Exploring Salinity Tolerance Mechanisms in Diverse Wheat Genotypes Using Physiological, Anatomical, Agronomic and Gene Expression Analyses
Abstract
:1. Introduction
2. Results
2.1. Response of Biochemical Compounds to Salinity Stress
2.2. Response of Enzyme Activities to Salinity Stress
2.3. Anatomical Measurements under Salinity Stress
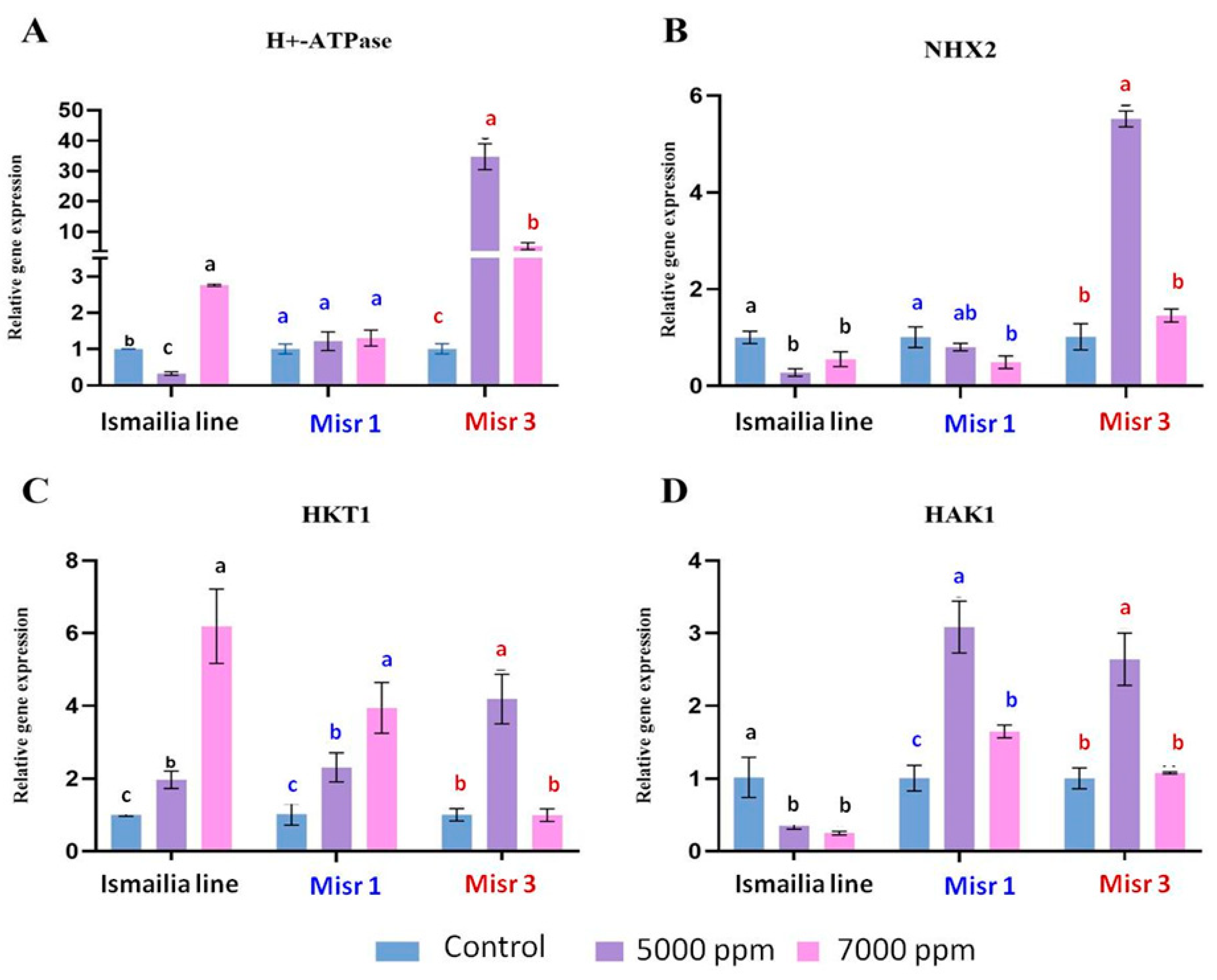
2.4. Gene Expression under Salinity Stress
2.5. Agronomic Response to Salinity Stress
2.6. Na+ and K+ Contents and K+/Na+ Ratio in Wheat Leaves
3. Discussion
4. Materials and Methods
4.1. Experimental Setup and Plant Material
4.2. Studied Characters
4.2.1. Physiological Parameters
4.2.2. Anatomical Measurements
4.2.3. RNA Extraction and qPCR Analysis
4.2.4. Agronomic Traits
4.3. Analysis of Variances
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raman, H.; Stodart, B.; Cavanagh, C.; Mackay, M.; Morell, M.; Milgate, A.; Martin, P. Molecular diversity and genetic structure of modern and traditional landrace cultivars of wheat (Triticum aestivum L.). Crop Pasture Sci. 2010, 61, 222–229. [Google Scholar] [CrossRef]
- FAOSTAT. Food and Agriculture Organization of the United Nations Statistics Database. 2023. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 21 June 2023).
