Structure-Guided Identification of Critical Residues in the Vacuolar Cation/Proton Antiporter NHX1 from Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
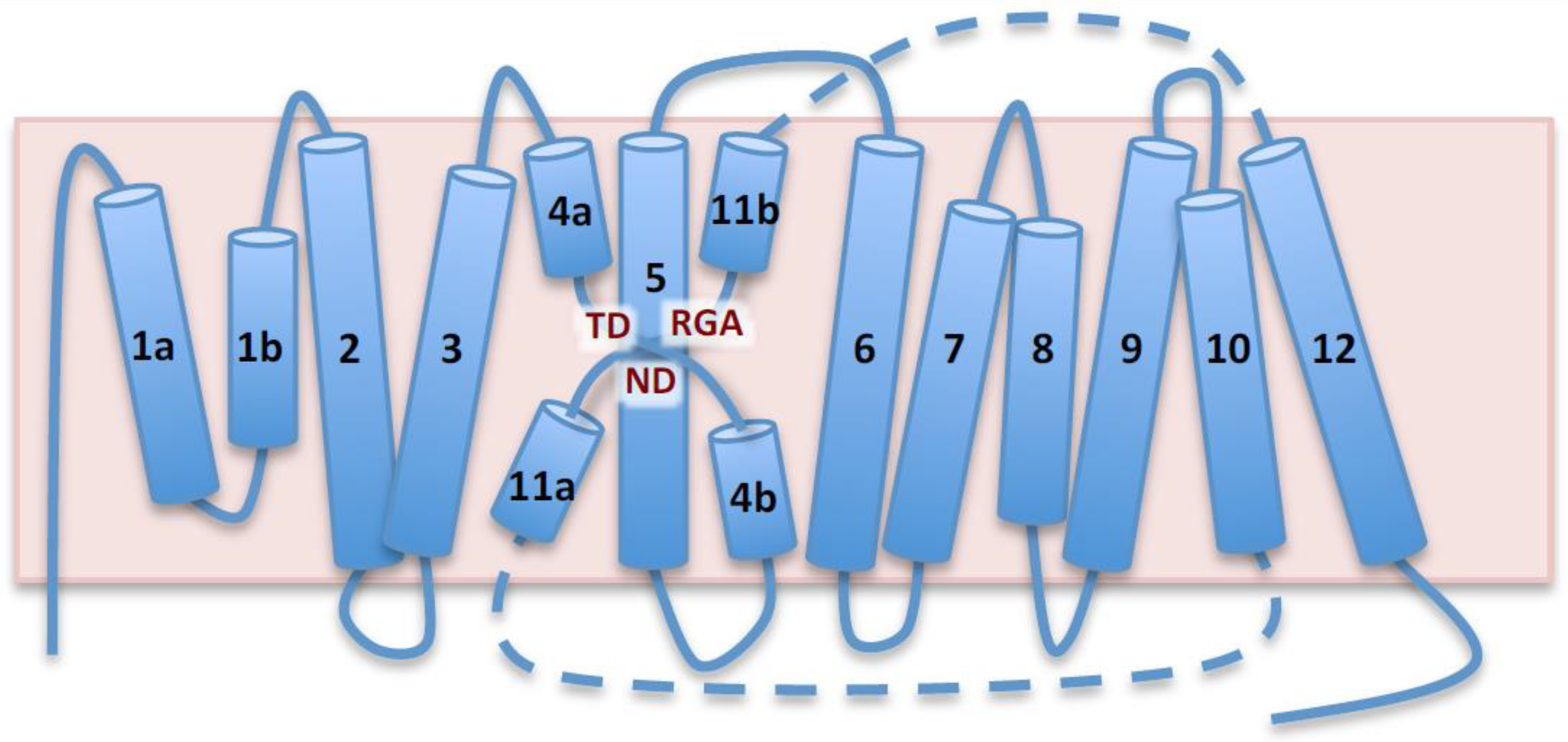
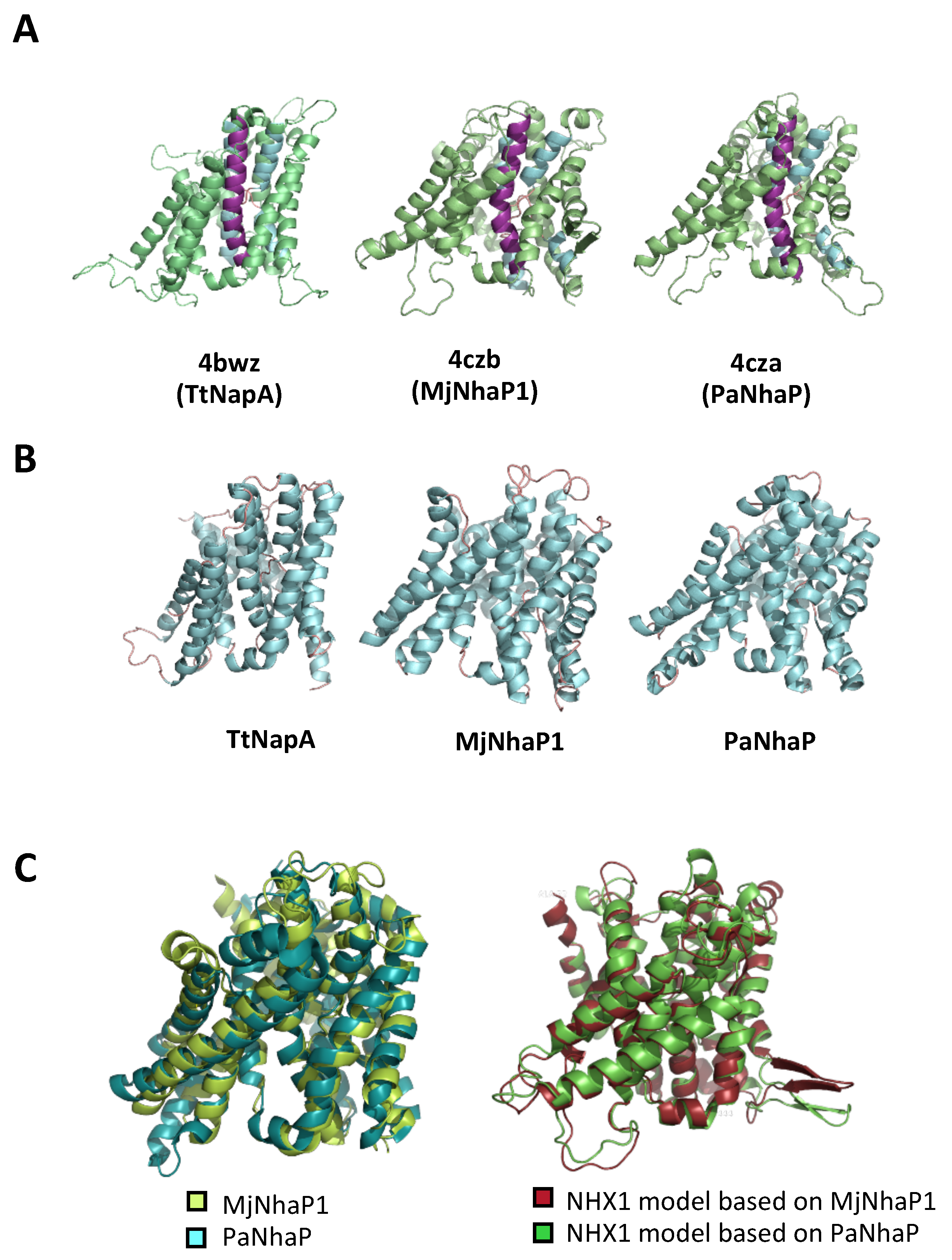
2.1. Topological and Ternary Structure Models
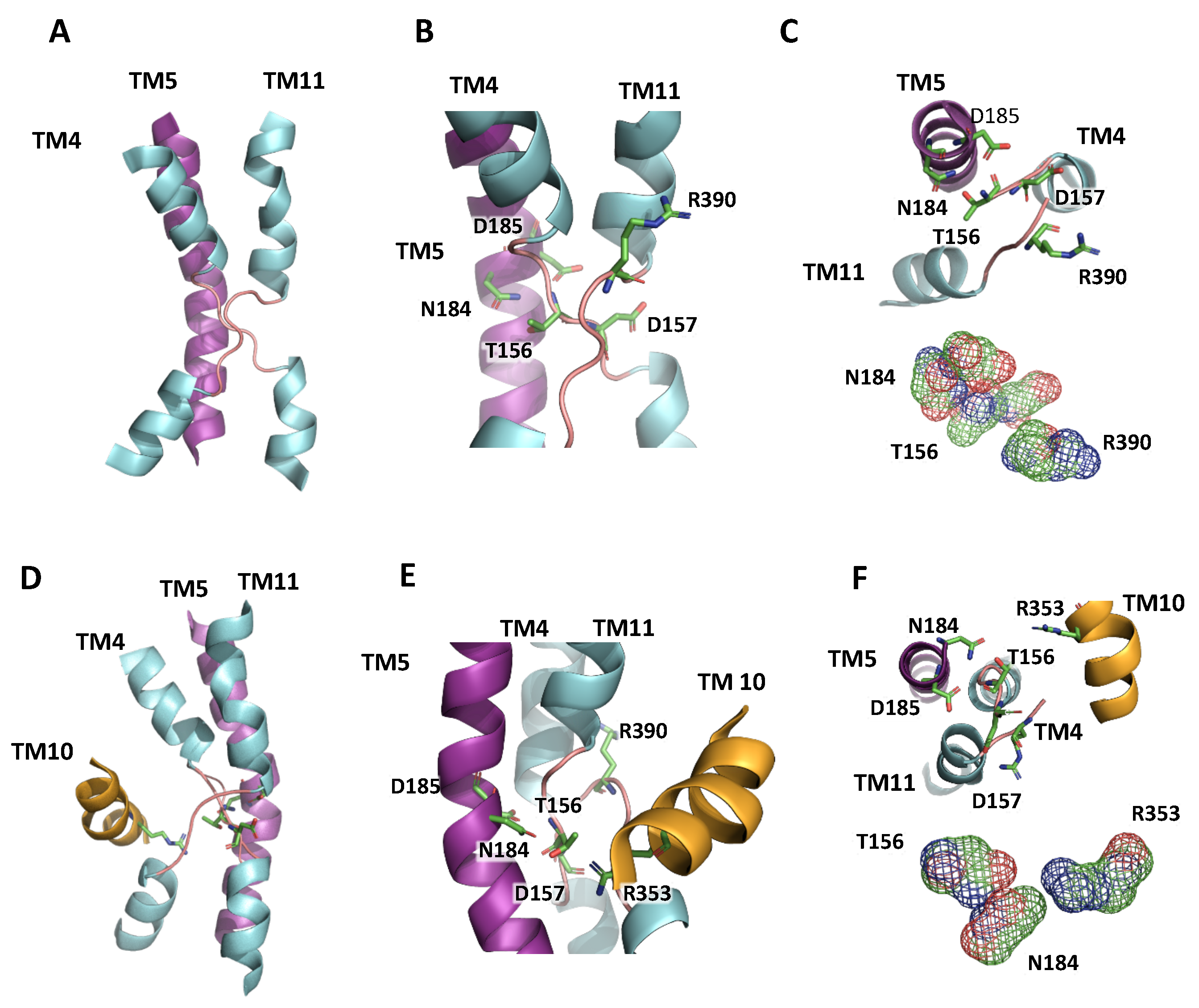
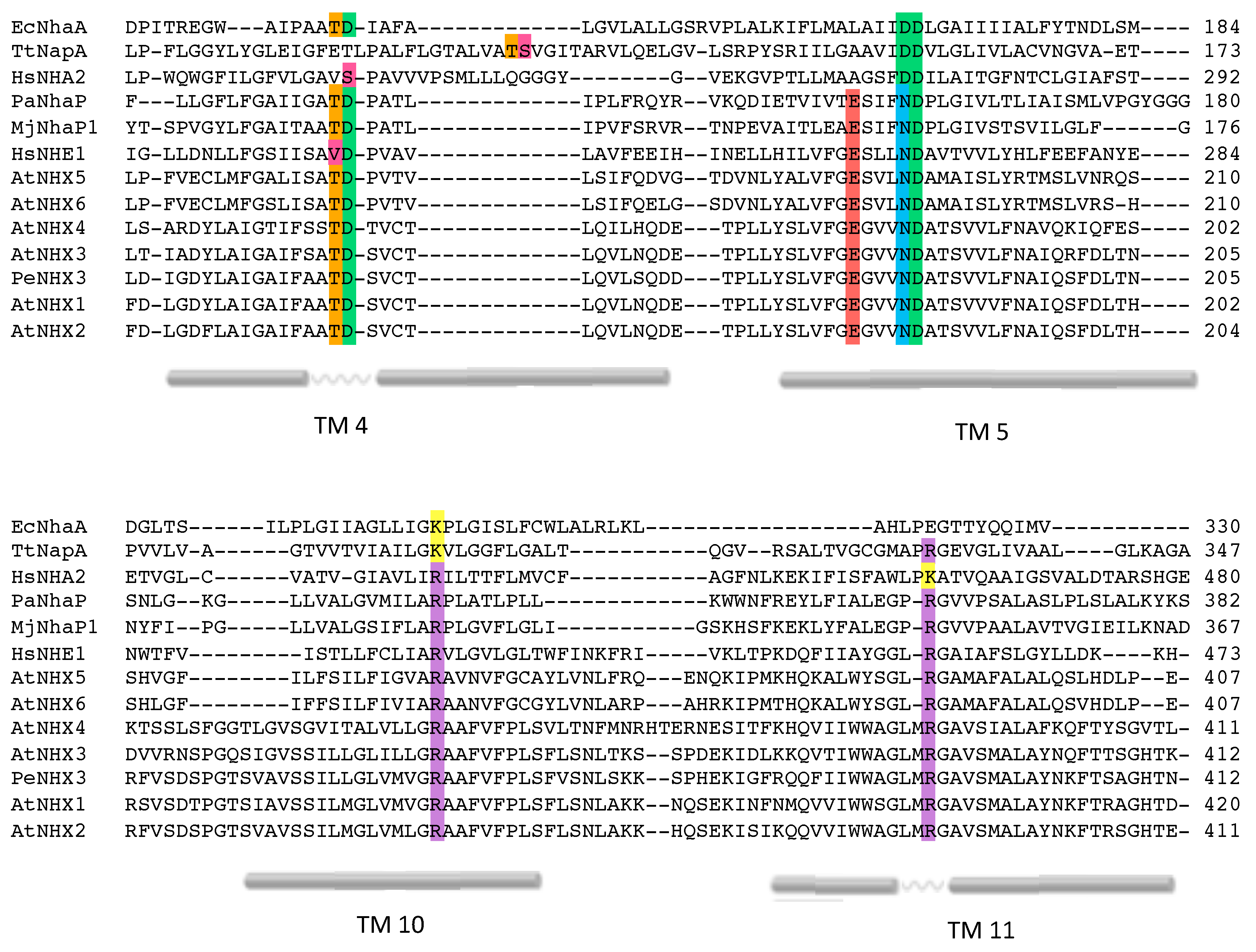
2.2. Conserved Residues in the Pore-Forming Domain
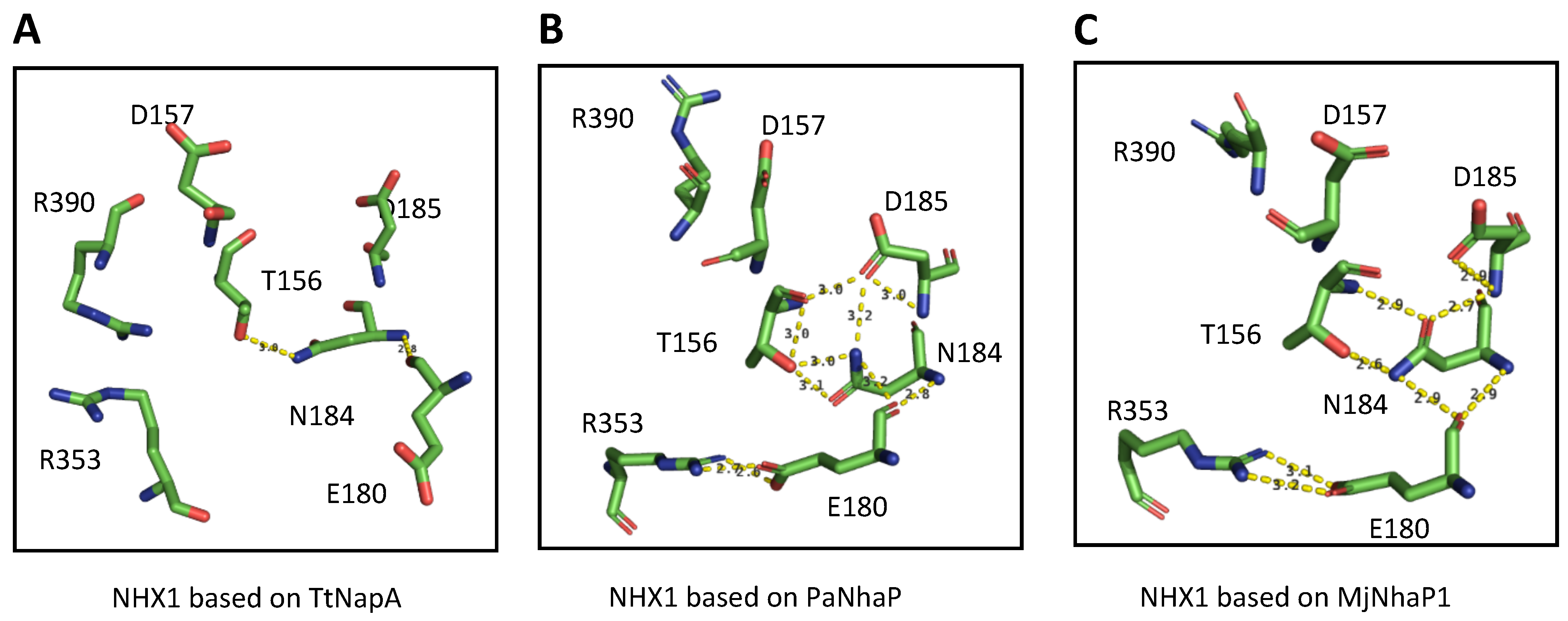
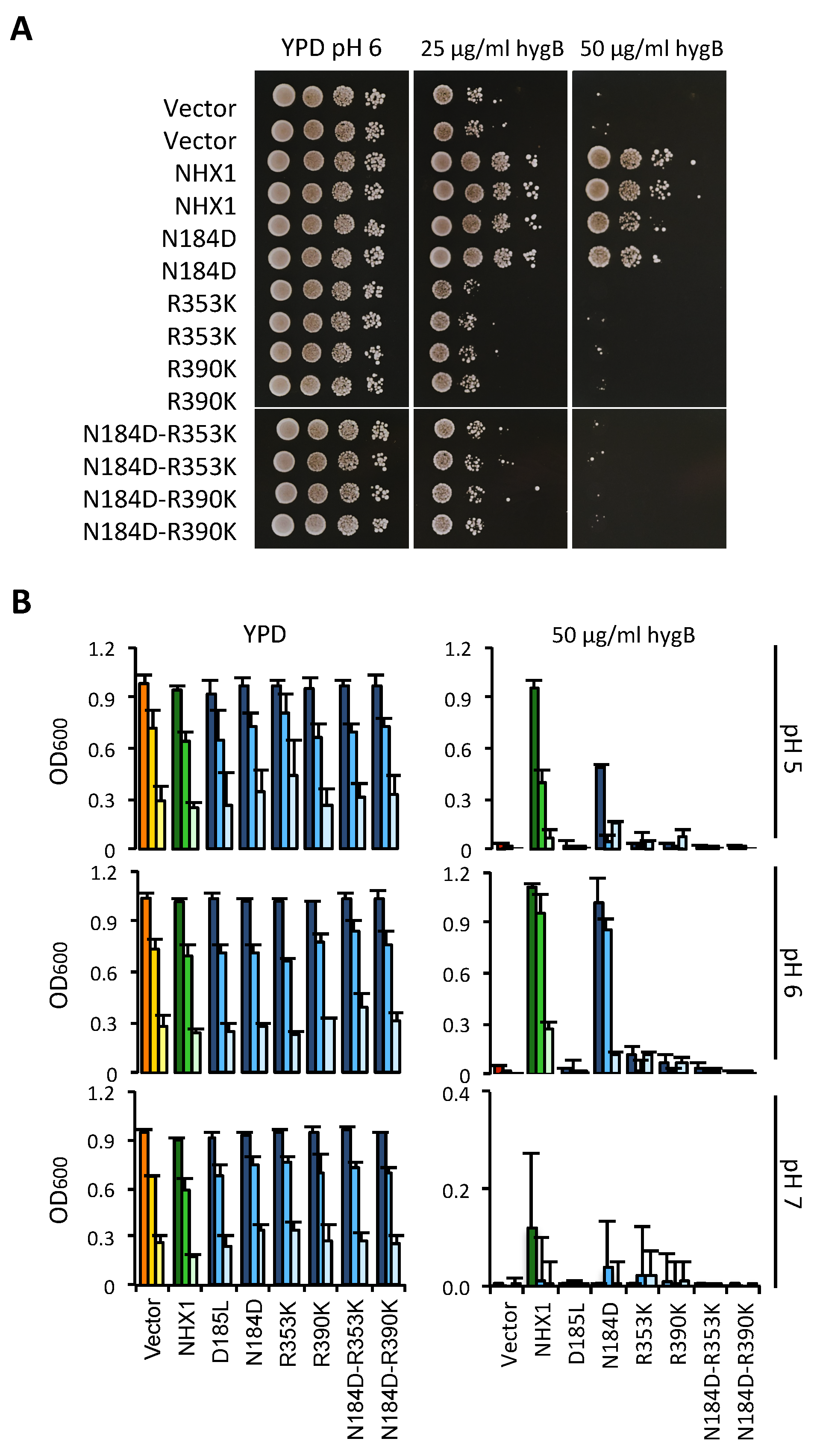
2.3. Conserved Residues with Functional Roles
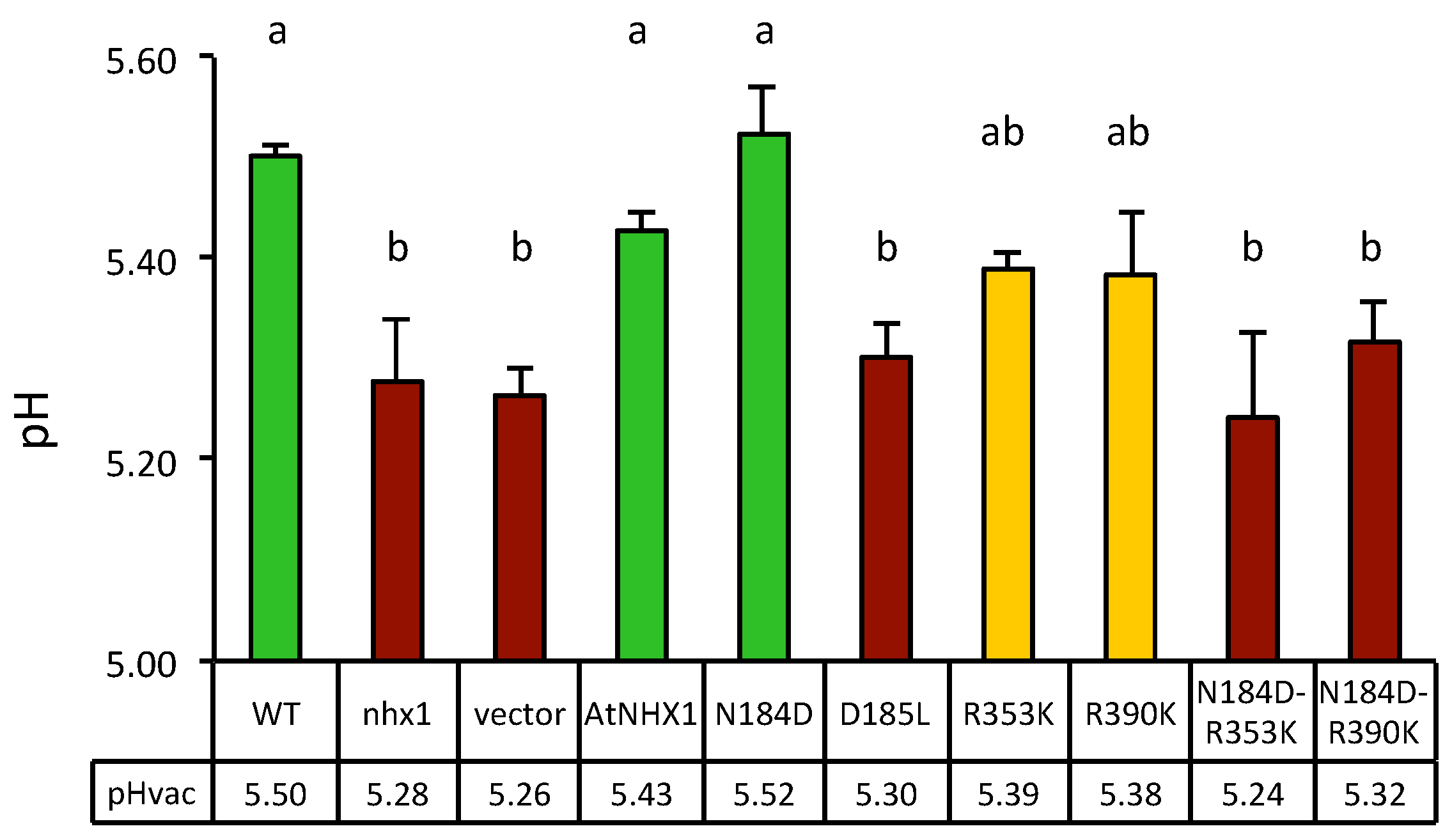
2.4. Vacuolar pH and AtNHX1 Mutants
3. Discussion
3.1. Generation of Topological and Tridimensional Models Based on Phylogenetic Relatedness
3.2. Identification of Conserved Residues with Functional Roles
3.3. Regulation of Vacuolar pH by AtNHX1
4. Materials and Methods
4.1. Structural Models for AtNHX1 and In Silico Validation
4.2. Plasmid Constructs
4.3. Complementation and Growth Assays
4.4. Measurements of Vacuolar pH
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Masrati, G.; Dwivedi, M.; Rimon, A.; Gluck-Margolin, Y.; Kessel, A.; Ashkenazy, H.; Mayrose, I.; Padan, E.; Ben-Tal, N. Broad phylogenetic analysis of cation/proton antiporters reveals transport determinants. Nat. Commun. 2018, 9, 4205. [Google Scholar] [CrossRef]
- Călinescu, O.; Dwivedi, M.; Patiño-Ruiz, M.; Padan, E.; Fendler, K. Lysine 300 is essential for stability but not for electrogenic transport of the Escherichia coli NhaA Na+/H+ antiporter. J. Biol. Chem. 2017, 292, 7932–7941. [Google Scholar] [CrossRef] [PubMed]
- Hunte, C.; Screpanti, E.; Venturi, M.; Rimon, A.; Padan, E.; Michel, H. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 2005, 435, 1197–1202. [Google Scholar] [CrossRef] [PubMed]
