Genotype × Environment Interaction and Stability Analysis of Selected Cassava Cultivars in South Africa
Abstract
1. Introduction
2. Results
2.1. AMMI Analysis of G × E Interaction
2.2. The GEI Patterns of Traits and Genotypes Based on GGE Biplot Analysis
2.3. Fresh Root Yield
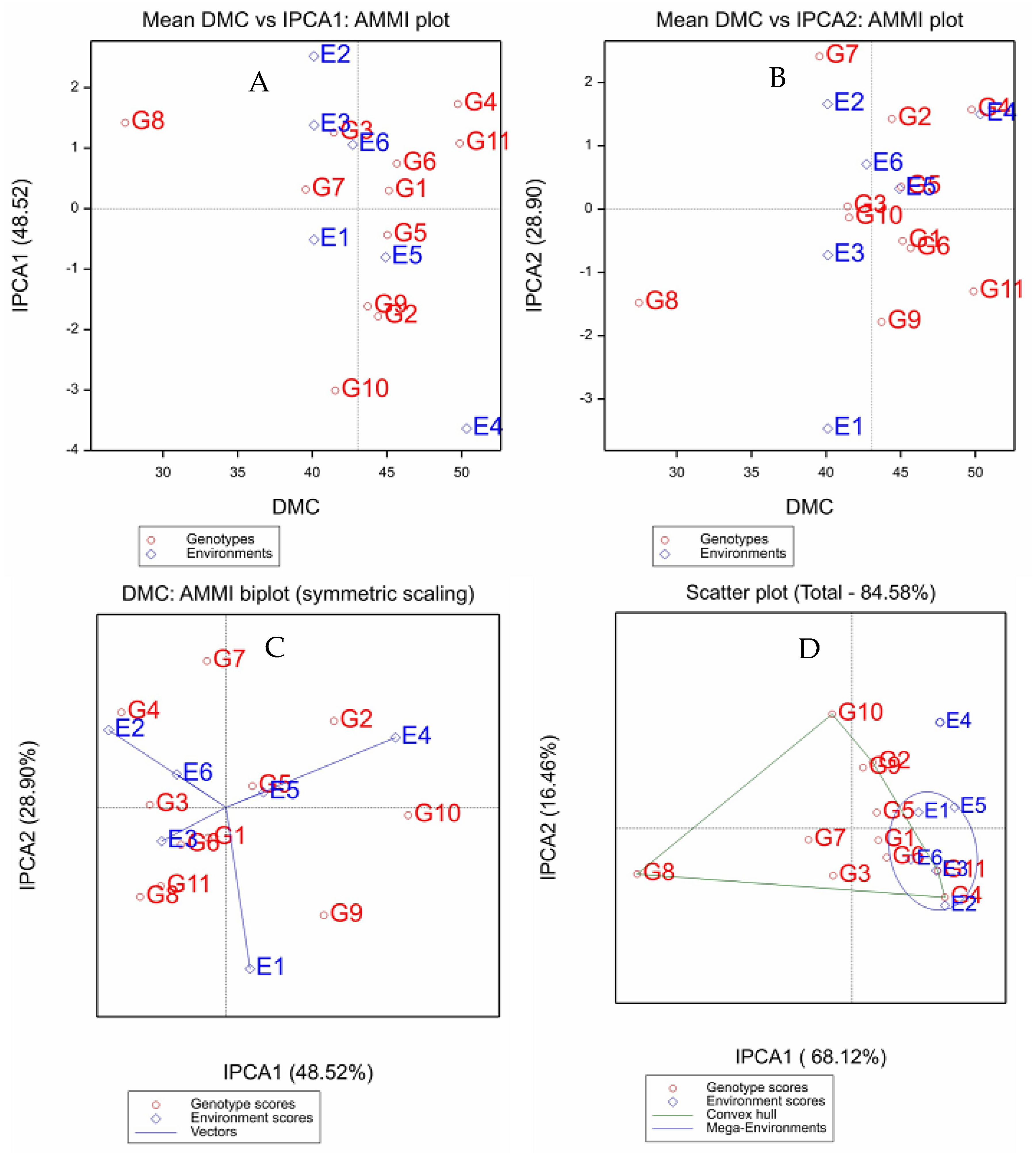
2.4. Dry Matter Content
2.5. Stability Analysis Using AMMI Model
3. Discussion
4. Materials and Methods
4.1. Trial Site Description
4.2. Planting Material and Experimental Design
4.3. Data Collected and Preparation of Samples
4.4. Data Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tonukari, N.J. Cassava and the future of starch. Electron. J. Biotechnol. 2004, 7, 5–8. [Google Scholar] [CrossRef]
- Amelework, A.B.; Bairu, M.W.; Maema, O.; Venter, S.L.; Laing, M. Adoption and Promotion of Resilient Crops for Climate Risk Mitigation and Import Substitution: A Case Analysis of Cassava for South African Agriculture. Front. Sustain. Food Syst. 2021, 5, 617783. [Google Scholar] [CrossRef]
- Giller, K.E.; Wilson, K.J. Nitrogen Fixation in Tropical Cropping System; CAB International: Wallingford, UK, 1991. [Google Scholar]
- Fotso, A.K.; Hanna, R.; Kulakow, P.; Parkes, E.; Iluebbey, P.; Ngome, F.A.; Suh, C.; Massussi, J.; Choutnji, I.; Wirnkar, V.L. AMMI analysis of cassava response to contrasting environments: Case study of genotype by environment effect on pests and diseases, root yield, and carotenoids content in Cameroon. Euphytica 2018, 214, 155. [Google Scholar] [CrossRef]
- Bustos-Korts, D.; Romagosa, I.; Borràs-Gelonch, G.; Casas, A.M.; Slafer, G.A.; van Eeuwijk, F. Genotype by environment interaction and adaptation. In Encyclopaedia of Sustainability Science and Technology; Meyers, R.A., Ed.; Springer Nature: Cham, Switzerland, 2018; pp. 1–44. [Google Scholar]
- Fox, P.N.; Crossa, J.; Romagosa, I. Multi-environment testing and genotype × environment interaction. In Statistical Methods for Plant Variety Evaluation; Kempton, R.A., Fox, P.N., Cerezo, M., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 117–138. [Google Scholar]
- Ebem, E.C.; Afuape, S.O.; Chukwu, S.C.; Ubi, B.E. Genotype × Environment Interaction and Stability Analysis for Root Yield in Sweet Potato [Ipomoea batatas (L.) Lam]. Front. Agron. 2021, 3, 665564. [Google Scholar] [CrossRef]
- Ceballos, H.; Iglesias, C.A.; Pérez, J.C.; Dixon, A.G. Cassava breeding: Opportunities and challenges. Plant Mol. Biol. 2004, 56, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Purchase, J.L.; Hatting, H.; van Deventer, C.S. Genotype × environment interaction of winter wheat (Triticum aestivum L.) in South Africa: II. Stability analysis of yield performance. S. Afr. J. Plant Soil 2000, 17, 101–107. [Google Scholar] [CrossRef]
- Yan, W. GGE Biplot vs. AMMI Graphs for genotype-by-environment data analysis. J. Ind. Soc. Agric. Stat. 2011, 65, 181–193. [Google Scholar]