- Gander, M.J.; Singh, G. Climate change is increasing global salt pollution. Water Int. 2023, 1–8. [Google Scholar] [CrossRef]
- Desoky, E.-S.M.; Merwad, A.-R.M.; Abo El-Maati, M.F.; Mansour, E.; Arnaout, S.M.; Awad, M.F.; Ramadan, M.F.; Ibrahim, S.A. Physiological and biochemical mechanisms of exogenously applied selenium for alleviating destructive impacts induced by salinity stress in bread wheat. Agronomy 2021, 11, 926. [Google Scholar] [CrossRef]
- Awaad, H.A.; Mansour, E.; Akrami, M.; Fath, H.E.S.; Javadi, A.A.; Negm, A. Availability and feasibility of water desalination as a non-conventional resource for agricultural irrigation in the mena region: A review. Sustainability 2020, 12, 7592. [Google Scholar] [CrossRef]
- Islam, R.; Ahmed, R.; Dey, B.; Haque, M.S.; Aktar, S.; Bhuiyan, M.S.; Arif, M.S.; Ador, M.A.H.; Haque, M.M.U.; Saha, N. Salinity hazard drives the alteration of occupation, land use and ecosystem service in the coastal areas: Evidence from the south-western coastal region of Bangladesh. Heliyon 2023, 9, e18512. [Google Scholar] [CrossRef]
- ElSayed, A.I.; Mohamed, A.H.; Rafudeen, M.S.; Omar, A.A.; Awad, M.F.; Mansour, E. Polyamines mitigate the destructive impacts of salinity stress by enhancing photosynthetic capacity, antioxidant defense system and upregulation of calvin cycle-related genes in rapeseed (Brassica napus L.). Saudi J. Biol. Sci. 2022, 29, 3675–3686. [Google Scholar] [CrossRef]
- Paz, A.M.; Amezketa, E.; Canfora, L.; Castanheira, N.; Falsone, G.; Gonçalves, M.C.; Gould, I.; Hristov, B.; Mastrorilli, M.; Ramos, T. Salt-affected soils: Field-scale strategies for prevention, mitigation, and adaptation to salt accumulation. Ital. J. Agron. 2023, 18, 2166. [Google Scholar] [CrossRef]
- Qadir, M.; Quillérou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J.; Drechsel, P.; Noble, A.D. Economics of salt-induced land degradation and restoration. Nat. Resour. Forum 2014, 38, 282–295. [Google Scholar] [CrossRef]
- EL Sabagh, A.; Islam, M.S.; Skalicky, M.; Ali Raza, M.; Singh, K.; Anwar Hossain, M.; Hossain, A.; Mahboob, W.; Iqbal, M.A.; Ratnasekera, D. Salinity stress in wheat (Triticum aestivum L.) in the changing climate: Adaptation and management strategies. Front. Agron. 2021, 3, 661932. [Google Scholar] [CrossRef]
- Oyiga, B.C.; Sharma, R.; Shen, J.; Baum, M.; Ogbonnaya, F.; Léon, J.; Ballvora, A. Identification and characterization of salt tolerance of wheat germplasm using a multivariable screening approach. J. Agron. Crop Sci. 2016, 202, 472–485. [Google Scholar] [CrossRef]
- Ellis, R.; Forster, B.; Gordon, D.; Handley, L.; Keith, R.; Lawrence, P.; Meyer, R.; Powell, W.; Robinson, D.; Scrimgeour, C. Phenotype/genotype associations for yield and salt tolerance in a barley mapping population segregating for two dwarfing genes. J. Exp. Bot. 2002, 53, 1163–1176. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Shabala, S.; Shabala, L.; Fan, Y.; Zhou, M.X. Evaluating Predictive Values of Various Physiological Indices for Salinity Stress Tolerance in Wheat. J. Agron. Crop Sci. 2016, 202, 115–124. [Google Scholar] [CrossRef]
- Hu, P.; Zheng, Q.; Luo, Q.; Teng, W.; Li, H.; Li, B.; Li, Z. Genome-wide association study of yield and related traits in common wheat under salt-stress conditions. BMC Plant Biol. 2021, 21, 27. [Google Scholar] [CrossRef] [PubMed]
- El-Hendawy, S.E.; Hassan, W.M.; Al-Suhaibani, N.A.; Refay, Y.; Abdella, K.A. Comparative performance of multivariable agro-physiological parameters for detecting salt tolerance of wheat cultivars under simulated saline field growing conditions. Front. Plant Sci. 2017, 8, 435. [Google Scholar] [CrossRef]
- Kamara, M.M.; Rehan, M.; Mohamed, A.M.; El Mantawy, R.F.; Kheir, A.M.S.; Abd El-Moneim, D.; Safhi, F.A.; Alshamrani, S.M.; Hafez, E.M.; Behiry, S.I.; et al. Genetic potential and inheritance patterns of physiological, agronomic and quality traits in bread wheat under normal and water deficit conditions. Plants 2022, 11, 952. [Google Scholar] [CrossRef]
- Junghans, U.; Polle, A.; Düchting, P.; Weiler, E.; Kuhlman, B.; Gruber, F.; Teichmann, T. Adaptation to high salinity in poplar involves changes in xylem anatomy and auxin physiology. Plant Cell Environ. 2006, 29, 1519–1531. [Google Scholar] [CrossRef] [PubMed]