- Uzdavinys, P.; Coinçon, M.; Nji, E.; Ndi, M.; Winkelmann, I.; von Ballmoos, C.; Drew, D. Dissecting the proton transport pathway in electrogenic Na(+)/H(+) antiporters. Proc. Natl. Acad. Sci. USA 2017, 114, E1101–E1110. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Sakaguchi, M. Topogenic Properties of Transmembrane Segments of Arabidopsis thaliana NHX1 Reveal a Common Topology Model of the Na+/H+ Exchanger Family. J. Biochem. 2005, 138, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Apse, M.P.; Shi, H.Z.; Blumwald, E. Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc. Natl. Acad. Sci. USA 2003, 100, 12510–12515. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, X.; Zhao, H.; Wang, L.; An, L.; Qiu, Q.S. Functional Analysis of the Na+, K+/H+ Antiporter PeNHX3 from the Tree Halophyte Populus euphratica in Yeast by Model-Guided Mutagenesis. PLoS ONE 2014, 9, e104147. [Google Scholar] [CrossRef] [PubMed]
- Czerny, D.D.; Padmanaban, S.; Anishkin, A.; Venema, K.; Riaz, Z.; Sze, H. Protein architecture and core residues in unwound α-helices provide insights to the transport function of plant AtCHX17. Biochim. Biophys. Acta Biomembr. 2016, 1858, 1983–1998. [Google Scholar] [CrossRef]
- Ashnest, J.R.; Huynh, D.L.; Dragwidge, J.M.; Ford, B.A.; Gendall, A.R. Arabidopsis Intracellular NHX-Type Sodium-Proton Antiporters are Required for Seed Storage Protein Processing. Plant Cell Physiol. 2015, 56, 2220–2233. [Google Scholar] [CrossRef]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository—New features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Wöhlert, D.; Kühlbrandt, W.; Yildiz, O. Structure and substrate ion binding in the sodium/proton antiporter PaNhaP. eLife 2014, 3, e03579. [Google Scholar] [CrossRef]
- Goswami, P.; Paulino, C.; Hizlan, D.; Vonck, J.; Yildiz, O.; Kühlbrandt, W.; Yildiz, Ö.; Kühlbrandt, W. Structure of the archaeal Na+/H+ antiporter NhaP1 and functional role of transmembrane helix 1. EMBO J. 2011, 30, 439–449. [Google Scholar] [CrossRef]