- Nowosad, K.; Liersch, A.; Popławska, W.; Bocianowski, J. Genotype by environment interaction for seed yield in rapeseed (Brassica napus L.) using additive main effects and multiplicative interaction model. Euphytica 2016, 208, 187–194. [Google Scholar] [CrossRef]
- Yan, W.; Kang, M.S.; Ma, B.; Woods, S.; Cornelius, P.L. GGE Biplot vs. AMMI Analysis of Genotype-by-Environment Data. Crop Sci. 2007, 47, 643–653. [Google Scholar] [CrossRef]
- Annicchiarico, P. Genotype × Environment Interactions: Challenges and Opportunities for Plant Breeding and Cultivar Recommendations; FAO Plant Production and Protection Paper, 0259-2517, 174; Food and Agriculture Organization of the United Nations: Rome, Italy, 2022; pp. 1–126. [Google Scholar]
- Jiwuba, L.; Danquah, A.; Asante, I.; Blay, E.; Onyeka, J.; Danquah, E.; Egesi, C. Genotype by Environment Interaction on Resistance to Cassava Green Mite Associated Traits and Effects on Yield Performance of Cassava Genotypes in Nigeria. Front. Plant Sci. 2020, 11, 572200. [Google Scholar] [CrossRef]
- Chipeta, M.M.; Melis, R.; Shanahan, P.; Sibiya, J.; Benesi, I.R.M. Genotype × environment interaction and stability analysis of cassava genotypes at different harvest times. J. Anim. Plant Sci. 2017, 27, 901–919. [Google Scholar]
- Egesi, C.N.; Ilona, P.; Ogbe, F.O.; Akoroda, M.; Dixon, A. Genetic Variation and Genotype × Environment Interaction for Yield and Other Agronomic Traits in Cassava in Nigeria. Agron. J. 2007, 99, 1137–1142. [Google Scholar] [CrossRef]
- Akinwale, M.G.; Akinyele, B.O.; Dixon, A.G.O.; Odiyi, A.C. Genetic variability among forty-three cassava genotypes in there agro-ecological zones of Nigeria. J. Plant Breed. Crop Sci. 2010, 2, 104–109. [Google Scholar]
- Adjebeng-Danquah, J.; Manu-Aduening, J.; Gracen, V.E.; Asante, I.K.; Offei, S.K. AMMI Stability Analysis and Estimation of Genetic Parameters for Growth and Yield Components in Cassava in the Forest and Guinea Savannah Ecologies of Ghana. Int. J. Agron. 2017, 2017, 8075846. [Google Scholar] [CrossRef]
- Rotich, D.C.; Kiplagat, O.K.; Were, V.W. Genotype by environment interaction on yield components and stability analysis of elite cassava genotypes. Int. J. Plant Breed. Crop Sci. 2018, 5, 361–369. [Google Scholar]
- Bakare, M.A.; Kayondo, S.I.; Aghogho, C.I.; Wolfe, M.D.; Parkes, E.Y.; Kulakow, P.; Egesi, C.; Rabbi, I.Y.; Jannink, J.-L. Exploring genotype by environment interaction on cassava yield and yield related traits using classical statistical methods. PLoS ONE 2022, 17, e0268189. [Google Scholar] [CrossRef] [PubMed]
- Aina, O. Additive Main Effects and Multiplicative Interaction (AMMI) Analysis for Yield of Cassava in Nigeria. J. Biol. Sci. 2007, 7, 796–800. [Google Scholar] [CrossRef]