- Solomon, M.; Gedalovich, E.; Mayer, A.; Poljakoff-Mayber, A. Changes induced by salinity to the anatomy and morphology of excised pea roots in culture. Ann. Bot. 1986, 57, 811–818. [Google Scholar] [CrossRef]
- Syvertsen, J.; Lloyd, J.; McConchie, C.; Kriedemann, P.; Farquhar, G. On the relationship between leaf anatomy and CO2 diffusion through the mesophyll of hypostomatous leaves. Plant Cell Environ. 1995, 18, 149–157. [Google Scholar] [CrossRef]
- Terletskaya, N.; Rysbekova, A.; Iskakova, A.; Khailenko, N.; Polimbetova, F. Saline stress response of plantlets of common wheat (Triticum aestivum) and its wild congeners. J. Agric. Sci. Technol. B 2011, 6, 198–204. [Google Scholar]
- Niinemets, Ü.; Sack, L. Structural determinants of leaf light-harvesting capacity and photosynthetic potentials. In Progress in Botany; Springer: Berlin/Heidelberg, Germany, 2006; pp. 385–419. [Google Scholar] [CrossRef]
- Mondal, R.; Kumar, A.; Gnanesh, B.N. Crop germplasm: Current challenges, physiological-molecular perspective, and advance strategies towards development of climate-resilient crops. Heliyon 2023, 9, e12973. [Google Scholar] [CrossRef]
- Al-Faifi, S.A.; Migdadi, H.M.; Algamdi, S.S.; Khan, M.A.; Ammar, M.H.; Al-Obeed, R.S.; Al-Thamra, M.I.; El-Harty, E.H.; Jakse, J. Development, characterization and use of genomic SSR markers for assessment of genetic diversity in some Saudi date palm (Phoenix dactylifera L.) cultivars. Electron. J. Biotechnol. 2016, 21, 18–25. [Google Scholar] [CrossRef]
- Kronzucker, H.J.; Coskun, D.; Schulze, L.M.; Wong, J.R.; Britto, D.T. Sodium as nutrient and toxicant. Plant Soil 2013, 369, 1–23. [Google Scholar] [CrossRef]
- Assaha, D.V.M.; Ueda, A.; Saneoka, H.; Al-Yahyai, R.; Yaish, M.W. The Role of Na+ and K+ Transporters in Salt Stress Adaptation in Glycophytes. Front. Physiol. 2017, 8, 509. [Google Scholar] [CrossRef]
- Mushke, R.; Yarra, R.; Kirti, P. Improved salinity tolerance and growth performance in transgenic sunflower plants via ectopic expression of a wheat antiporter gene (TaNHX2). Mol. Biol. Rep. 2019, 46, 5941–5953. [Google Scholar] [CrossRef] [PubMed]
- Borjigin, C.; Schilling, R.K.; Jewell, N.; Brien, C.; Sanchez-Ferrero, J.C.; Eckermann, P.J.; Watson-Haigh, N.S.; Berger, B.; Pearson, A.S.; Roy, S.J. Identifying the genetic control of salinity tolerance in the bread wheat landrace Mocho de Espiga Branca. Funct. Plant Biol. 2021, 48, 1148–1160. [Google Scholar] [CrossRef] [PubMed]
- Neang, S.; Goto, I.; Skoulding, N.S.; Cartagena, J.A.; Kano-Nakata, M.; Yamauchi, A.; Mitsuya, S. Tissue-specific expression analysis of Na+ and Cl− transporter genes associated with salt removal ability in rice leaf sheath. BMC Plant Biol. 2020, 20, 502. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Y.; Li, D.; Feng, S.; Yang, J.; Zhang, J.; Zhang, J.; Wang, D.; Gan, Y. Improving salt tolerance in potato through overexpression of AtHKT1 gene. BMC Plant Biol. 2019, 19, 357. [Google Scholar] [CrossRef] [PubMed]
- Yousefirad, S.; Soltanloo, H.; Ramezanpour, S.S.; Zaynali Nezhad, K.; Shariati, V. The RNA-seq transcriptomic analysis reveals genes mediating salt tolerance through rapid triggering of ion transporters in a mutant barley. PLoS ONE 2020, 15, e0229513. [Google Scholar] [CrossRef]
- Mansour, E.; Moustafa, E.S.; Abdul-Hamid, M.I.; Ash-shormillesy, S.M.; Merwad, A.-R.M.; Wafa, H.A.; Igartua, E. Field responses of barley genotypes across a salinity gradient in an arid Mediterranean environment. Agric. Water Manag. 2021, 258, 107206. [Google Scholar] [CrossRef]
- Chen, W.; Jin, M.; Ferré, T.P.A.; Liu, Y.; Xian, Y.; Shan, T.; Ping, X. Spatial distribution of soil moisture, soil salinity, and root density beneath a cotton field under mulched drip irrigation with brackish and fresh water. Field Crops Res. 2018, 215, 207–221. [Google Scholar] [CrossRef]
- Hickey, L.T.; Hafeez, A.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Mansour, E.; Moustafa, E.S.; Desoky, E.-S.M.; Ali, M.M.; Yasin, M.A.; Attia, A.; Alsuhaibani, N.; Tahir, M.U.; El-Hendawy, S. Multidimensional evaluation for detecting salt tolerance of bread wheat genotypes under actual saline field growing conditions. Plants 2020, 9, 1324. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, H.; Song, C.; Zhu, J.-K.; Shabala, S. Mechanisms of Plant Responses and Adaptation to Soil Salinity. Innovation 2020, 1, 100017. [Google Scholar] [CrossRef] [PubMed]
