Antagonistic Activity of Streptomyces alfalfae 11F against Fusarium Wilt of Watermelon and Transcriptome Analysis Provides Insights into the Synthesis of Phenazine-1-Carboxamide
Abstract
:1. Introduction
2. Results
2.1. Antimicrobial Activity Assay
2.2. Strain 11F Enhanced Watermelon Plant Growth and Disease Resistance
2.3. Transcriptome Sequencing, Data Quality, and Transcript Assembly
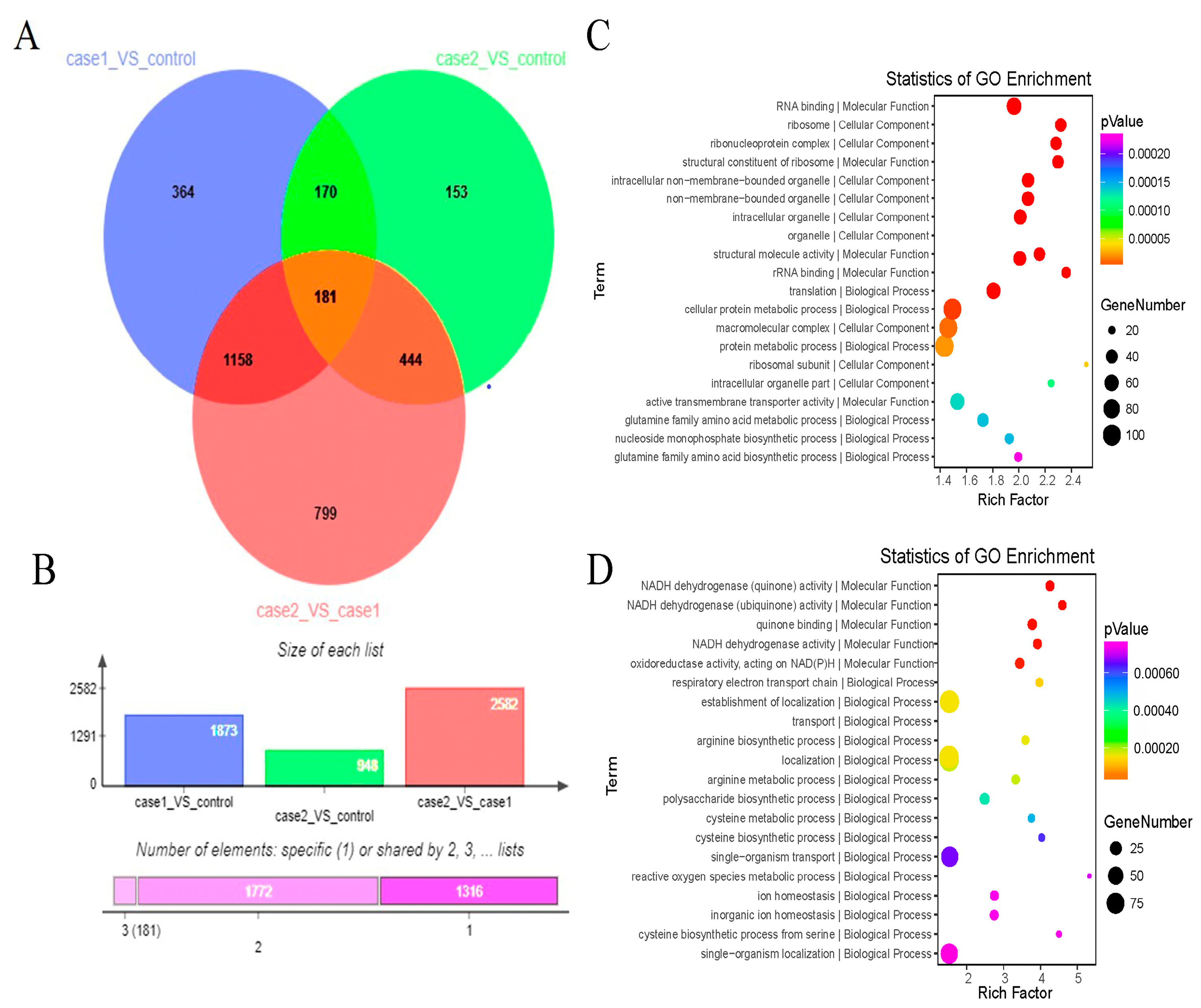
2.4. Differential Gene Expression Analysis
2.5. Verification of DEG Expression Levels by Quantitative Real-Time PCR Analysis
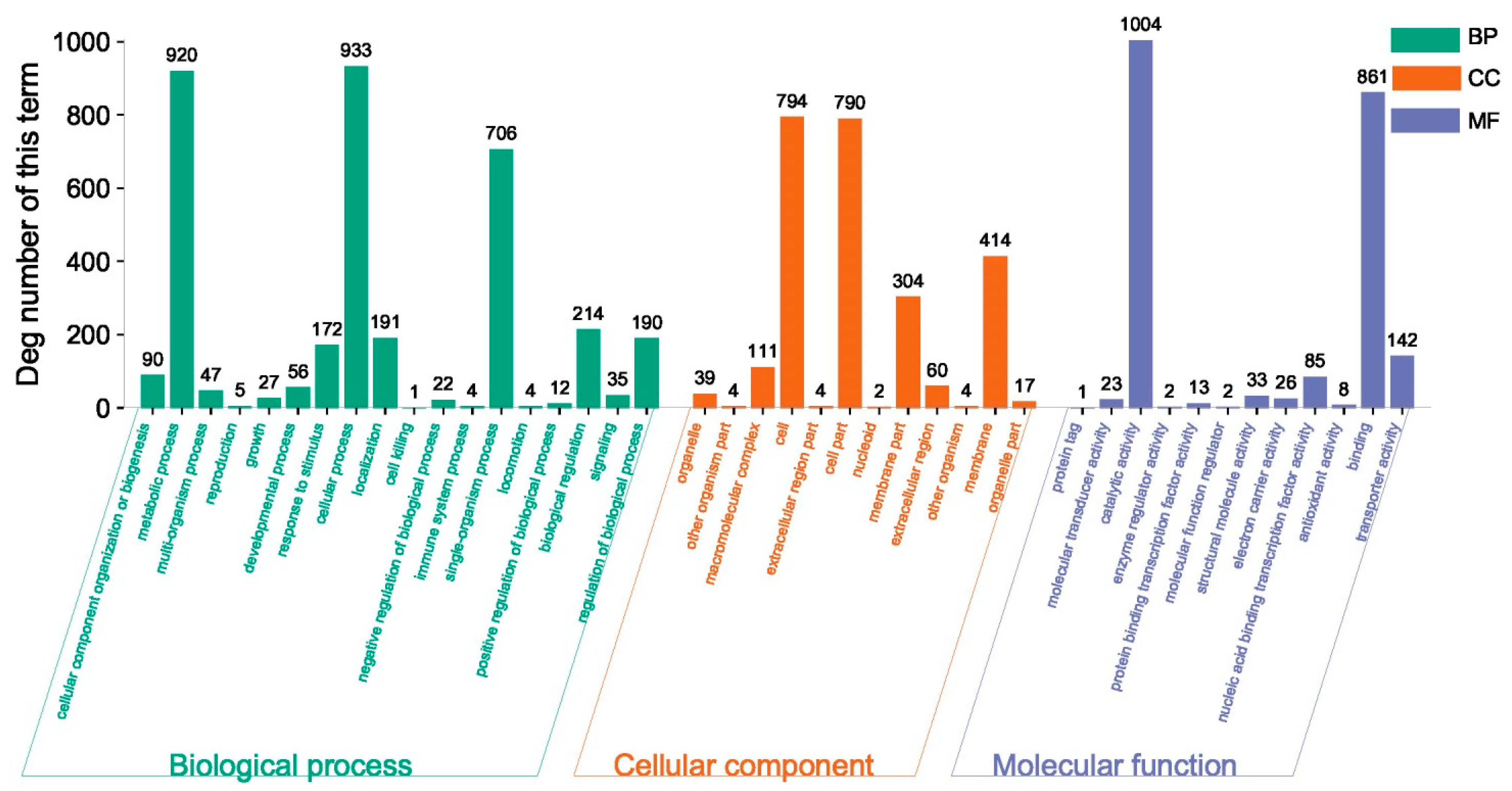
2.6. Functional Annotation and Enrichment Analysis of the DEGs
2.7. Expression of phzF and Characterization of a Phenazine Compound in Strain 11F
3. Discussion
4. Materials and Methods
4.1. Microbe Strains and Culture Conditions
4.2. Assay of the S. alfalfae 11F Antimicrobial Spectrum
4.3. Pot Experiment
4.4. Sample Collection and Library Construction
4.5. Illumina HiSeq Sequencing and Analysis
4.6. Quantitative Real-Time PCR Analysis of DEGs
4.7. Isolation, Purification, and Determination of the Antifungal Metabolite
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Strub, C.; Dieye, C.A.T.; Nguyen, P.A.; Constancias, F.; Durand, N.; Guendouz, S.; Pratlong, M.; Fontana, A.; Schorr-Galindo, S. Transcriptomes of the interaction between Fusarium verticillioides and a Streptomyces strain reveal the fungal defense strategy under the pressure of a potential biocontrol agent. Fungal Biol. 2021, 125, 78–88. [Google Scholar] [CrossRef]
- Nguyen, P.A.; Strub, C.; Lagrée, M.; Bertrand-Michel, J.; Schorr-Galindo, S.; Fontana, A. Study of in vitro interaction between Fusarium verticillioides and Streptomyces sp. using metabolomics. Folia Microbiol. 2020, 65, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Todd, E.V.; Black, M.A.; Gemmell, N.J. The power and promise of RNA-seq in ecology and evolution. Mol. Ecol. 2016, 25, 1224–1241. [Google Scholar] [CrossRef]
