Retrieval of Leaf Chlorophyll Contents (LCCs) in Litchi Based on Fractional Order Derivatives and VCPA-GA-ML Algorithms
Abstract
:1. Introduction
2. Results
2.1. Correlation Analysis between LCC and FOD Spectra
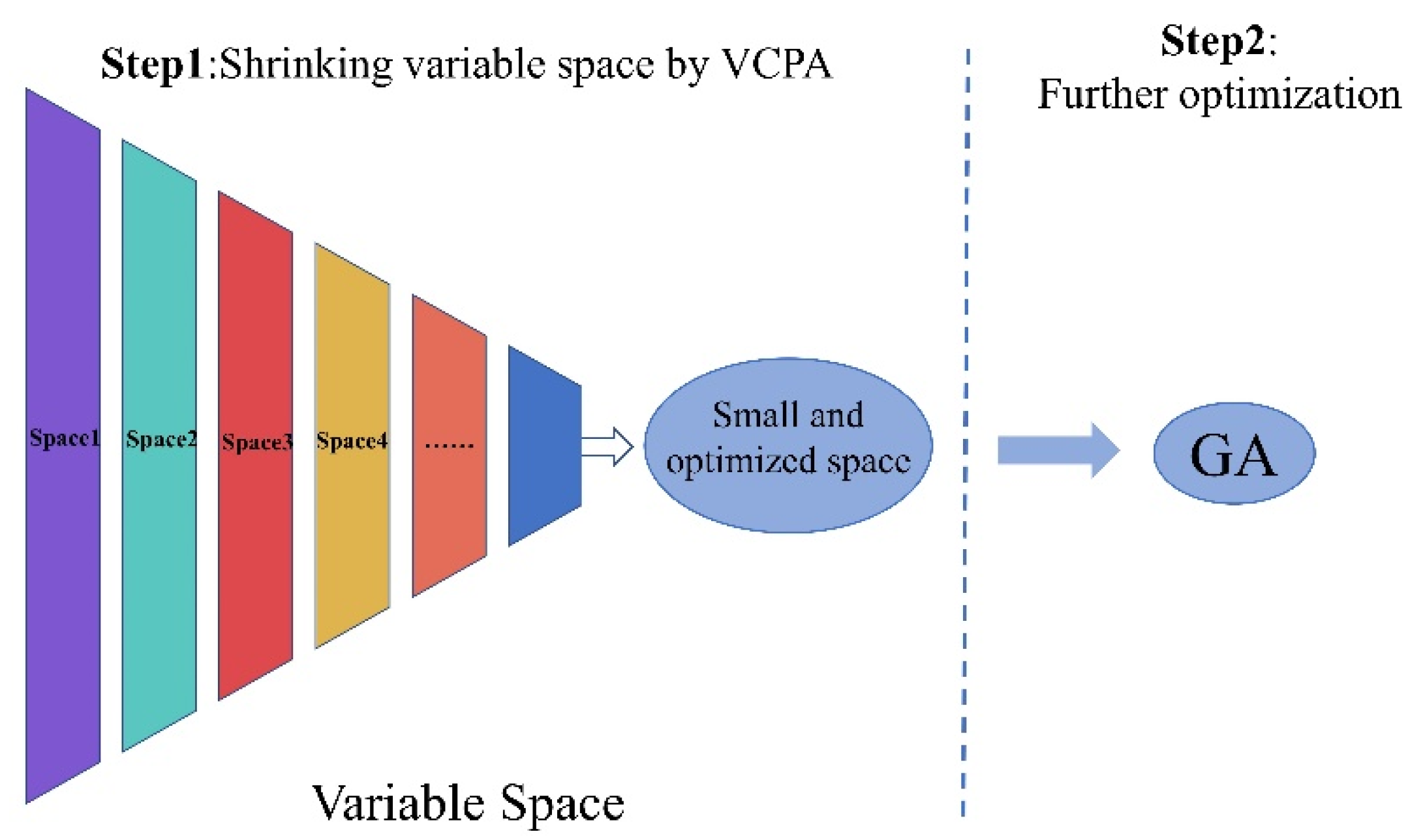
2.2. Performance of VCPA-GA Hybrid Strategy for Variable Selection
2.3. MLRAs for Estimating the LCC of Litchi
3. Materials and Methods
3.1. Study Area
3.2. Hyperspectral Measurements and Preprocessing
3.3. Determination of the LCC
3.4. VCPA-GA Hybrid Strategy for Variable Selection
3.5. The Evaluation of the Proposed MLRMs
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dan, L.; Wang, C.; Hao, J.; Peng, Z.; Yang, J.; Su, Y.; Song, J.; Chen, S. Monitoring litchi canopy foliar phosphorus content using hyperspectral data. Comput. Electron. Agric. 2018, 154, 176–186. [Google Scholar]
- Gitelson, A.A.; Merzlyak, M.N. Remote estimation of chlorophyll content in higher plant leaves. Int. J. Remote Sens. 1997, 18, 2691–2697. [Google Scholar] [CrossRef]
- Croft, H.; Chen, J.M.; Luo, X.; Bartlett, P.; Chen, B.; Staebler, R.M. Leaf chlorophyll content as a proxy for leaf photosynthetic capacity. Glob. Chang. Biol. 2017, 23, 3513–3524. [Google Scholar] [CrossRef] [Green Version]
- Fridley, J.D. Extended leaf phenology and the autumn niche in deciduous forest invasions. Nature 2012, 485, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Sonobe, R.; Wang, Q. Hyperspectral indices for quantifying leaf chlorophyll concentrations performed differently with different leaf types in deciduous forests. Ecol. Inform. 2017, 37, 1–9. [Google Scholar] [CrossRef]
- Clevers, J.; Kooistra, L. Using Hyperspectral Remote Sensing Data for Retrieving Canopy Chlorophyll and Nitrogen Content. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 574–583. [Google Scholar] [CrossRef]
- Schlemmer, M.; Gitelson, A.; Schepers, J.; Ferguson, R.; Peng, Y.; Shanahan, J.; Rundquist, D. Remote estimation of nitrogen and chlorophyll contents in maize at leaf and canopy levels. Int. J. Appl. Earth Obs. Geoinf. 2013, 25, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Gitelson, A.A.; Peng, Y.; Arkebauer, T.J.; Schepers, J. Relationships between gross primary production, green LAI, and canopy chlorophyll content in maize: Implications for remote sensing of primary production. Remote Sens. Environ. 2014, 144, 65–72. [Google Scholar] [CrossRef]
