The Overexpression of Peanut (Arachis hypogaea L.) AhALDH2B6 in Soybean Enhances Cold Resistance
Abstract
:1. Introduction
2. Results
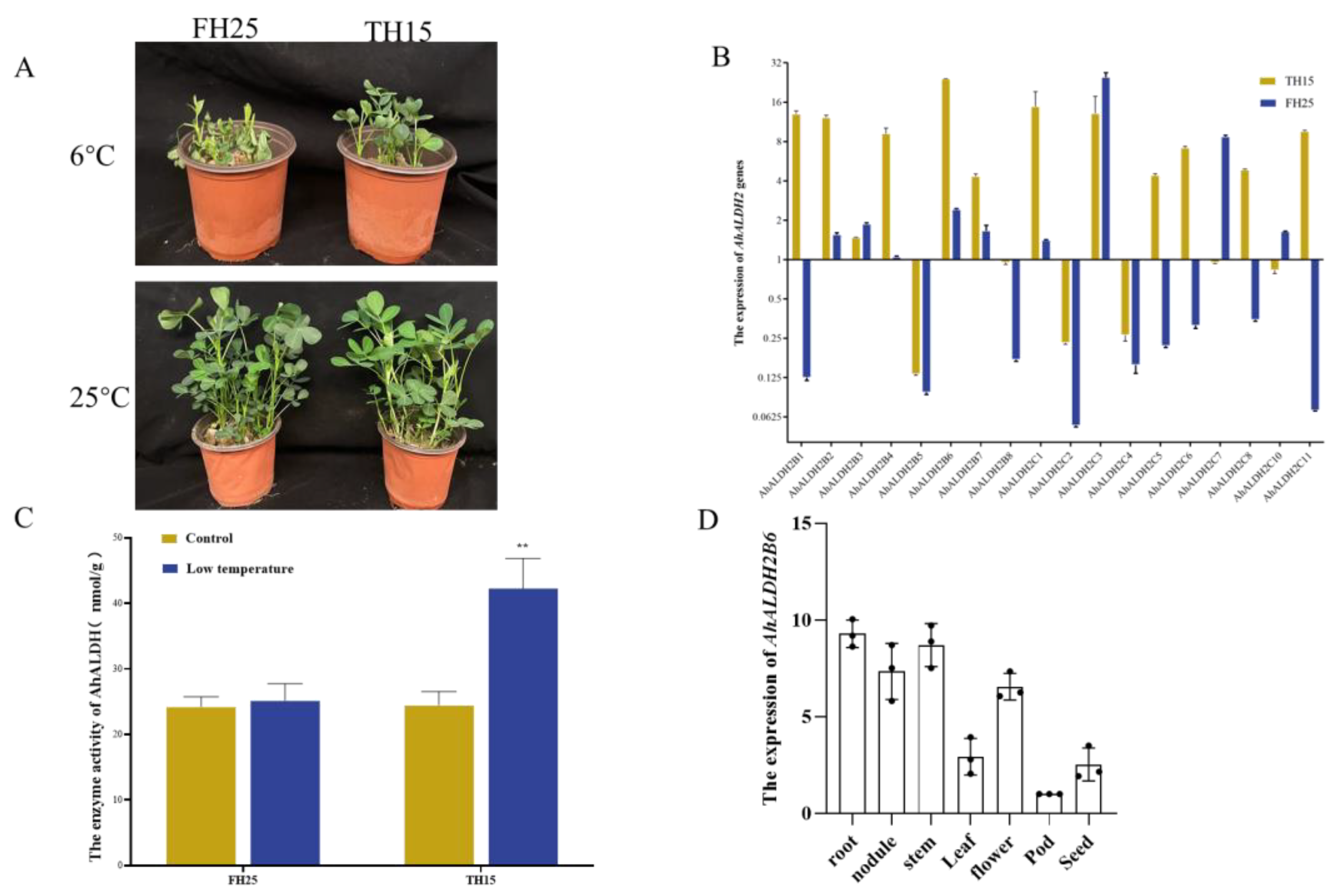
2.1. Expression of AhALDH2 Genes in Peanut Plants under Low-Temperature Stress
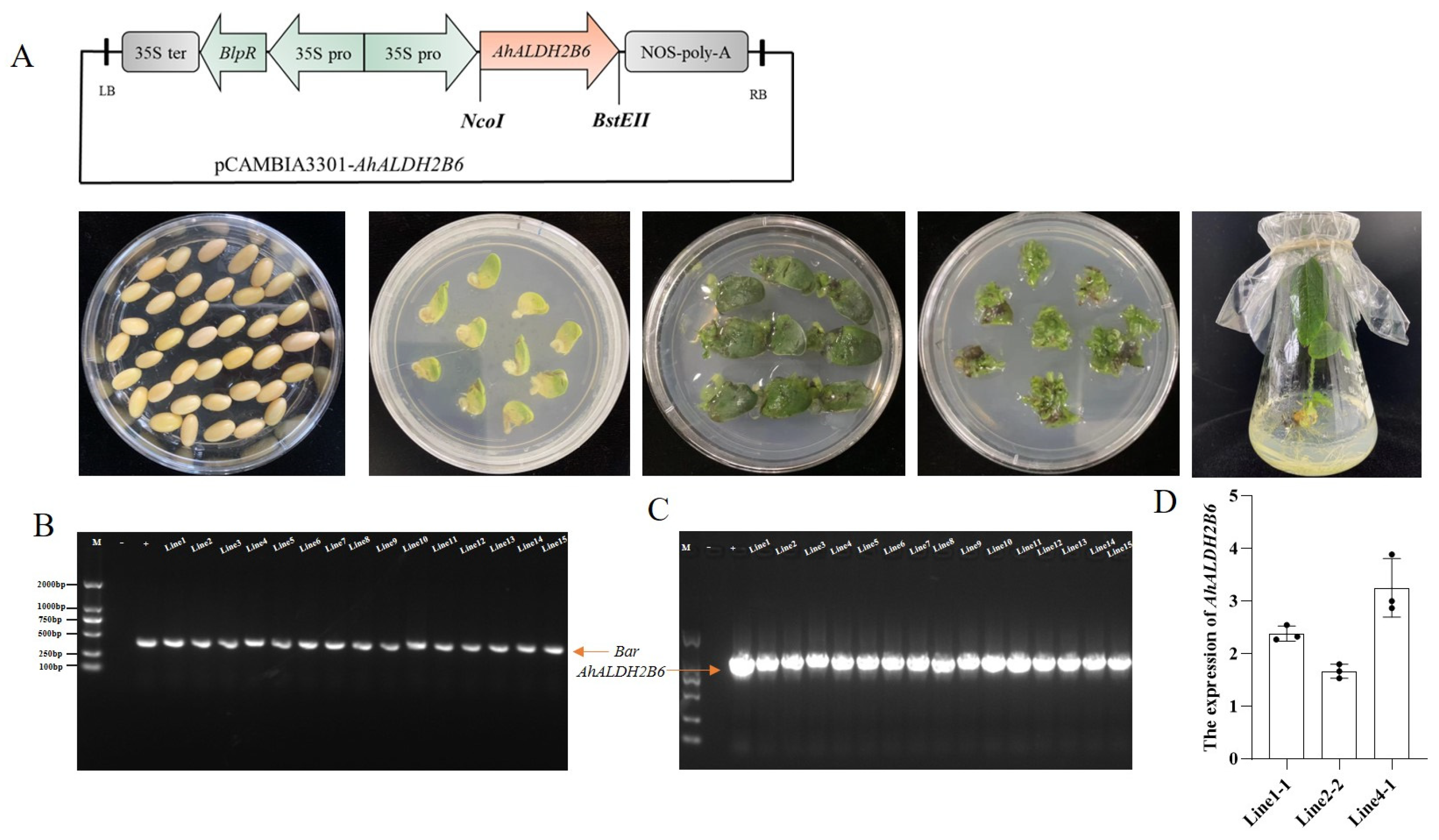
2.2. Soybean Transformation and Positive Line Identification
2.3. AhALDH2B6 Increases Soybean Low-Temperature Tolerance
2.4. Overexpression of AhALDH2B6 Reduced Lipid Peroxidation and Increased Protective Enzyme Activity
2.5. RNA-Seq Analysis of Transgenic Lines and P3 Soybeans
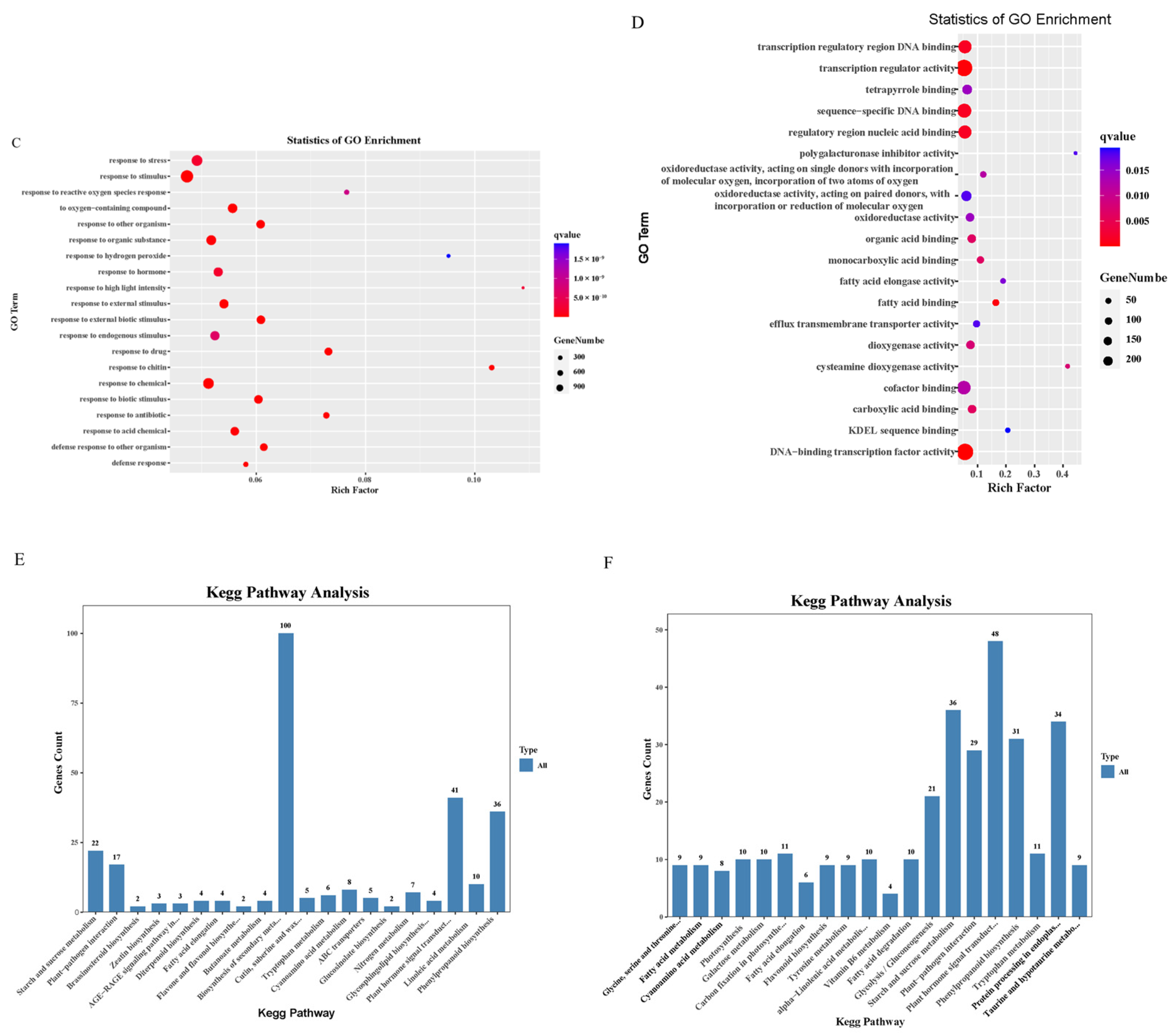
2.6. Gene Ontology and KEGG Enrichment Analysis of Differentially Expressed Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Vector Construction
4.3. Genetic Transformation and Progeny Identification in Soybean
4.4. Yeast Transformation and Low-Temperature Treatment
4.5. Low-Temperature Stress Treatment
4.6. RNA Library Construction
4.7. qRT-PCR Validation of Gene Expression
4.8. Quality Control and Reads Mapping
4.9. Gene Ontology and KEGG Pathway Analysis
4.10. Data Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qin, P.; Wang, T.; Luo, Y. A review on plant-based proteins from soybean: Health benefits and soy product development. J. Agric. Food Res. 2022, 7, 100265. [Google Scholar] [CrossRef]
- Bhat, K.A.; Mahajan, R.; Pakhtoon, M.M.; Urwat, U.; Bashir, Z.; Shah, A.A.; Agrawal, A.; Bhat, B.; Sofi, P.A.; Masi, A.; et al. Low Temperature Stress Tolerance: An Insight Into the Omics Approaches for Legume Crops. Front. Plant Sci. 2022, 13, 888710. [Google Scholar] [CrossRef]