- Ssemakula, G.; Dixon, A. Genotype × environment interaction, stability and agronomic performance of carotenoid-rich cassava clones. Sci. Res. Essays 2007, 2, 390–399. [Google Scholar]
- Egesi, C.N.; Onyeka, T.J.; Asiedu, R. Environmental stability of resistance to anthracnose and virus diseases of water yam (Dioscorea alata). Afr. J. Agric. Res. 2009, 4, 113–118. [Google Scholar] [CrossRef]
- Aina, O.O.; Dixon, A.G.O.; Paul, I.; Akinrinde, E.A. G × E interaction effects on yield and yield components of cassava (landraces and improved) genotypes in the savannah regions of Nigeria. Afr. J. Biotechnol. 2009, 8, 4933–4945. [Google Scholar]
- Benesi, I.; Labuschagne, M.; Dixon, A.; Mahungu, N. Genotype × enviroment interaction effects on native cassava starch quality and potential for starch use in the commercial sector. Afr. Crop. Sci. J. 2004, 12, 205–216. [Google Scholar] [CrossRef]
- Boakye, B.; Kwadwo, O.; Asante, I.; Parkes, E.Y. Performance of nine cassava (Manihot esculanta Crantz) clones across three environments. J. Plant Breed. Crop Sci. 2013, 5, 48–53. [Google Scholar] [CrossRef]
- Tumuhimbise, R.; Melis, R.; Shanahan, P.; Kawuki, R. Genotype × environment interaction effects on early fresh storage root yield and related traits in cassava. Crop. J. 2014, 2, 329–337. [Google Scholar] [CrossRef]
- Nduwumuremyi, A.; Melis, R.; Shanahan, P.; Theodore, A. Interaction of genotype and environment effects on important traits of cassava (Manihot esculenta Crantz). Crop. J. 2017, 5, 373–386. [Google Scholar] [CrossRef]
- Farshadfar, E.; Mahmodi, N.; Yaghotipoor, A. AMMI stability value and simultaneous estimation of yield and yield stability in bread wheat (Triticum aestivum L.). Aust. J. Crop. Sci. 2011, 5, 1837–1844. [Google Scholar]
- Hagos, H.G.; Abay, F. AMMI and GGE biplot analysis of bread wheat genotypes in the northern part of Ethiopia. J. Plant Breed. Genet. 2013, 1, 12–18. [Google Scholar]
- Yan, W.; Tinker, N.A. Biplot analysis of multi-environment trial data: Principles and applications. Can. J. Plant Sci. 2006, 86, 623–645. [Google Scholar] [CrossRef]
- Gauch, H.G. Statistical Analysis of Yield Trials by AMMI and GGE. Crop. Sci. 2006, 46, 1488–1500. [Google Scholar] [CrossRef]
- Murphy, S.E.; Lee, E.A.; Woodrow, L.; Seguin, P.; Kumar, J.; Rajcan, I.; Ablett, G.R. Genotype × Environment Interaction and Stability for Isoflavone Content in Soybean. Crop. Sci. 2009, 49, 1313–1321. [Google Scholar] [CrossRef]
- Khan, M.H.; Rafii, M.Y.; Ramlee, S.I.; Jusoh, M.; Mamun, A. AMMI and GGE biplot analysis for yield performance and stability assessment of selected Bambara groundnut (Vigna subterranea L. Verdc.) genotypes under the multi-environmental trials (METs). Sci. Rep. 2021, 11, 22791. [Google Scholar] [CrossRef]
- Gauch, H.G., Jr.; Zobel, R.W. Identifying Mega-Environments and Targeting Genotypes. Crop. Sci. 1997, 37, 311–326. [Google Scholar] [CrossRef]