- Lee, C.; Kang, H.J.; von Ballmoos, C.; Newstead, S.; Uzdavinys, P.; Dotson, D.L.; Iwata, S.; Beckstein, O.; Cameron, A.D.; Drew, D. A two-domain elevator mechanism for sodium/proton antiport. Nature 2013, 501, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Yashiro, S.; Dotson, D.L.; Uzdavinys, P.; Iwata, S.; Sansom, M.S.P.; von Ballmoos, C.; Beckstein, O.; Drew, D.; Cameron, A.D. Crystal structure of the sodium–proton antiporter NhaA dimer and new mechanistic insights. J. Gen. Physiol. 2014, 144, 529–544. [Google Scholar] [CrossRef] [PubMed]
- Whiteman, S.-A.; Serazetdinova, L.; Jones, A.M.E.; Sanders, D.; Rathjen, J.; Peck, S.C.; Maathuis, F.J.M. Identification of novel proteins and phosphorylation sites in a tonoplast enriched membrane fraction of Arabidopsis thaliana. Proteomics 2008, 8, 3536–3547. [Google Scholar] [CrossRef] [PubMed]
- Pabuayon, I.C.M.; Jiang, J.; Qian, H.; Chung, J.-S.; Shi, H. Gain-of-function mutations of AtNHX1 suppress sos1 salt sensitivity and improve salt tolerance in Arabidopsis. Stress Biol. 2021, 1, 14. [Google Scholar] [CrossRef]
- Hernandez, A.; Jiang, X.; Cubero, B.; Nieto, P.M.; Bressan, R.A.; Hasegawa, P.M.; Pardo, J.M. Mutants of the Arabidopsis thaliana Cation/H+ Antiporter AtNHX1 Conferring Increased Salt Tolerance in Yeast: The Endosome/Prevacuolar Compartment is a Target for Salt Toxicity. J. Biol. Chem. 2009, 284, 14276–14285. [Google Scholar] [CrossRef]
- Venema, K.; Quintero, F.J.; Pardo, J.M.; Donaire, J.P. The Arabidopsis Na+/H+ exchanger AtNHX1 catalyzes low affinity Na+ and K+ transport in reconstituted liposomes. J. Biol. Chem. 2002, 277, 2413–2418. [Google Scholar] [CrossRef]
- Landau, M.; Herz, K.; Padan, E.; Ben-Tal, N. Model structure of the Na+/H+ exchanger 1 (NHE1): Functional and clinical implications. J. Biol. Chem. 2007, 282, 37854–37863. [Google Scholar] [CrossRef]
- Schushan, M.; Xiang, M.; Bogomiakov, P.; Padan, E.; Rao, R.; Ben-Tal, N. Model-guided mutagenesis drives functional studies of human NHA2, implicated in hypertension. J. Mol. Biol. 2010, 396, 1181–1196. [Google Scholar] [CrossRef] [PubMed]