- Parida, A.K.; Das, A.B. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef]
- Hernandez, J.; Olmos, E.; Corpas, F.; Sevilla, F.; Del Rio, L. Salt-induced oxidative stress in chloroplasts of pea plants. Plant Sci. 1995, 105, 151–167. [Google Scholar] [CrossRef]
- Singh, A.; Singh, R.; Kumar, S. Influence of salinity on seedling growth and metabolism in maize genoytypes. Indian J. Plant Physiol. 2008, 13, 95–99. [Google Scholar] [CrossRef]
- Verma, S.; Mishra, S.N. Putrescine alleviation of growth in salt stressed Brassica juncea by inducing antioxidative defense system. J. Plant Physiol. 2005, 162, 669–677. [Google Scholar] [CrossRef]
- Agastian, P.; Kingsley, S.; Vivekanandan, M. Effect of salinity on photosynthesis and biochemical characteristics in mulberry genotypes. Photosynthetica 2000, 38, 287–290. [Google Scholar] [CrossRef]
- Mitra, A.; Banerjee, K. Pigments of Heritiera fomes seedlings under different salinity conditions: Perspective sea level rise. Mesopotamian J. Mar. Sci. 2010, 25, 1–10. [Google Scholar] [CrossRef]
- Khatkar, D.; Kuhad, M. Short-term salinity induced changes in two wheat cultivars at different growth stages. Biol. Plant. 2000, 43, 629–632. [Google Scholar] [CrossRef]
- Ahmad, M.S.A.; Ashraf, M.; Ali, Q. Soil salinity as a selection pressure is a key determinant for the evolution of salt tolerance in Blue Panicgrass (Panicum antidotale Retz.). Flora-Morphol. Distrib. Funct. Ecol. Plants 2010, 205, 37–45. [Google Scholar] [CrossRef]
- Siddique, A.; Kandpal, G.; Kumar, P. Proline accumulation and its defensive role under diverse stress condition in plants: An overview. J. Pure Appl. Microbiol. 2018, 12, 1655–1659. [Google Scholar] [CrossRef]
- Mangal, V.; Lal, M.K.; Tiwari, R.K.; Altaf, M.A.; Sood, S.; Kumar, D.; Bharadwaj, V.; Singh, B.; Singh, R.K.; Aftab, T. Molecular Insights into the Role of Reactive Oxygen, Nitrogen and Sulphur Species in Conferring Salinity Stress Tolerance in Plants. J. Plant Growth Regul. 2023, 42, 554–574. [Google Scholar] [CrossRef]
- Khaleghi, A.; Naderi, R.; Brunetti, C.; Maserti, B.E.; Salami, S.A.; Babalar, M. Morphological, physiochemical and antioxidant responses of Maclura pomifera to drought stress. Sci. Rep. 2019, 9, 19250. [Google Scholar] [CrossRef] [PubMed]
- Pastuszak, J.; Kopeć, P.; Płażek, A.; Gondek, K.; Szczerba, A.; Hornyák, M.; Dubert, F. Antioxidant activity as a response to cadmium pollution in three durum wheat genotypes differing in salt-tolerance. Open Chem. 2020, 18, 1230–1241. [Google Scholar] [CrossRef]
- Kumar, S.; Abedin, M.M.; Singh, A.K.; Das, S. Role of phenolic compounds in plant-defensive mechanisms. Plant Phenolics Sustain. Agric. 2020, 1, 517–532. [Google Scholar] [CrossRef]
- Sairam, R.; Srivastava, G.; Agarwal, S.; Meena, R. Differences in antioxidant activity in response to salinity stress in tolerant and susceptible wheat genotypes. Biol. Plant. 2005, 49, 85–91. [Google Scholar] [CrossRef]
- Muthukumarasamy, M.; Gupta, S.D.; Panneerselvam, R. Enhancement of peroxidase, polyphenol oxidase and superoxide dismutase activities by triadimefon in NaCl stressed Raphanus sativus L. Biol. Plant. 2000, 43, 317–320. [Google Scholar] [CrossRef]
- Jaleel, C.A.; Gopi, R.; Manivannan, P.; Panneerselvam, R. Antioxidative potentials as a protective mechanism in Catharanthus roseus (L.) G. Don. plants under salinity stress. Turk. J. Bot. 2007, 31, 245–251. [Google Scholar]
- Datir, S.; Singh, N.; Joshi, I. Effect of NaCl-induced salinity stress on growth, osmolytes and enzyme activities in wheat genotypes. Bull. Environ. Contam. Toxicol. 2020, 104, 351–357. [Google Scholar] [CrossRef]
- Dionisio-Sese, M.L.; Tobita, S. Antioxidant responses of rice seedlings to salinity stress. Plant Sci. 1998, 135, 1–9. [Google Scholar] [CrossRef]
- Abdel Latef, A. Changes of antioxidative enzymes in salinity tolerance among different wheat cultivars. Cereal Res. Commun. 2010, 38, 43–55. [Google Scholar] [CrossRef]