- Niu, Z.F.; Su, D.L.H.; Ma, S.; Hu, L.; Hou, X.C.; Zhang, T.T.; Dong, D.; Zhang, D.P.; Lu, C.G.; Fan, X.F.; et al. The characterization of Streptomyces alfalfae strain 11F and its effect on seed germination and growth promotion in switchgrass. Biomass Bioenerg. 2022, 158, 106360. [Google Scholar] [CrossRef]
- Chen, J.L.; Sun, S.Z.; Miao, C.P.; Wu, K.; Chen, Y.W.; Xu, L.H.; Guan, H.L.; Zhao, L.X. Endophytic Trichoderma gamsii YIM PH30019: A promising biocontrol agent with hyperosmolar, mycoparasitism, and antagonistic activities of induced volatile organic compounds on root-rot pathogenic fungi of Panax notoginseng. J. Ginseng Res. 2016, 40, 315–324. [Google Scholar] [CrossRef]
- Laskin, A.I.; Lechevalier, H.A. CRC Handbook of Microbiology: Microbial Phenazines; CRC Press: Boca Raton, FL, USA, 1984; pp. 573–576. [Google Scholar]
- Toohey, J.I.; Nelson, C.D.; Krotkov, G. Toxicity of phenazine carboxylic acid to some bacteria, algae, higher plants, and animals. Can. J. Bot. 1965, 43, 1151–1155. [Google Scholar] [CrossRef]
- Ingram, J.M.; Blackwood, A.C. Microbial production of phenazines. Adv. Microb. Physiol. 1970, 13, 267. [Google Scholar]
- Sunish-Kumar, R.; Ayyadurai, N.; Pandiaraja, P.; Reddy, A.V.; Venkateswarlu, Y.; Prakash, O.; Sakthivel, N. Characterization of antifungal metabolite produced by a new strain Pseudomonas aeruginosa PUPa3 that exhibits broad-spectrum antifungal activity and biofertilizing traits. J. Appl. Microbiol. 2005, 98, 145–154. [Google Scholar] [CrossRef]
- Chen, L.; Zhao, M.; Ye, Y.; Guo, D.; Ding, L.S.; Zhou, Y. Chemical constituents of Aspergillus fumigatus from Acorus calamus L. Chin. J. Appl. Environ. Biol. 2017, 23, 918–923. [Google Scholar]
- Bérdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Manivasagan, P.; Venkatesan, J.; Sivakumar, K.; Kim, S.K. Pharmaceutically active secondary metabolites of marine actinobacteria. Microbiol. Res. 2014, 169, 262–278. [Google Scholar] [CrossRef]
- Takemoto, D.; Tanaka, A.; Scott, B. A p67 Phox-like regulator is recruited to control hyphal branching in a fungal-grass mutualistic symbiosis. Plant Cell 2006, 18, 2807–2821. [Google Scholar] [CrossRef]