- Cui, B.; Zhao, Q.; Huang, W.; Song, X.; Ye, H.; Zhou, X. A New Integrated Vegetation Index for the Estimation of Winter Wheat Leaf Chlorophyll Content. Remote Sens. 2019, 11, 974. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.Q.; Bu, Z.J.; Lu, S.; Omasa, K. A General Algorithm of Leaf Chlorophyll Content Estimation for a Wide Range of Plant Species. IEEE Trans. Geosci. Remote Sens. 2022, 60, 4406814. [Google Scholar] [CrossRef]
- Ustin, S.L.; Gitelson, A.A.; Jacquemoud, S.; Schaepman, M.; Asner, G.P.; Gamon, J.A.; Zarco-Tejada, P. Retrieval of foliar information about plant pigment systems from high resolution spectroscopy. Remote Sens. Environ. 2009, 113, S67–S77. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Yun, Y.H.; Liang, Y.Z.; Xie, G.X.; Li, H.D.; Cao, D.S.; Xu, Q.S. A perspective demonstration on the importance of variable selection in inverse calibration for complex analytical systems. Analyst 2013, 138, 6412–6421. [Google Scholar] [CrossRef] [PubMed]
- Filho, H.; Galvo, R.; Araújo, M.C.U.; Silva, E.C.D.; Rohwedder, J.J.R. A strategy for selecting calibration samples for multivariate modeling. Chemom. Intell. Lab. Syst. 2004, 72, 83–91. [Google Scholar] [CrossRef]
- Cai, W.; Li, Y.; Shao, X. A variable selection method based on uninformative variable elimination for multivariate calibration of near-infrared spectra. Chemom. Intell. Lab. Syst. 2008, 90, 188–194. [Google Scholar] [CrossRef]
- Li, H.; Liang, Y.; Xu, Q.; Cao, D. Key wavelengths screening using competitive adaptive reweighted sampling method for multivariate calibration. Anal. Chim. Acta 2009, 648, 77–84. [Google Scholar] [CrossRef]
- Yun, Y.H.; Wang, W.T.; Deng, B.C.; Lai, G.B.; Liu, X.B.; Ren, D.B.; Liang, Y.Z.; Fan, W.; Xu, Q.S. Using variable combination population analysis for variable selection in multivariate calibration. Anal. Chim. Acta 2015, 862, 14–23. [Google Scholar] [CrossRef]
- Norgaard, L.; Saudland, A.; Wagner, J.; Nielsen, G.P.; Engelsen, S.B. Interval Partial Least-Squares Regression (iPLS): A Comparative Chemometric Study with an Example from Near-Infrared Spectroscopy. Appl. Spectrosc. 2000, 54, 413–419. [Google Scholar] [CrossRef]
- Yun, Y.H.; Li, H.D.; Wood, L.E.; Fan, W.; Wang, J.J.; Cao, D.S.; Xu, Q.S.; Liang, Y.Z. An efficient method of wavelength interval selection based on random frog for multivariate spectral calibration. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 111, 31–36. [Google Scholar] [CrossRef]
- Lin, Y.W.; Deng, B.C.; Wang, L.L.; Xu, Q.S.; Liu, L.; Liang, Y.Z. Fisher optimal subspace shrinkage for block variable selection with applications to NIR spectroscopic analysis. Chemom. Intell. Lab. Syst. 2016, 159, 196–204. [Google Scholar] [CrossRef]