- Gradogna, A.; Scholz-Starke, J.; Pardo, J.M.; Carpaneto, A. Beyond the patch-clamp resolution: Functional activity of nonelectrogenic vacuolar NHX proton/potassium antiporters and inhibition by phosphoinositides. New Phytol. 2021, 229, 3026–3036. [Google Scholar] [CrossRef] [PubMed]
- Paulino, C.; Wöhlert, D.; Kapotova, E.; Yildiz, Ö.; Kühlbrandt, W. Structure and transport mechanism of the sodium/proton antiporter MjNhaP1. eLife 2014, 3, e03583. [Google Scholar] [CrossRef]
- Quintero, F.J.; Ohta, M.; Shi, H.Z.; Zhu, J.K.; Pardo, J.M. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA 2002, 99, 9061–9066. [Google Scholar] [CrossRef] [PubMed]
- Bowers, K.; Levi, B.P.; Patel, F.I.; Stevens, T.H. The sodium/proton exchanger Nhx1p is required for endosomal protein trafficking in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 2000, 11, 4277–4294. [Google Scholar] [CrossRef] [PubMed]
- Hellmer, J.; Teubner, A.; Zeilinger, C. Conserved arginine and aspartate residues are critical for function of MjNhaP1, a Na+/H+ antiporter of M. jannaschii. FEBS Lett. 2003, 542, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Maes, M.; Rimon, A.; Kozachkov-Magrisso, L.; Friedler, A.; Padan, E. Revealing the ligand binding site of NhaA Na+/H+ antiporter and its pH dependence. J. Biol. Chem. 2012, 287, 38150–38157. [Google Scholar] [CrossRef]
- Călinescu, O.; Linder, M.; Wöhlert, D.; Yildiz, Ö.; Kühlbrandt, W.; Fendler, K. Electrogenic Cation Binding in the Electroneutral Na+/H+ Antiporter of Pyrococcus abyssi. J. Biol. Chem. 2016, 291, 26786–26793. [Google Scholar] [CrossRef]
- Kozachkov, L.; Herz, K.; Padan, E. Functional and structural interactions of the transmembrane domain X of NhaA, Na+/H+ antiporter of Escherichia coli, at physiological pH. Biochemistry 2007, 46, 2419–2430. [Google Scholar] [CrossRef]
- Călinescu, O.; Paulino, C.; Kühlbrandt, W.; Fendler, K. Keeping it simple, transport mechanism and pH regulation in Na+/H+ exchangers. J. Biol. Chem. 2014, 289, 13168–13176. [Google Scholar] [CrossRef]