- Scavo, A.; Mauromicale, G.; Ierna, A. Dissecting the Genotype × Environment Interaction for Potato Tuber Yield and Components. Agronomy 2023, 13, 101. [Google Scholar] [CrossRef]
- Hauser, S.; Wairegi, L.; Asadu, C.L.A.; Asawalam, D.O.; Jokthan, G.; Ugbe, U. Cassava System cropping guide. Africa Soil Health Consortium: Nairobi, Kenya; CAB International: Wallingford, UK, 2014. [Google Scholar]
- GenStat. GenStat for Windows, 19th ed.; VSN International Ltd.: Oxford, UK, 2020. [Google Scholar]
- Jalata, Z. GGE-biplot Analysis of Multi-environment Yield Trials of Barley (Hordeium vulgare L.) Genotypes in Southeastern Ethiopia Highlands. Int. J. Plant Breed. Genet. 2011, 5, 59–75. [Google Scholar] [CrossRef]
- Maroya, N.G.; Kulakow, P.; Dixon, A.G.O.; Maziya-Dixon, B.B. Genotype × Environment Interaction of Mosaic Disease, Root Yields and Total Carotene Concentration of Yellow-Fleshed Cassava in Nigeria. Int. J. Agron. 2012, 2012, 434675. [Google Scholar] [CrossRef]
- Gabriel, K.R. The biplot graphic display of matrices with application to principal component analysis. Biometrika 1971, 58, 453–467. [Google Scholar] [CrossRef]
- Yan, W. GGEbiplot—A Windows Application for Graphical Analysis of Multienvironment Trial Data and Other Types of Two-Way Data. Agron. J. 2002, 93, 1111–1118. [Google Scholar] [CrossRef]

| Source | DF | Fresh Root Yield | Dry Matter Content | ||||
|---|---|---|---|---|---|---|---|
| SS | MS | % SS Explained | SS | MS | % SS Explained | ||
| Treatments | 65 | 173,736 | 2673 | 12,784 | 197 | ||
| Genotypes | 10 | 31,367 | 3137 *** | 18.05 | 6640 | 664 *** | 51.94 |
| Environments | 5 | 63,219 | 12,644 *** | 36.39 | 2726 | 545 *** | 21.32 |
| Interactions (GEI) | 50 | 79,150 | 1583 *** | 45.56 | 3418 | 68 *** | 26.74 |
| IPCA 1 | 14 | 39,273 | 2805 *** | 49.62 | 1658 | 118 *** | 48.51 |
| IPCA 2 | 12 | 19,846 | 1654 *** | 25.07 | 988 | 82 *** | 28.91 |
| Residuals | 24 | 20,031 | 835 | 772 | 32 | ||
| Error | 120 | 16,722 | 139 | 939 | 7.8 | ||
| Genotype | Mean | RY | IPCA1 | IPCA2 | ASV | RASV | GSI | RGSI |
|---|---|---|---|---|---|---|---|---|
| G1 | 86.77 | 1 | 3.14 | −0.55 | 6.24 | 8 | 9 | 2 |
| G2 | 71.31 | 7 | 1.77 | 1.79 | 3.94 | 4 | 11 | 4 |
| G3 | 85.91 | 2 | 2.31 | −7.40 | 8.70 | 9 | 11 | 6 |
| G4 | 50.22 | 11 | 1.56 | 1.91 | 3.63 | 3 | 14 | 8 |
| G5 | 72.13 | 5 | −1.78 | −1.95 | 4.03 | 5 | 10 | 3 |
| G6 | 84.84 | 3 | 1.04 | 2.21 | 3.02 | 1 | 4 | 1 |
| G7 | 78.51 | 4 | −3.14 | 0.54 | 6.23 | 7 | 11 | 5 |
| G8 | 52.8 | 10 | −1.72 | 1.03 | 3.56 | 2 | 12 | 7 |
| G9 | 65.11 | 8 | 5.47 | 2.62 | 11.13 | 10 | 18 | 11 |
| G10 | 55.91 | 9 | −2.15 | −1.21 | 4.43 | 6 | 15 | 9 |
| G11 | 71.37 | 6 | −6.49 | 1.02 | 12.88 | 11 | 17 | 10 |
| Genotype | Mean | RY | IPCA1 | IPCA2 | ASV | RASV | GSI | RGSI |