- Zhao, Y.; Selvaraj, J.N.; Xing, F.; Zhou, L.; Wang, Y.; Song, H.; Tan, X.; Sun, L.; Sangare, L.; Folly, Y.M.E.; et al. Antagonistic action of Bacillus subtilis strain SG6 on Fusarium graminearum. PLoS ONE 2014, 9, e92486. [Google Scholar] [CrossRef]
- Banerjee, A.; Singh, A.; Roychoudhury, A. De novo RNA-Seq analysis in sensitive rice cultivar and comparative transcript profiling in contrasting genotypes reveal genetic biomarkers for fluoride-stress response. Environ. Pollut. 2020, 267, 115378. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Sun, J.; Wu, B.; Gao, Y.; Nie, H.; Nie, Z.; Quan, S.; Wang, Y.; Cao, X.; Li, S. CPSF30-L-mediated recognition of mRNA m6A modification controls alternative polyadenylation of nitrate signaling-related gene transcripts in Arabidopsis. Mol. Plant 2021, 14, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Gouveia, E.R.; Baptista-Neto, A.; Badino, A.C.; Hokka, C.O. Optimisation of medium composition for clavulanic acid production by Streptomyces clavuligerus. Biotechnol. Lett. 2001, 23, 157–161. [Google Scholar] [CrossRef]
- Wang, Y.H.; Yang, B.; Ren, J.; Dong, M.L.; Liang, D.; Xu, A.L. Optimization of medium composition for the production of clavulanic acid by Streptomyces clavuligerus. Process Biochem. 2005, 40, 1161–1166. [Google Scholar] [CrossRef]
- Higo, A.; Hara, H.; Horinouchi, S.; Ohnishi, Y. Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res. 2012, 19, 259–273. [Google Scholar] [CrossRef]
- Plabutong, N.; Ekronarongchai, S.; Niwetbowornchai, N.; Edwards, S.W.; Virakul, S.; Chiewchengchol, D.; Thammahong, A. The inhibitory effect of validamycin A on Aspergillus flavus. Int. J. Microbiol. 2020, 2020, 3972415. [Google Scholar] [CrossRef]
- Méndez, C.; Salas, J.A. ABC transporters in antibiotic-producing actinomycetes. FEMS Microbiol. Lett. 1998, 158, 1–8. [Google Scholar] [CrossRef]
