [Google Scholar] [CrossRef]
- Gibon, Y.; Blaesing, O.E.; Hannemann, J.; Carillo, P.; Höhne, M.; Hendriks, J.H.M.; Palacios, N.; Cross, J.; Selbig, J.; Stitt, M. A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: Comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 2004, 16, 3304–3325. [Google Scholar] [CrossRef] [Green Version]
- Fernie, A.; Roessner, U.; Trethewey, R.; Willmitzer, L. The contribution of plastidial phosphoglucomutase to the control of starch synthesis within the potato tuber. Planta 2001, 213, 418–426. [Google Scholar] [CrossRef]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 2006, 1, 387–396. [Google Scholar] [CrossRef]
- Cross, J.; Von Korf, M.; Altmann, T.; Bartzetko, L.; Sulpice, R.; Gibon, Y.; Palacios, N.; Stitt, M. Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions. Plant Physiol. 2006, 142, 1574–1588. [Google Scholar] [CrossRef] [Green Version]
- Nunes-Nesi, A.; Carrari, F.; Gibon, Y.; Sulpice, R.; Lytovchenko, A.; Fisahn, J.; Graham, J.; Ratcliffe, R.; Sweetlove, L.; Fernie, A. Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J. 2007, 50, 1093–1106. [Google Scholar] [CrossRef]
- Osorio, S.; Alba, R.; Nikoloski, Z.; Kochevenko, A.; Fernie, A.R.; Giovannoni, J.J. Integrative comparative analyses of transcript and metabolite profiles from pepper and tomato ripening and development stages uncovers species-specific patterns of network regulatory behavior. Plant Physiol. 2012, 159, 1713–1729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopka, J.; Schauer, N.; Krueger, S.; Birkemeyer, C.; Usadel, B.; Bergmüller, E.; Dörmann, P.; Weckwerth, W.; Gibon, Y.; Stitt, M.; et al. GMD@CSB. DB: The Golm metabolome database. Bioinformatics 2005, 21, 1635–1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]