|---|---|---|---|---|---|---|---|---|
| G1 | 45.13 | 4 | 0.30 | −0.50 | 0.71 | 1 | 5 | 1 |
| G2 | 44.41 | 6 | −1.78 | 1.43 | 3.31 | 10 | 16 | 9 |
| G3 | 41.44 | 9 | 1.26 | 0.04 | 2.11 | 4 | 13 | 6 |
| G4 | 49.75 | 2 | 1.73 | 1.57 | 3.30 | 9 | 11 | 5 |
| G5 | 45.03 | 5 | −0.44 | 0.35 | 0.81 | 2 | 7 | 4 |
| G6 | 45.66 | 3 | 0.74 | −0.62 | 1.39 | 3 | 6 | 2 |
| G7 | 39.56 | 10 | 0.32 | 2.41 | 2.47 | 6 | 16 | 8 |
| G8 | 27.46 | 11 | 1.42 | −1.48 | 2.80 | 7 | 18 | 10 |
| G9 | 43.71 | 7 | −1.62 | −1.78 | 3.25 | 8 | 15 | 7 |
| G10 | 41.54 | 8 | −3.01 | −0.13 | 5.05 | 11 | 19 | 11 |
| G11 | 49.88 | 1 | 1.08 | −1.30 | 2.23 | 5 | 6 | 3 |
| Code | Location | District | Province | Soil Type | GPS Coordinate |
|---|---|---|---|---|---|
| E1 | Nseleni | Empangeni | KZN | Sandy | −28.634120, 31.912331 |
| E2 | Mabuyeni | King Cetshwayo | KZN | Silt | −28.853811, 31.961901 |
| E3 | Masibekela | Ehlanzeni | Mpumalanga | Sandy loam | −25.870814, 31.825738 |
| E4 | Shatale | Ehlanzeni | Mpumalanga | Sandy loam | −24.747785, 31.035320 |
| E5 | Mandlakazi | Mopani | Limpopo | Sandy loam | −23.801784, 30.377987 |
| E6 | Mutale | Vhembe | Limpopo | Clay | −22.721418, 30.572238 |
| Code | Type | Source | Trait | |
|---|---|---|---|---|
| G1 | 98/0002 | Released cultivar | IITA | CMD resistance |
| G2 | 98/0505 | Released cultivar | IITA | CMD resistance |
| G3 | MSFA2 | Landrace | ARC | High FRY/Low DMC |
| G4 | P1/19 | Breeding line | IITA | High DMC |
| G5 | P4/10 | Breeding line | IITA | High DMC |
| G6 | UKF3 | Breeding line | Kenya | High SC |
| G7 | UKF4 | Breeding line | Kenya | High SC |
| G8 | UKF5 | Breeding line | Kenya | High SC |
| G9 | UKF7 | Breeding line | Kenya | High SC |
| G10 | UKF8 | Breeding line | Kenya | High SC |
| G11 | UKF9 | Breeding line | Kenya | High SC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amelework, A.B.; Bairu, M.W.; Marx, R.; Laing, M.; Venter, S.L. Genotype × Environment Interaction and Stability Analysis of Selected Cassava Cultivars in South Africa. Plants 2023, 12, 2490. https://doi.org/10.3390/plants12132490
Amelework AB, Bairu MW, Marx R, Laing M, Venter SL. Genotype × Environment Interaction and Stability Analysis of Selected Cassava Cultivars in South Africa. Plants. 2023; 12(13):2490. https://doi.org/10.3390/plants12132490
Chicago/Turabian StyleAmelework, Assefa B., Michael W. Bairu, Roelene Marx, Mark Laing, and Sonja L. Venter. 2023. "Genotype × Environment Interaction and Stability Analysis of Selected Cassava Cultivars in South Africa" Plants 12, no. 13: 2490. https://doi.org/10.3390/plants12132490
APA StyleAmelework, A. B., Bairu, M. W., Marx, R., Laing, M., & Venter, S. L. (2023). Genotype × Environment Interaction and Stability Analysis of Selected Cassava Cultivars in South Africa. Plants, 12(13), 2490. https://doi.org/10.3390/plants12132490