- Guo, S.; Guo, E.; Zhang, Z.; Dong, M.; Wang, X.; Fu, Z.; Guan, K.; Zhang, W.; Zhang, W.; Zhao, J.; et al. Impacts of mean climate and extreme climate indices on soybean yield and yield components in Northeast China. Sci. Total Environ. 2022, 838, 156284. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.; Browse, J. Cold comfort farm: The acclimation of plants to freezing temperatures. Plant Cell Environ. 2001, 23, 893–902. [Google Scholar] [CrossRef]
- Lyons, J.M.; Raison, J.K.; Steponkus, P.L. The Plant Membrane in Response to Low Temperature: An Overview. In Low Temperature Stress in Crop Plants; Elsevier: Amsterdam, The Netherlands, 1979. [Google Scholar]
- Maqbool, A.; Shafiq, S.; Lake, L. Radiant frost tolerance in pulse crops—A review. Euphytica 2009, 172, 1–12. [Google Scholar] [CrossRef]
- Duke, S.H.; Schrader, L.E.; Miller, M.G. Low Temperature Effects on Soybean (Glycine max [L.] Merr. cv. Wells) Mitochondrial Respiration and Several Dehydrogenases during Imbibition and Germination. Plant Physiol. 1977, 60, 716–722. [Google Scholar] [CrossRef]
- Zhang, W.B.; Qiu, P.C.; Jiang, H.W.; Liu, C.Y.; Xin Da, W.; Li, C.D.; Hu, G.H.; Chen, Q.S. Dissection of genetic overlap of drought and low-temperature tolerance QTLs at the germination stage using backcross introgression lines in soybean. Mol. Biol. Rep. 2012, 39, 6087–6094. [Google Scholar] [CrossRef] [PubMed]
- Lyu, J.; Cai, Z.; Li, Y.; Suo, H.; Yi, R.; Zhang, S.; Nian, H. The Floral Repressor GmFLC-like Is Involved in Regulating Flowering Time Mediated by Low Temperature in Soybean. Int. J. Mol. Sci. 2020, 21, 1322. [Google Scholar] [CrossRef]
- Ohnishi, S.; Miyoshi, T.; Shirai, S. Low temperature stress at different flower developmental stages affects pollen development, pollination, and pod set in soybean. Environ. Exp. Bot. 2010, 69, 56–62. [Google Scholar] [CrossRef]