- Andrés, Z.; Pérez-Hormaeche, J.; Leidi, E.O.; Schlücking, K.; Steinhorst, L.; McLachlan, D.H.; Schumacher, K.; Hetherington, A.M.; Kudla, J.; Cubero, B.; et al. Control of vacuolar dynamics and regulation of stomatal aperture by tonoplast potassium uptake. Proc. Natl. Acad. Sci. USA 2014, 111, E1806–E1814. [Google Scholar] [CrossRef]
- Coonrod, E.M.; Graham, L.A.; Carpp, L.N.; Carr, T.M.; Stirrat, L.; Bowers, K.; Bryant, N.J.; Stevens, T.H. Homotypic vacuole fusion in yeast requires organelle acidification and not the V-ATPase membrane domain. Dev. Cell 2013, 27, 462–468. [Google Scholar] [CrossRef]
- Brett, C.L.; Kallay, L.; Hua, Z.; Green, R.; Chyou, A.; Zhang, Y.; Graham, T.R.; Donowitz, M.; Rao, R. Genome-wide analysis reveals the vacuolar pH-stat of Saccharomyces cerevisiae. PLoS ONE 2011, 6, e17619. [Google Scholar] [CrossRef] [PubMed]
- Plant, P.J.; Manolson, M.F.; Grinstein, S.; Demaurex, N. Alternative mechanisms of vacuolar acidification in H(+)-ATPase-deficient yeast. J. Biol. Chem. 1999, 274, 37270–37279. [Google Scholar] [CrossRef] [PubMed]
- Brett, C.L.; Tukaye, D.N.; Mukherjee, S.; Rao, R.J. The yeast endosomal Na+(K+)/H+ exchanger Nhx1 regulates cellular pH to control vesicle trafficking. Mol. Biol. Cell 2005, 16, 1396–1405. [Google Scholar] [CrossRef] [PubMed]
- Ali, R.; Brett, C.L.; Mukherjee, S.; Rao, R. Inhibition of sodium/proton exchange by a Rab-GTPase-activating protein regulates endosomal traffic in yeast. J. Biol. Chem. 2004, 279, 4498–4506. [Google Scholar] [CrossRef] [PubMed]
- James-Kracke, M.R. Quick and accurate method to convert BCECF fluorescence to pHi: Calibration in three different types of cell preparations. J. Cell. Physiol. 1992, 151, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Brett, C.L.; Donowitz, M.; Rao, R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol.-Cell Physiol. 2005, 288, C223–C239. [Google Scholar] [CrossRef]
- Sze, H.; Chanroj, S. Plant Endomembrane Dynamics: Studies of K(+)/H(+) Antiporters Provide Insights on the Effects of pH and Ion Homeostasis. Plant Physiol. 2018, 177, 875–895. [Google Scholar] [CrossRef] [PubMed]