- Liu, L.; Chen, X. RNA quality control as a key to suppressing RNA silencing of endogenous genes in plants. Mol. Plant 2016, 9, 826–836. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Álvarez, R.; Rodríguez-García, A.; Santamarta, I.; Pérez-Redondo, R.; Prieto-Domínguez, A.; Martínez-Burgo, Y.; Liras, P. Transcriptomic analysis of Streptomyces clavuligerus ΔccaR:tsr: Effects of the cephamycin C-clavulanic acid cluster regulator CcaR on global regulation. Microb. Biotechnol. 2014, 7, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, R.C.; Glaring, M.A.; Olsson, S.; Stougaard, P. Transcriptomic profiling of microbe-microbe interactions reveals the specific response of the biocontrol strain P. fluorescens In5 to the phytopathogen Rhizoctonia solani. BMC Res. Notes 2017, 10, 376. [Google Scholar] [CrossRef]
- Li, S.; Wang, W.; Li, X.; Fan, K.; Yang, K. Genome-wide identification and characterization of reference genes with different transcript abundances for Streptomyces coelicolor. Sci Rep. 2015, 5, 15840. [Google Scholar] [CrossRef] [PubMed]
- Arseneault, T.; Roquigny, R.; Novinscak, A.; Goyer, C.; Filion, M. Phenazine-1-carboxylic acid-producing Pseudomonas synxantha LBUM223 alters the transcriptome of Streptomyces scabies, the causal agent of potato common scab. Physiol. Mol. Plant Pathol. 2020, 110, 101480. [Google Scholar] [CrossRef]
- Chin, T.F.C.; Bloemberg, G.V.; Bij, A.J.V.D.; Drift, K.M.G.M.; Schripsema, J.; Kroon, B.; Scheffer, R.J.; Keel, C.; Bakker, P.A.H.M.; Tichy, H.; et al. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicislycopersici. Mol. Plant-Microbe Interac. 1998, 11, 1069–1077. [Google Scholar] [CrossRef]
- Xu, M.; Wang, Y.; Zhao, Z.; Gao, G.; Huang, S.X.; Kang, Q.; He, X.; Lin, S.; Pang, X.; Deng, Z.; et al. Functional genome mining for metabolites encoded by large gene clusters through heterologous expression of a whole-genome bacterial artifificial chromosome library in Streptomyces spp. Appl. Environ. Microb. 2016, 82, 5795–5805. [Google Scholar] [CrossRef]
- Buzón-Durán, L.; Martín-Gil, J.; Pérez-Lebeña, E.; Ruano-Rosa, D.; Revuelta, J.L.; Casanova-Gascón, J.; Ramos-Sánchez, M.C.; Martín-Ramos, P. Antifungal agents based on chitosan oligomers, ε-polylysine and Streptomyces spp. secondary metabolites against three botryosphaeriaceae species. Antibiotics 2019, 8, 99. [Google Scholar] [CrossRef]