- Lokdarshi, A.; Guan, J.; Urquidi Camacho, R.A.; Cho, S.K.; Morgan, P.W.; Leonard, M.; Shimono, M.; Day, B.; von Arnim, A.G. Light Activates the Translational Regulatory Kinase GCN2 via Reactive Oxygen Species Emanating from the Chloroplast. Plant Cell 2020, 32, 1161–1178. [Google Scholar] [CrossRef]
- Farmer, E.E.; Mueller, M.J. ROS-mediated lipid peroxidation and RES-activated signaling. Annu. Rev. Plant Biol. 2013, 64, 429–450. [Google Scholar] [CrossRef]
- Ahmed Laskar, A.; Younus, H. Aldehyde toxicity and metabolism: The role of aldehyde dehydrogenases in detoxification, drug resistance and carcinogenesis. Drug Metab. Rev. 2019, 51, 42–64. [Google Scholar] [CrossRef]
- Stiti, N.; Giarola, V.; Bartels, D. From algae to vascular plants: The multistep evolutionary trajectory of the ALDH superfamily towards functional promiscuity and the emergence of structural characteristics. Environ. Exp. Bot. 2021, 185, 104376. [Google Scholar] [CrossRef]
- Wang, W.; Jiang, W.; Liu, J.; Li, Y.; Gai, J.; Li, Y. Genome-wide characterization of the aldehyde dehydrogenase gene superfamily in soybean and its potential role in drought stress response. BMC Genom. 2017, 18, 518. [Google Scholar] [CrossRef] [PubMed]
- Levitt, J. Responses of Plants to Environmental Stresses. In Chilling Freezing & High Temperature Stress; Academic Press: Cambridge, MA, USA, 1980. [Google Scholar]
- Xiaoming Zhang, J.Z.; Cao, L.; Ren, C.; Yu, G.; Gu, Y.; Ruan, J.; Zhao, S.; Wang, L.; Ru, H.; Cheng, L.; et al. Genome-wide characterization of aldehyde dehydrogenase gene family members in groundnut (Arachis hypogaea) and the analysis under saline-alkali stress. Front. Plant Sci. 2023, 14, 1097001. [Google Scholar] [CrossRef]