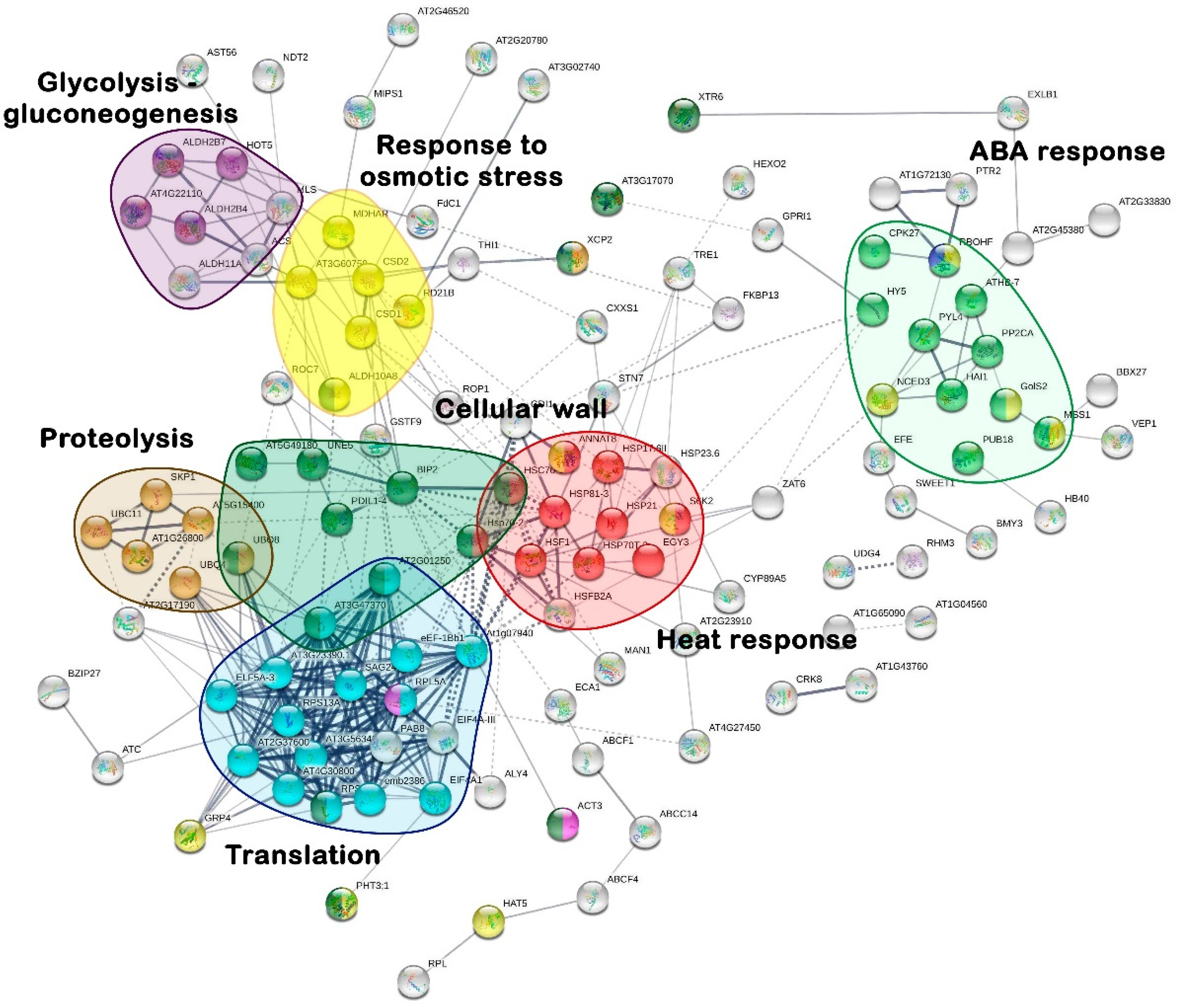

| GO Domains | ||||
|---|---|---|---|---|
| Biological Process | Molecular Function | Cellular Component | Total | |
| Enriched functions by up-regulated genes. | 44 | 28 | 12 | 84 |
| Enriched functions by down-regulated genes. | 162 | 99 | 27 | 288 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avico, E.H.; Acevedo, R.M.; Duarte, M.J.; Rodrigues Salvador, A.; Nunes-Nesi, A.; Ruiz, O.A.; Sansberro, P.A. Integrating Transcriptional, Metabolic, and Physiological Responses to Drought Stress in Ilex paraguariensis Roots. Plants 2023, 12, 2404. https://doi.org/10.3390/plants12132404
Avico EH, Acevedo RM, Duarte MJ, Rodrigues Salvador A, Nunes-Nesi A, Ruiz OA, Sansberro PA. Integrating Transcriptional, Metabolic, and Physiological Responses to Drought Stress in Ilex paraguariensis Roots. Plants. 2023; 12(13):2404. https://doi.org/10.3390/plants12132404
Chicago/Turabian StyleAvico, Edgardo H., Raúl M. Acevedo, María J. Duarte, Acácio Rodrigues Salvador, Adriano Nunes-Nesi, Oscar A. Ruiz, and Pedro A. Sansberro. 2023. "Integrating Transcriptional, Metabolic, and Physiological Responses to Drought Stress in Ilex paraguariensis Roots" Plants 12, no. 13: 2404. https://doi.org/10.3390/plants12132404
APA StyleAvico, E. H., Acevedo, R. M., Duarte, M. J., Rodrigues Salvador, A., Nunes-Nesi, A., Ruiz, O. A., & Sansberro, P. A. (2023). Integrating Transcriptional, Metabolic, and Physiological Responses to Drought Stress in Ilex paraguariensis Roots. Plants, 12(13), 2404. https://doi.org/10.3390/plants12132404