- Riahi, K.; Hosni, K.; Raies, A.; Oliveira, R. Unique secondary metabolites of a Streptomyces strain isolated from extreme salty wetland show antioxidant and antibacterial activities. J. Appl. Microbiol. 2019, 127, 1727–1740. [Google Scholar] [CrossRef]
- Bezert, G.; Chappe, P.; Mourey, A.; Loubinoux, B. Action de bacillus et d’actinomycètes sur les champignons du bleuissement du bois. Bull. Acad. Soc. Lorraines Sci. 1996, 35, 3. [Google Scholar]
- Dhaouadi, S.; Rouissi, W.; Mougou-Hamdane, A.; Nasraoui, B. Evaluation of biocontrol potential of Achromobacter xylosoxidans against Fusarium wilt of melon. Eur. J. Plant Pathol. 2019, 154, 179–188. [Google Scholar] [CrossRef]
- Shaunese, L.; Brenda, L. A major QTL associated with Fusarium oxysporum race 1 resistance identified in genetic populations derived from closely related watermelon lines using selective genotyping and genotyping-by-sequencing for SNP discovery. Theor. Appl. Genet. 2014, 127, 2105–2115. [Google Scholar]
- Geoffrey, M.; Cecilia, M. Genotyping by sequencing for SNP discovery and genetic mapping of resistance to race 1 of Fusarium oxysporum in watermelon. Sci. Hortic. 2016, 209, 31–40. [Google Scholar]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R. TopHat 2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]







| Treatment (v/v) | Plant Height/cm | Leaf Length | Leaf Width/cm | Fresh Weight/g | Dry Weight/g |
|---|---|---|---|---|---|
| Control | 8.75 ± 0.18 b | 2.90 ± 0.17 c | 3.35 ± 0.11 c | 0.82 ± 0.01 c | 0.05 ± 0.00 c |
| 11F (1:30) | 9.20 ± 0.08 a | 3.47 ± 0.13 b | 3.45 ± 0.18 b | 0.89 ± 0.19 b | 0.07± 0.00 b |
| 11F (1:60) | 9.51 ± 0.13 a | 4.08 ± 0.24 a | 3.98 ± 0.19 a | 1.01 ± 0.01 a | 0.08 ± 0.01 a |
| 11F (1:100) | 8.32 ± 0.11 c | 3.15 ± 0.03 b c | 3.12 ± 0.10 b c | 0.84 ± 0.02 c | 0.06 ± 0.00 c |
| Treatment | Incidence (%) | Disease Index | Control Effect (%) |
|---|---|---|---|