- Arif, Y.; Singh, P.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salinity induced physiological and biochemical changes in plants: An omic approach towards salt stress tolerance. Plant Physiol. Biochem. 2020, 156, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Nassar, R.M.; Kamel, H.A.; Ghoniem, A.E.; Alarcón, J.J.; Sekara, A.; Ulrichs, C.; Abdelhamid, M.T. Physiological and anatomical mechanisms in wheat to cope with salt stress induced by seawater. Plants 2020, 9, 237. [Google Scholar] [CrossRef]
- Atabayeva, S.; Nurmahanova, A.; Minocha, S.; Ahmetova, A.; Kenzhebayeva, S.; Aidosova, S.; Nurzhanova, A.; Zhardamalieva, A.; Asrandina, S.; Alybayeva, R. The effect of salinity on growth and anatomical attributes of barley seedling (Hordeum vulgare L.). Afr. J. Biotechnol. 2013, 12, 2366. [Google Scholar] [CrossRef]
- Hu, Y.; Fromm, J.; Schmidhalter, U. Effect of salinity on tissue architecture in expanding wheat leaves. Planta 2005, 220, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Hameed, M.; Ashraf, M.; Naz, N. Anatomical adaptations to salinity in cogon grass [Imperata cylindrica (L.) Raeuschel] from the Salt Range, Pakistan. Plant Soil 2009, 322, 229–238. [Google Scholar] [CrossRef]
- Gupta, A.; Shaw, B.P.; Sahu, B.B.; Munns, R. Post-translational regulation of the membrane transporters contributing to salt tolerance in plants. Funct. Plant Biol. 2021, 48, 1199–1212. [Google Scholar] [CrossRef]
- Almeida, D.M.; Oliveira, M.M.; Saibo, N.J. Regulation of Na+ and K+ homeostasis in plants: Towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 2017, 40, 326–345. [Google Scholar] [CrossRef]
- Zhang, M.; Liang, X.; Wang, L.; Cao, Y.; Song, W.; Shi, J.; Lai, J.; Jiang, C. A HAK family Na+ transporter confers natural variation of salt tolerance in maize. Nat. Plants 2019, 5, 1297–1308. [Google Scholar] [CrossRef]
- Shazia, I.; Sajid, H.; Muhammad Abdul, Q.; Muhammad, A.; Saifullah. The Response of Maize Physiology under Salinity Stress and Its Coping Strategies. In Plant Stress Physiology; Akbar, H., Ed.; IntechOpen: Rijeka, Croatia, 2020; p. 3. [Google Scholar]
- Munns, R.; James, R.A. Screening methods for salinity tolerance: A case study with tetraploid wheat. Plant Soil 2003, 253, 201–218. [Google Scholar] [CrossRef]
- Yousfi, S.; Serret, M.D.; Márquez, A.J.; Voltas, J.; Araus, J.L. Combined use of δ13C, δ18O and δ15N tracks nitrogen metabolism and genotypic adaptation of durum wheat to salinity and water deficit. New Phytol. 2012, 194, 230–244. [Google Scholar] [CrossRef] [PubMed]
- Chaurasia, S.; Singh, A.K.; Kumar, A.; Songachan, L.; Yadav, M.C.; Kumar, S.; Kumari, J.; Bansal, R.; Sharma, P.C.; Singh, K. Genome-wide association mapping reveals key genomic regions for physiological and yield-related traits under salinity stress in wheat (Triticum aestivum L.). Genomics 2021, 113, 3198–3215. [Google Scholar] [CrossRef]
- Genc, Y.; Oldach, K.; Verbyla, A.P.; Lott, G.; Hassan, M.; Tester, M.; Wallwork, H.; McDonald, G.K. Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress. Theor. Appl. Genet. 2010, 121, 877–894. [Google Scholar] [CrossRef]
- Moustafa, E.S.; Ali, M.M.; Kamara, M.M.; Awad, M.F.; Hassanin, A.A.; Mansour, E. Field screening of wheat advanced lines for salinity tolerance. Agronomy 2021, 11, 281. [Google Scholar] [CrossRef]
- Hasan, A.; Hafiz, H.R.; Siddiqui, N.; Khatun, M.; Islam, R.; Mamun, A.-A. Evaluation of wheat genotypes for salt tolerance based on some physiological traits. J. Crop Sci. Biotechnol. 2015, 18, 333–340. [Google Scholar] [CrossRef]
- Nadeem, M.; Tariq, M.N.; Amjad, M.; Sajjad, M.; Akram, M.; Imran, M.; Shariati, M.A.; Gondal, T.A.; Kenijz, N.; Kulikov, D. Salinity-induced changes in the nutritional quality of bread wheat (Triticum aestivum L.) genotypes. AGRIVITA J. Agric. Sci. 2020, 42, 1–12. [Google Scholar] [CrossRef]
- Khokhar, I.M.; Hussain, M.; Anwar, J.; Zulkiffal, M.; Iqbal, M.M.; Khan, B.S.; Khan, A.M.; Qayyum, A.; Sabir, W.; Mehmood, S. Correlation and path analysis for yield and yield contributing characters in wheat (Triticum aestivum L.). Acta Agric. Serbica 2010, 15, 19–24. [Google Scholar] [CrossRef]
- von Wettstein, D. Chlorophyll-letale und der submikroskopische Formwechsel der Plastiden. Exp. Cell Res. 1957, 12, 427–506. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.A.; Teare, I. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef]
- Aebi, H. Catalase in vitro. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1984; Volume 105, pp. 121–126. [Google Scholar]