- Islam, M.S.; Ghosh, A. Evolution, family expansion, and functional diversification of plant aldehyde dehydrogenases. Gene 2022, 829, 146522. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Mohtasim, M.; Islam, T.; Ghosh, A. Aldehyde dehydrogenase superfamily in sorghum: Genome-wide identification, evolution, and transcript profiling during development stages and stress conditions. BMC Plant Biol. 2022, 22, 316. [Google Scholar] [CrossRef]
- Xing, W.; Rajashekar, C.B. Glycine betaine involvement in freezing tolerance and water stress in Arabidopsis thaliana. Environ. Exp. Bot. 2001, 46, 21–28. [Google Scholar] [CrossRef]
- Gao, C.; Jiang, B.; Wang, Y.; Liu, G.; Yang, C. Overexpression of a heat shock protein (ThHSP18.3) from Tamarix hispida confers stress tolerance to yeast. Mol. Biol. Rep. 2012, 39, 4889–4897. [Google Scholar] [CrossRef]
- Singh, S.; Brocker, C.; Koppaka, V.; Chen, Y.; Jackson, B.C.; Matsumoto, A.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic. Biol. Med. 2013, 56, 89–101. [Google Scholar] [CrossRef]
- Nakazono, M.; Tsuji, H.; Li, Y.; Saisho, D.; Arimura, S.; Tsutsumi, N.; Hirai, A. Expression of a gene encoding mitochondrial aldehyde dehydrogenase in rice increases under submerged conditions. Plant Physiol. 2000, 124, 587–598. [Google Scholar] [CrossRef]
- Guo, J.; Sun, W.; Liu, H.; Chi, J.; Odiba, A.S.; Li, G.; Jin, L.; Xin, C. Aldehyde dehydrogenase plays crucial roles in response to lower temperature stress in Solanum tuberosum and Nicotiana benthamiana. Plant Sci. 2020, 297, 110525. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, S.; Bashir, K.; Kim, J.M.; Ando, M.; Tanaka, M.; Seki, M. The modulation of acetic acid pathway genes in Arabidopsis improves survival under drought stress. Sci. Rep. 2018, 8, 7831. [Google Scholar] [CrossRef]