- Hamaji, K.; Nagira, M.; Yoshida, K.; Ohnishi, M.; Oda, Y.; Uemura, T.; Goh, T.; Sato, M.H.; Morita, M.T.; Tasaka, M.; et al. Dynamic aspects of ion accumulation by vesicle traffic under salt stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2023–2033. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Inoue, H.; Noumi, T.; Tsuchiya, T.; Kanazawa, H. Essential aspartic acid residues, Asp-133, Asp-163 and Asp-164, in the transmembrane helices of a Na+/H+antiporter (NhaA) from Escherichia coli. FEBS Lett. 1995, 363, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Galili, L.; Rothman, A.; Kozachkov, L.; Rimon, A.; Padan, E. Trans membrane domain IV is involved in ion transport activity and pH regulation of the NhaA-Na+/H+ antiporter of Escherichia coli. Biochemistry 2002, 41, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Rimon, A.; Dwivedi, M.; Friedler, A.; Padan, E. Asp133 Residue in NhaA Na+/H+ Antiporter Is Required for Stability Cation Binding and Transport. J. Mol. Biol. 2018, 430, 867–880. [Google Scholar] [CrossRef] [PubMed]
- Barragan, V.; Leidi, E.; Andres, Z.; Rubio, L.; De Luca, A.; Fernandez, J.; Cubero, B.; Pardo, J. Ion Exchangers NHX1 and NHX2 Mediate Active Potassium Uptake into Vacuoles to Regulate Cell Turgor and Stomatal Function in Arabidopsis. Plant Cell 2012, 24, 1127–1142. [Google Scholar] [CrossRef] [PubMed]
- Bassil, E.; Zhang, S.; Gong, H.; Tajima, H.; Blumwald, E. Cation Specificity of Vacuolar NHX-Type Cation/H(+) Antiporters. Plant Physiol. 2019, 179, 616–629. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.Z.; Quintero, F.J.; Pardo, J.M.; Zhu, J.K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 2002, 14, 465–477. [Google Scholar] [CrossRef]
- Qiu, Q.S.; Barkla, B.J.; Vera-Estrella, R.; Zhu, J.K.; Schumaker, K.S. Na+/H+ exchange activity in the plasma membrane of Arabidopsis. Plant Physiol. 2003, 132, 1041–1052. [Google Scholar] [CrossRef]