- Singh, P.; Kumar, V.; Sharma, J.; Saini, S.; Sharma, P.; Kumar, S.; Sinhmar, Y.; Kumar, D.; Sharma, A. Silicon supplementation alleviates the salinity stress in wheat plants by enhancing the plant water status, photosynthetic pigments, proline content and antioxidant enzyme activities. Plants 2022, 11, 2525. [Google Scholar] [CrossRef]
- Fuerst, E.P.; Anderson, J.V.; Morris, C.F. Polyphenol oxidase in wheat grain: Whole kernel and bran assays for total and soluble activity. Cereal Chem. 2006, 83, 10–16. [Google Scholar] [CrossRef]
- Madhu; Kaur, A.; Tyagi, S.; Shumayla; Singh, K.; Upadhyay, S.K. Exploration of glutathione reductase for abiotic stress response in bread wheat (Triticum aestivum L.). Plant Cell Rep. 2022, 41, 639–654. [Google Scholar] [CrossRef] [PubMed]
- Nassar, R.; Boghdady, M.S.; Ahmed, Y.M. Botanical studies on Phaseolus vulgaris L. II-Anatomy of vegetative and reproductive organs. J. Am. Sci. 2010, 6, 217–229. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-M.; Zhang, H.-M.; Liu, Z.-H.; Li, H.-C.; Guo, X.-L.; Li, G.-L. The wheat NHX antiporter gene TaNHX2 confers salt tolerance in transgenic alfalfa by increasing the retention capacity of intracellular potassium. Plant Mol. Biol. 2015, 87, 317–327. [Google Scholar] [CrossRef]
- Bharti, N.; Pandey, S.S.; Barnawal, D.; Patel, V.K.; Kalra, A. Plant growth promoting rhizobacteria Dietzia natronolimnaea modulates the expression of stress responsive genes providing protection of wheat from salinity stress. Sci. Rep. 2016, 6, 34768. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhao, Y.-J.; Zhao, B.-C.; Ge, R.-C.; Li, M.; Shen, Y.-Z.; Huang, Z.-J. Cloning and functional analysis of wheat VH+-ATPase subunit genes. Plant Mol. Biol. 2009, 69, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Dudziak, K.; Sozoniuk, M.; Szczerba, H.; Kuzdraliński, A.; Kowalczyk, K.; Börner, A.; Nowak, M. Identification of stable reference genes for qPCR studies in common wheat (Triticum aestivum L.) seedlings under short-term drought stress. Plant Methods 2020, 16, 58. [Google Scholar] [CrossRef]


| Studied Factor | Chlorophyll a (mg/g D.W) | Chlorophyll b (mg/g D.W) | Carotenoids (mg/g D.W) | Proline (mg/100 gmFW) | |
|---|---|---|---|---|---|
| Wheat genotypes (G) | |||||
| Ismailia line | 2.32 a | 4.09 a | 3.73 a | 12.84 a | |
| Misr 1 | 2.19 b | 3.73 c | 3.41 c | 12.05 a | |
| Misr 3 | 2.21 b | 3.97 b | 3.64 b | 11.68 b | |
| Salinity levels (S) | |||||
| Control | 2.28 a | 4.05 a | 3.72 a | 5.14 c | |
| 5000 ppm | 2.25 a | 3.88 b | 3.55 b | 15.04 b | |
| 7000 ppm | 2.19 a | 3.86 b | 3.52 b | 16.39 a | |
| Interaction (G × S) | |||||
| Ismailia line | Control | 2.41 a | 4.21 a | 3.81 a | 5.11 h |
| 5000 | 2.30 ab | 4.13 ab | 3.66 a | 15.61g | |
| 7000 | 2.24 ab | 3.93 bc | 3.73 a | 17.80 a | |
| Misr 1 | Control | 2.13 b | 3.95 bc | 3.65 a | 5.80 f |
| 5000 | 2.24 ab | 3.71 d | 3.21 c | 14.80 e | |
| 7000 | 2.20 ab | 3.55 e | 3.37 b | 15.55 c | |
| Misr 3 | Control | 2.31 ab | 4.00 bc | 3.70 a | 4.52 i |
| 5000 | 2.20 ab | 3.80 cd | 3.76 c | 14.72 d | |
| 7000 | 2.13 b | 4.10 ab | 3.46 b | 15.80 b | |
| ANOVA | df | p-Value | |||
| G | 2 | <0.001 | <0.001 | <0.001 | <0.001 |
| S | 2 | <0.001 | <0.001 | <0.001 | <0.001 |
| G × S | 4 | <0.001 | <0.001 | <0.001 | <0.001 |
| Studied Factor | Epidermis Thickness of Root (µm) | Cortex Thickness of Root (µm) | Vascular Bundle Thickness of Root (µm) | No. of Xylem Vessels | Pith Thickness (µm) | |
|---|---|---|---|---|---|---|
| Wheat genotypes (G) | ||||||
| Ismailia line | 18.7 a | 222.1 a | 225.2 a | 15.3 a | 180.9 a | |
| Misr 1 | 18.8 a | 92.8 b | 159.4 b | 14.0 b | 139.7 b | |
| Misr 3 | 18.1 b | 217.0 a | 167.0 b | 11.7 c | 85.7 c | |
| Salinity levels (S) | ||||||
| Control | 21.4 a | 238.10 a | 244.23 a | 14.67 a | 211.43 a | |
| 5000 ppm | 20.4 a | 201.3 b | 162.23 b | 13.67 a | 100.00 b | |
| 7000 ppm | 13.3 b | 92.41 b | 145.13 c | 11.67 b | 94.77 c | |
| Interaction (G × S) | ||||||
| Ismailia line | Control | 21.4 b | 300.0 a | 340.0 a | 18.0 a | 300.0 a |
| 5000 | 21.4 b | 252.0 b | 178.6 c | 15.0 b | 128.6 c | |
| 7000 | 13.3 d | 114.0 c | 157.0 e | 13.0 d | 114.0 d | |
| G2 | Control | 22.1 a | 114.3 c | 214.1 b | 15.0 b | 220.0 b |
| 5000 | 20.0 b | 100.0 d | 142.8 f | 15.0 b | 100.0 e | |