- Deng, B.C.; Yun, Y.H.; Ma, P.; Lin, C.C.; Liang, Y.Z. A new method for wavelength interval selection that intelligently optimizes the locations, widths and combinations of the intervals. Analyst 2015, 140, 1876–1885. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Huang, Y.; Tian, K.; Song, X.; Min, S. A new spectral variable selection pattern using competitive adaptive reweighted sampling combined with successive projections algorithm. Analyst 2014, 139, 4894–4902. [Google Scholar] [CrossRef]
- Kong, Q.M.; Su, Z.B.; Shen, W.Z.; Zhang, B.F.; Wang, J.B.; Ji, N.; Ge, H.F. Research of straw biomass based on NIR by wavelength selection of IPLS-SPA. Spectrosc. Spectr. Anal. 2015, 35, 1233–1238. [Google Scholar]
- Zheng, K.; Li, Q.; Wang, J.; Geng, J.; Peng, C.; Tao, S.; Xuan, W.; Du, Y. Stability competitive adaptive reweighted sampling (SCARS) and its applications to multivariate calibration of NIR spectra. Chemom. Intell. Lab. Syst. 2012, 112, 48–54. [Google Scholar] [CrossRef]
- Bin, J.; Ai, F.; Fan, W.; Zhou, J.; Li, X.; Tang, W.; Liang, Y. An efficient variable selection method based on variable permutation and model population analysis for multivariate calibration of NIR spectra. Chemom. Intell. Lab. Syst. 2016, 158, 1–13. [Google Scholar] [CrossRef]
- Datt, B. Remote Sensing of Chlorophyll a, Chlorophyll b, Chlorophyll a+b, and Total Carotenoid Content in Eucalyptus Leaves. Remote Sens. Environ. 1998, 66, 111–121. [Google Scholar] [CrossRef]
- Curran, P.J.; Dungan, J.L.; Peterson, D.L. Estimating the foliar biochemical concentration of leaves with reflectance spectrometry. Remote Sens. Environ. 2001, 76, 349–359. [Google Scholar] [CrossRef]
- Jin, J.; Wang, Q. Selection of Informative Spectral Bands for PLS Models to Estimate Foliar Chlorophyll Content Using Hyperspectral Reflectance. IEEE Trans. Geosci. Remote Sens. 2019, 57, 3064–3072. [Google Scholar] [CrossRef]
- Zhang, W.W.; Yang, K.M.; Xia, T.; Liu, C.; Sun, T.T. Correlation analysis on spectral fractional-order differential and the content of heavy metal copper in corn leaves. Sci. Technol. Eng. 2017, 17, 33–38. [Google Scholar]
- Zhang, Y.Q.; Chen, J.M.; John, R.M.; Thomas, L.N. Leaf chlorophyll content retrieval from airborne hyperspectral remote sensing imagery. Remote Sens. Environ. 2008, 112, 3234–3247. [Google Scholar] [CrossRef]
- Tao, Z.; Ning, L.; Li, W.; Li, M.; Sun, H.; Zhang, Q.; Wu, J. Estimation of Chlorophyll Content in Potato Leaves Based on Spectral Red Edge Position. IFAC-PapersOnLine 2018, 51, 602–606. [Google Scholar]