- An, R.; Chen, Q.J.; Chai, M.F.; Lu, P.L.; Su, Z.; Qin, Z.X.; Chen, J.; Wang, X.C. AtNHX8, a member of the monovalent cation: Proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li+/H+ antiporter. Plant J. 2007, 49, 718–728. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Aharon, G.S.; Sottosanto, J.B.; Blumwald, E. Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+- and pH-dependent manner. Proc. Natl. Acad. Sci. USA 2005, 102, 16107–16112. [Google Scholar] [CrossRef]
- Xu, K.; Zhang, H.; Blumwald, E.; Xia, T. A novel plant vacuolar Na+/H+ antiporter gene evolved by DNA shuffling confers improved salt tolerance in yeast. J. Biol. Chem. 2010, 285, 22999–23006. [Google Scholar] [CrossRef] [PubMed]
- Bassil, E.; Tajima, H.; Liang, Y.C.; Ohto, M.A.; Ushijima, K.; Nakano, R.; Esumi, T.; Coku, A.; Belmonte, M.; Blumwald, E. The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 2011, 23, 3482–3497. [Google Scholar] [CrossRef] [PubMed]
- Reguera, M.; Bassil, E.; Tajima, H.; Wimmer, M.; Chanoca, A.; Otegui, M.S.; Paris, N.; Blumwald, E. pH Regulation by NHX-Type Antiporters Is Required for Receptor-Mediated Protein Trafficking to the Vacuole in Arabidopsis. Plant Cell 2015, 27, 1200–1217. [Google Scholar] [CrossRef] [PubMed]
- Haro, R.; Garciadeblas, B.; Rodriguez-Navarro, A. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 1991, 291, 189–191. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Wallin, E.; von Heijne, G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. A Publ. Protein Soc. 1998, 7, 1029–1038. [Google Scholar] [CrossRef]
- Landau, M.; Mayrose, I.; Rosenberg, Y.; Glaser, F.; Martz, E.; Pupko, T.; Ben-Tal, N. ConSurf 2005: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005, 33, W299–W302. [Google Scholar] [CrossRef]
- Bairoch, A.; Apweiler, R.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. The Universal Protein Resource (UniProt). Nucleic Acids Res. 2005, 33, D154–D159. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Eddy, S.R. Hidden Markov models. Curr. Opin. Struct. Biol. 1996, 6, 361–365. [Google Scholar] [CrossRef]