- Huang, W.; Ma, X.; Wang, Q.; Gao, Y.; Xue, Y.; Niu, X.; Yu, G.; Liu, Y. Significant improvement of stress tolerance in tobacco plants by overexpressing a stress-responsive aldehyde dehydrogenase gene from maize (Zea mays). Plant Mol. Biol. 2008, 68, 451–463. [Google Scholar] [CrossRef]
- Jiang, J.; Hou, R.; Yang, N.; Li, L.; Deng, J.; Qin, G.; Ding, D. Physiological and TMT-labeled proteomic analyses reveal important roles of sugar and secondary metabolism in Citrus junos under cold stress. J. Proteom. 2021, 237, 104145. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Miao, Y.; Shao, L.; Hu, T.; Yang, P. De novo transcriptome sequencing and gene expression profiling of Elymus nutans under cold stress. BMC Genom. 2016, 17, 870. [Google Scholar] [CrossRef]
- Bahrman, N.; Hascoet, E.; Jaminon, O.; Depta, F.; Hu, J.F.; Bouchez, O.; Lejeune-Henaut, I.; Delbreil, B.; Legrand, S. Identification of Genes Differentially Expressed in Response to Cold in Pisum sativum Using RNA Sequencing Analyses. Plants 2019, 8, 288. [Google Scholar] [CrossRef] [PubMed]
- Thapa, R.; Tabien, R.E.; Johnson, C.D.; Septiningsih, E.M. Comparative transcriptomic analysis of germinating rice seedlings to individual and combined anaerobic and cold stress. BMC Genom. 2023, 24, 185. [Google Scholar] [CrossRef] [PubMed]
- Berens, M.L.; Wolinska, K.W.; Spaepen, S.; Ziegler, J.; Nobori, T.; Nair, A.; Kruler, V.; Winkelmuller, T.M.; Wang, Y.; Mine, A.; et al. Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc. Natl. Acad. Sci. USA 2019, 116, 2364–2373. [Google Scholar] [CrossRef]