| 7000 | 14.3 c | 64.20 f | 121.4 g | 12.0 e | 99.0 e | |
| G3 | Control | 20.0 b | 300.0 a | 178.6 c | 14.0 c | 114.3 d |
| 5000 | 20.0 b | 252.0 b | 165.3 d | 11.0 f | 71.4 f | |
| 7000 | 14.3 c | 99.0 e | 157.0 e | 10.0 g | 71.3 f | |
| ANOVA | df | p-Value | ||||
| G | 2 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| S | 2 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| G × S | 4 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Studied Factor | Mesophyll Thickness (µm) | Main Vascular Bundle Thickness (µm) | Sclerenchyma Tissue Upper Vascular Bundle Thickness (µm) | Sclerenchyma Tissue Lower Vascular Bundle Thickness (µm) | |
|---|---|---|---|---|---|
| Wheat genotypes (G) | |||||
| Ismailia line | 100.0 a | 64.0 a | 47.6 a | 42.9 a | |
| Misr 1 | 73.7 c | 57.4 c | 33.3 b | 31.0 c | |
| Misr 3 | 85.4 b | 61.8 b | 33.1 b | 35.2 b | |
| Salinity levels (S) | |||||
| Control | 102.4 a | 69.1 a | 28.3 c | 30.5 c | |
| 5000 ppm | 80.9 b | 59.4 b | 38.1 b | 35.7 b | |
| 7000 ppm | 75.8 c | 54.7c | 47.6 a | 42.8 a | |
| Interaction (G × S) | |||||
| Ismailia line | Control | 128.6 a | 77.6 a | 35.7 c | 35.7 c |
| 5000 | 85.7 c | 64.3 b | 35.7 c | 28.6 d | |
| 7000 | 85.7 c | 50.0 d | 71.4 a | 64.3 a | |
| Misr 1 | Control | 85.7 c | 65.3 b | 21.4 e | 28.6 d |
| 5000 | 71.4 d | 50.0 d | 42.9 b | 35.7 c | |
| 7000 | 64.1 e | 57.0 c | 35.7 b | 28.6 d | |
| Misr 3 | Control | 92.9 b | 64.3 b | 27.93 d | 27.2 d |
| 5000 | 85.7 c | 64.0 b | 35.7 c | 42.9 b | |
| 7000 | 77.6 d | 57.0 c | 35.67 c | 35.37 c | |
| ANOVA | df | p-Value | |||
| G | 2 | <0.001 | <0.001 | <0.001 | <0.001 |
| S | 2 | <0.001 | <0.001 | <0.001 | <0.001 |
| G × S | 4 | <0.001 | <0.001 | <0.001 | <0.001 |
| Studied Factor | Days to 50% Heading | Plant Height (cm) | No. of Effective Tillers | No. of Spikelets/Spike | |||||
|---|---|---|---|---|---|---|---|---|---|
| 2020 | 2021 | 2020 | 2021 | 2020 | 2021 | 2020 | 2021 | ||
| Wheat genotypes (G) | |||||||||
| Ismailia line | 78.6 a | 80.3 a | 91.6 a | 89.9 a | 2.4 a | 2.8 a | 16.11 a | 16.2 a | |
| Misr 1 | 73.2 b | 74.6 b | 66.1 b | 64.7 b | 2.2 a | 2.6 a | 15.3 a | 15.6 a | |
| Misr 3 | 71.4 c | 72.7 c | 64.1 b | 63.0 b | 1.8 b | 2.1 b | 13.9 b | 13.9 b | |
| Salinity level (S) | |||||||||
| Control | 80.0 a | 81.3 a | 98.1 a | 96.3 a | 3.2 a | 3.6 a | 18.7 a | 18.9 a | |
| 5000 | 71.8 b | 73.3 b | 64.33 b | 63.0 b | 1.6 b | 2.0 b | 13.7 b | 14.1 b | |
| 7000 | 71.4 b | 72.9 b | 59.4 b | 58.3 b | 1.6 b | 1.9 b | 12.7 b | 12.7 c | |
| RD% | 10.75 | 10.33 | 39.45 | 39.46 | 50.0 | 47.22 | 32.09 | 32.8 | |
| Interaction (G × S) | |||||||||
| Ismailia line | control | 89.3 a | 90.7 a | 117.3 a | 115.0 a | 3.3 a | 3.7 a | 20.7 a | 20.7 a |
| 5000 | 76.3 b | 77.7 b | 90.0 b | 88.8 b | 3.3 a | 3.7 a | 17.3 b | 18.0 b | |
| 7000 | 74.3 c | 75.7 c | 87.0 bc | 85.3 bc | 3.0 ab | 3.3 a | 18.3 b | 18.0 b | |
| Misr 1 | control | 73.3 c | 75.0 c | 81.7 bc | 80.0 bc | 1.7 bc | 2.0 b | 14.3 c | 15.0 c |
| 5000 | 71.7 d | 73.0 d | 59.0 d | 57.8 d | 2.0 a–c | 2.3 b | 14.7 c | 15.0 c | |
| 7000 | 70.7 d | 72.0 d | 52.3 de | 51.3 de | 1.3 c | 1.3 b | 12.3 cd | 12.3 cd | |
| Misr 3 | control | 73.3 c | 75.3 c | 76.0 c | 74.5 c | 1.7 bc | 2.0 b | 13.3 c | 13.0 cd |
| 5000 | 71.7 d | 73.0 d | 43.3 e | 42.5 e | 2.0 a–c | 2.3 b | 14.0 c | 13.7 cd | |
| 7000 | 69.3 e | 70.3 e | 59.0 d | 57.8 d | 1.3 c | 1.7 b | 11.0 d | 11.3 d | |
| ANOVA | df | p-Value | |||||||
| G | 2 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| S | 2 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| G × S | 4 | <0.001 | <0.001 | 0.034 | <0.001 | 0.0013 | <0.001 | <0.001 | <0.001 |
| Studied Factor | Spike Length (cm) | No. of Grains/Spike | 1000-Grain Weight (g) | Grain Yield/Plant (g) | |||||
|---|---|---|---|---|---|---|---|---|---|
| 2020 | 2021 | 2020 | 2021 | 2020 | 2021 | 2020 | 2021 | ||
| Wheat genotypes (G) | |||||||||
| Ismailia line | 11.2 a | 11.4 a | 31.3 a | 34.0 a | 32.6 c | 32.4 b | 46.13 a | 46.55 a | |
| Misr 1 | 10.3 b | 10.7 b | 29.2 b | 31.2 b | 35.6 b | 35.6 a | 41.05 b | 41.55 b | |
| Misr 3 | 9.5 c | 9.4 c | 25.1 c | 28.4 c | 37.5 a | 36.5 a | 34.91 c | 35.11 c | |
| Salinity levels (S) | |||||||||
| Control | 12.2 a | 12.2 a | 41.3 a | 43.8 a | 43.2 a | 42.1 a | 61.44 a | 62.11 a | |