- Sun, J.; Shi, S.; Yang, J.; Chen, B.; Gong, W. Estimating leaf chlorophyll status using hyperspectral lidar measurements by PROSPECT model inversion. Remote Sens. Environ. 2018, 212, 1–7. [Google Scholar] [CrossRef]
- Gaurav, S.; Babankumar, B.; Lini, M.; Jonali, G.; Goswami, B.U.; Choudhury, P.L.N. Chlorophyll estimation using multi-spectral unmanned aerial system based on machine learning techniques. Remote Sens. Appl. Soc. Environ. 2019, 15, 100235. [Google Scholar]
- Mao, Z.H.; Deng, L.; Duan, F.Z.; Li, X.J.; Qiao, D.Y. Angle effects of vegetation indices and the influence on prediction of SPAD values in soybean and maize. Int. J. Appl. Earth Obs. Geoinf. 2020, 93, 102198. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, J.; Chen, D.; Huang, Y.; Huang, W. Assessment of Leaf Chlorophyll Content Models for Winter Wheat Using Landsat-8 Multispectral Remote Sensing Data. Remote Sens. 2020, 12, 2574. [Google Scholar] [CrossRef]
- Zhu, W.; Sun, Z.; Gong, H.; Li, J.; Zhu, K. Estimating leaf chlorophyll content of crops via optimal unmanned aerial vehicle hyperspectral data at multi-scales. Comput. Electron. Agric. 2020, 178, 105786. [Google Scholar] [CrossRef]
- Li, Y.; Ma, Q.; Chen, J.M.; Croft, H.; Liu, J. Fine-scale leaf chlorophyll distribution across a deciduous forest through two-step model inversion from Sentinel-2 data. Remote Sens. Environ. 2021, 264, 112618. [Google Scholar] [CrossRef]
- Yang, Z.; Tian, J.; Feng, K.; Gong, X.; Liu, J. Application of a hyperspectral imaging system to quantify leaf-scale chlorophyll, nitrogen and chlorophyll fluorescence parameters in grapevine. Plant Physiol. Biochem. 2021, 166, 723–737. [Google Scholar] [CrossRef] [PubMed]
- Filella, I.; Penuelas, J. The red edge position and shape as indicators of plant chlorophyll content, biomass and hydric status. Int. J. Remote Sens. 1994, 15, 1459–1470. [Google Scholar] [CrossRef]
- Zhao, D.; Wang, J.; Miao, J.; Zhen, J.; Wang, J.; Gao, C.; Jiang, J.; Wu, G. Spectral features of Fe and organic carbon in estimating low and moderate concentration of heavy metals in mangrove sediments across different regions and habitat types. Geoderma 2022, 426, 116093. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Miller, J.R.; Mohammed, G.H.; Noland, T.L.; Sampson, P.H. Chlorophyll Fluorescence Effects on Vegetation Apparent Reflectance: I. Leaf-Level Measurements and Model Simulation. Remote Sens. Environ. 2000, 74, 582–595. [Google Scholar] [CrossRef]
- Benkhettou, N.; Brito da Cruz, A.M.C.; Torres, D.F.M. A fractional calculus on arbitrary time scales: Fractional differentiation and fractional integration. Signal Process. 2015, 107, 230–237. [Google Scholar] [CrossRef] [Green Version]