- Paz, M.M.; Martinez, J.C.; Kalvig, A.B.; Fonger, T.M.; Wang, K. Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation. Plant Cell Rep. 2006, 25, 206–213. [Google Scholar] [CrossRef]
- Zhang, L.; Li, T.; Wang, Y.; Zhang, Y.; Dong, Y.S. FvC5SD overexpression enhances drought tolerance in soybean by reactive oxygen species scavenging and modulating stress-responsive gene expression. Plant Cell Rep. 2019, 38, 1039–1051. [Google Scholar] [CrossRef] [PubMed]
- De Sena Brandine, G.; Smith, A.D. Falco: High-speed FastQC emulation for quality control of sequencing data. F1000Research 2019, 8, 1874. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Teng, Y.; Yue, T.; Wang, Z.; Feng, G.; Ruan, J.; Yan, S.; Zheng, Y.; Zhang, L.; Chen, Q.; et al. The Overexpression of Peanut (Arachis hypogaea L.) AhALDH2B6 in Soybean Enhances Cold Resistance. Plants 2023, 12, 2928. https://doi.org/10.3390/plants12162928
Yang M, Teng Y, Yue T, Wang Z, Feng G, Ruan J, Yan S, Zheng Y, Zhang L, Chen Q, et al. The Overexpression of Peanut (Arachis hypogaea L.) AhALDH2B6 in Soybean Enhances Cold Resistance. Plants. 2023; 12(16):2928. https://doi.org/10.3390/plants12162928
Chicago/Turabian StyleYang, Mingyu, Yuhan Teng, Tong Yue, Ziye Wang, Guanghui Feng, Jingwen Ruan, Shi Yan, Yuhong Zheng, Ling Zhang, Qingshan Chen, and et al. 2023. "The Overexpression of Peanut (Arachis hypogaea L.) AhALDH2B6 in Soybean Enhances Cold Resistance" Plants 12, no. 16: 2928. https://doi.org/10.3390/plants12162928
APA StyleYang, M., Teng, Y., Yue, T., Wang, Z., Feng, G., Ruan, J., Yan, S., Zheng, Y., Zhang, L., Chen, Q., & Meng, F. (2023). The Overexpression of Peanut (Arachis hypogaea L.) AhALDH2B6 in Soybean Enhances Cold Resistance. Plants, 12(16), 2928. https://doi.org/10.3390/plants12162928