| 5000 | 10.16 b | 10.5 b | 25.9 b | 28.3 b | 33.9 b | 33.9 b | 39.38 b | 39.67 b | |
| 7000 | 8.6 c | 8.8 c | 18.4 c | 21.5 c | 28.6 c | 28.6 c | 21.26 c | 21.44 c | |
| RD% | 29.51 | 27.87 | 55.45 | 50.91 | 33.8 | 32.07 | 65.40 | 65.48 | |
| Interaction (G × S) | |||||||||
| Ismailia line | control | 13.8 a | 14.2 a | 46.3 a | 48.7 a | 39.3 c | 39.0 b | 70.7 a | 71.4 a |
| 5000 | 12.0 b | 12.0 b | 38.0 b | 40.0 b | 32.3 d | 32.3 c | 42.8 d | 43.3 d | |
| 7000 | 10.8b c | 10.5 c | 39.7 b | 42.7 b | 26.0 e | 26.0 d | 24.9 f | 25.2 f | |
| Misr 1 | control | 10.8b c | 10.9 c | 27.3 c | 29.3 c | 42.7 b | 39.8 b | 67.0 b | 67.7 b |
| 5000 | 10.2 cd | 10.7 c | 27.3 c | 29.3 c | 37.0 c | 37.0 bc | 37.2 e | 37.5 e | |
| 7000 | 9.5 de | 9.8 cd | 23.0 d | 26.3 cd | 33.0 d | 33.0 c | 19.0 g | 19.2 g | |
| Misr 3 | control | 9.2 d–f | 9.2 d | 20.3 d | 24.0 d | 47.7 a | 47.6 a | 46.7 c | 47.0 c |
| 5000 | 8.7 ef | 9.3 d | 22.3 d | 24.3 d | 32.3 d | 32.3 c | 38.2 e | 38.7 e | |
| 7000 | 8.2 f | 8.0 e | 12.7 e | 16.3 e | 26.7 e | 26.6 d | 19.9 g | 20.1 g | |
| ANOVA | df | p-Value | |||||||
| G | 2 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| S | 2 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| G × S | 4 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Studied Factor | Na+ (%) | K+ (%) | K+/Na+ | |
|---|---|---|---|---|
| Wheat genotypes (G) | ||||
| Ismailia line (G1) | 8.37 c | 15.83 a | 2.41 a | |
| Misr 1(G2) | 9.56 b | 15.29 b | 1.92 b | |
| Misr 3 (G3) | 10.49 a | 13.32 c | 1.66 c | |
| Salinity levels (S) | ||||
| Control | 5.39 c | 19.11 a | 3.61 a | |
| 5000 ppm | 9.44 b | 15.29 b | 1.64 b | |
| 7000 ppm | 13.95 a | 10.03 c | 0.75 c | |
| RD% | 158.81 | 47.51 | ||
| Interaction (G × S) | ||||
| Ismailia line | Control | 4.54 h | 19.66 a | 4.33 a |
| 5000 | 5.78 g | 19.36 a | 3.32 b | |
| 7000 | 5.85 g | 18.30 b | 3.17 b | |
| Misr 1 | Control | 8.31 f | 16.37 c | 1.97 c |
| 5000 | 9.63 e | 16.20 c | 1.68 d | |
| 7000 | 10.39 d | 13.30 d | 1.28 e | |
| Misr 3 | Control | 12.27 c | 11.44 e | 0.93 f |
| 5000 | 13.20 b | 10.30 f | 0.78 f | |
| 7000 | 15.32 a | 8.35 g | 0.55 g | |
| ANOVA | df | p-Value | ||
| G | 2 | <0.001 | <0.001 | <0.001 |
| S | 2 | <0.001 | <0.001 | <0.001 |
| G × S | 4 | <0.001 | <0.001 | <0.001 |
| Primer Name | Sequences (5′-3′) | Accession | Reference |
|---|---|---|---|
| TaNHX2-F | CTCCAGAACTTCGATCCTAACC | AY040246 | [81] |
| TaNHX2-R | GCACTAAGCAATCCAGTAAACAC | ||
| TaHKT1-F | ATGGGCCGGGTGAAAAGATT | U16709 | [82] |
| TaHKT1-R | TCCAGAAGGGGTGAACATGC | ||
| Ta H+-ATPase (Sub E)-F | GAGGATGCAATGAAGGAACTCC | DQ272489 | [83] |
| Ta H+-ATPase (Sub E)-R | AAGCAGGCCCTGAACAACG | ||
| TaHAK1-F | ACGCTTACGGGATCTGTGTG | JF495466 | [82] |
| TaHAK1-R | GAGCCGAACACGACGTAGAA | ||
| TaActin-F | GGAGAAGCTCGCTTACGTG | AB181991 | [84] |
| TaActin-R | GGGCACCTGAACCTTTCTGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussein, M.A.A.; Alqahtani, M.M.; Alwutayd, K.M.; Aloufi, A.S.; Osama, O.; Azab, E.S.; Abdelsattar, M.; Hassanin, A.A.; Okasha, S.A. Exploring Salinity Tolerance Mechanisms in Diverse Wheat Genotypes Using Physiological, Anatomical, Agronomic and Gene Expression Analyses. Plants 2023, 12, 3330. https://doi.org/10.3390/plants12183330
Hussein MAA, Alqahtani MM, Alwutayd KM, Aloufi AS, Osama O, Azab ES, Abdelsattar M, Hassanin AA, Okasha SA. Exploring Salinity Tolerance Mechanisms in Diverse Wheat Genotypes Using Physiological, Anatomical, Agronomic and Gene Expression Analyses. Plants. 2023; 12(18):3330. https://doi.org/10.3390/plants12183330
Chicago/Turabian StyleHussein, Mohammed A. A., Mesfer M. Alqahtani, Khairiah M. Alwutayd, Abeer S. Aloufi, Omnia Osama, Enas S. Azab, Mohamed Abdelsattar, Abdallah A. Hassanin, and Salah A. Okasha. 2023. "Exploring Salinity Tolerance Mechanisms in Diverse Wheat Genotypes Using Physiological, Anatomical, Agronomic and Gene Expression Analyses" Plants 12, no. 18: 3330. https://doi.org/10.3390/plants12183330
APA StyleHussein, M. A. A., Alqahtani, M. M., Alwutayd, K. M., Aloufi, A. S., Osama, O., Azab, E. S., Abdelsattar, M., Hassanin, A. A., & Okasha, S. A. (2023). Exploring Salinity Tolerance Mechanisms in Diverse Wheat Genotypes Using Physiological, Anatomical, Agronomic and Gene Expression Analyses. Plants, 12(18), 3330. https://doi.org/10.3390/plants12183330