| Control | 0.00 ± 0.00 c | 0.00 ± 0.00 c | – |
| F. oxysporum f. sp. niveum | 100.00 ± 0.00 a | 73.59 ± 1.07 a | – |
| 11F | 0.00 ± 0.00 c | 0.00 ± 0.00 c | – |
| F. oxysporum f. sp. niveum + 11F | 25.14 ± 0.14 b | 27.57 ± 0.18 b | 62.54 |
| No. | δH (J in Hz) | δC |
|---|---|---|
| 1 | 131.0 | |
| 2 | 8.69, dd (7.0, 1.7) | 134.1 |
| 3 | 8.07, m | 130.3 |
| 4 | 8.44, dd (8.7, 1.7) | 133.0 |
| 4a | 142.8 | |
| 5a | 141.3 | |
| 6 | 8.42, m | 129.3 |
| 7 | 8.06, m | 132.0 |
| 8 | 8.04, m | 131.6 |
| 9 | 8.31, m | 129.2 |
| 9a | 142.6 | |
| 10a | 140.2 | |
| CONH2 | Ha: 9.73, s Hb: 8.09, br s | 165.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, D.; Li, M.; Zhang, T.; Niu, Z.; Xue, G.; Bai, H.; Zhao, W.; Yu, J.; Jiang, W.; Wu, H. Antagonistic Activity of Streptomyces alfalfae 11F against Fusarium Wilt of Watermelon and Transcriptome Analysis Provides Insights into the Synthesis of Phenazine-1-Carboxamide. Plants 2023, 12, 3796. https://doi.org/10.3390/plants12223796
Dong D, Li M, Zhang T, Niu Z, Xue G, Bai H, Zhao W, Yu J, Jiang W, Wu H. Antagonistic Activity of Streptomyces alfalfae 11F against Fusarium Wilt of Watermelon and Transcriptome Analysis Provides Insights into the Synthesis of Phenazine-1-Carboxamide. Plants. 2023; 12(22):3796. https://doi.org/10.3390/plants12223796
Chicago/Turabian StyleDong, Dan, Maoying Li, Taotao Zhang, Zhenfeng Niu, Guoping Xue, Hongmei Bai, Wenyu Zhao, Jiajia Yu, Wei Jiang, and Huiling Wu. 2023. "Antagonistic Activity of Streptomyces alfalfae 11F against Fusarium Wilt of Watermelon and Transcriptome Analysis Provides Insights into the Synthesis of Phenazine-1-Carboxamide" Plants 12, no. 22: 3796. https://doi.org/10.3390/plants12223796
APA StyleDong, D., Li, M., Zhang, T., Niu, Z., Xue, G., Bai, H., Zhao, W., Yu, J., Jiang, W., & Wu, H. (2023). Antagonistic Activity of Streptomyces alfalfae 11F against Fusarium Wilt of Watermelon and Transcriptome Analysis Provides Insights into the Synthesis of Phenazine-1-Carboxamide. Plants, 12(22), 3796. https://doi.org/10.3390/plants12223796