- Mayrose, I.; Mitchell, A.; Pupko, T. Site-specific evolutionary rate inference: Taking phylogenetic uncertainty into account. J. Mol. Evol. 2005, 60, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Grantham, R. Amino acid difference formula to help explain protein evolution. Science 1974, 185, 862–864. [Google Scholar] [CrossRef]
- Wang, W.; Malcolm, B.A. Two-Stage PCR Protocol Allowing Introduction of Multiple Mutations, Deletions and Insertions Using QuikChange TM Site-Directed Mutagenesis. BioTechniques 1999, 26, 680–682. [Google Scholar] [CrossRef]
- Elble, R. A simple and efficient procedure for transformation of yeasts. Biotechniques 1992, 13, 18–20. [Google Scholar] [PubMed]
- Rodriguez-Navarro, A.; Ramos, J. Dual system for potassium transport in Saccharomyces cerevisiae. J. Bacteriol. 1984, 159, 940–945. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rombolá-Caldentey, B.; Mendoza, I.; Quintero, F.J.; Pardo, J.M. Structure-Guided Identification of Critical Residues in the Vacuolar Cation/Proton Antiporter NHX1 from Arabidopsis thaliana. Plants 2023, 12, 2778. https://doi.org/10.3390/plants12152778
Rombolá-Caldentey B, Mendoza I, Quintero FJ, Pardo JM. Structure-Guided Identification of Critical Residues in the Vacuolar Cation/Proton Antiporter NHX1 from Arabidopsis thaliana. Plants. 2023; 12(15):2778. https://doi.org/10.3390/plants12152778
Chicago/Turabian StyleRombolá-Caldentey, Belén, Imelda Mendoza, Francisco J. Quintero, and José M. Pardo. 2023. "Structure-Guided Identification of Critical Residues in the Vacuolar Cation/Proton Antiporter NHX1 from Arabidopsis thaliana" Plants 12, no. 15: 2778. https://doi.org/10.3390/plants12152778
APA StyleRombolá-Caldentey, B., Mendoza, I., Quintero, F. J., & Pardo, J. M. (2023). Structure-Guided Identification of Critical Residues in the Vacuolar Cation/Proton Antiporter NHX1 from Arabidopsis thaliana. Plants, 12(15), 2778. https://doi.org/10.3390/plants12152778