- Markwell, J.; Ostermann, J.C.; Mitchell, J.L. Calibration of the Minolta SPAD-502 leaf chlorophyll meter. Photosynth. Res. 1995, 46, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Deng, B.C.; Yun, Y.H.; Liang, Y.Z.; Yi, L.Z. A novel variable selection approach that iteratively optimizes variable space using weighted binary matrix sampling. Analyst 2014, 139, 4836–4845. [Google Scholar] [CrossRef]
- Li, H.D.; Liang, Y.Z.; Xu, Q.S.; Cao, D.S. Model population analysis for variable selection. J. Chemom. 2010, 24, 418–423. [Google Scholar] [CrossRef]
- Abakar, K.; Yu, C. Application of Genetic Algorithm for Feature Selection in Optimisation of SVMR Model for Prediction of Yarn Tenacity. Fibres Text. East. Eur. 2013, 21, 95–99. [Google Scholar]
- Yun, Y.H.; Bin, J.; Liu, D.L.; Xu, L.; Yan, T.L.; Cao, D.S.; Xu, Q.S. A hybrid variable selection strategy based on continuous shrinkage of variable space in multivariate calibration. Anal. Chim. Acta 2019, 1058, 58–69. [Google Scholar] [CrossRef]
- Rumere, F.; Soemartojo, S.M.; Widyaningsih, Y. Restricted Ridge Regression estimator as a parameter estimation in multiple linear regression model for multicollinearity case. J. Phys. Conf. Ser. 2021, 1725, 012021. [Google Scholar] [CrossRef]
- Hoeppner, J.M.; Skidmore, A.K.; Darvishzadeh, R.; Heurich, M.; Gara, T.W. Mapping Canopy Chlorophyll Content in a Temperate Forest Using Airborne Hyperspectral Data. Remote Sens. 2020, 12, 3573. [Google Scholar] [CrossRef]
- Chen, T.Q.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; p. 785. [Google Scholar]
- Rasmussen, C.E.; Williams, C. Gaussian Processes for Machine Learning; MIT Press: Cambridge, MA, USA, 2005. [Google Scholar]
- Kusumo, B.H.; Hedley, M.J.; Hedley, C.B.; Hueni, A.; Tuohy, M.P. The use of Vis-NIR spectral reflectance for determining root density: Evaluation of ryegrass roots in a glasshouse trial. Eur. J. Soil Sci. 2010, 60, 22–32. [Google Scholar] [CrossRef]
- Cui, S.C.; Zhou, K.F.; Ding, R.F.; Cheng, Y.Y.; Jiang, G. Estimation of soil copper content based on fractional-order derivative spectroscopy and spectral characteristic band selection. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 275, 121190. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Wang, Q. Hyperspectral indices developed from the low order fractional derivative spectra can capture leaf dry matter content across a variety of species better. Agric. For. Meteorol. 2022, 322, 109007. [Google Scholar] [CrossRef]
- Sonobe, R.; Yamashita, H.; Mihara, H.; Morita, A.; Ikka, T. Estimation of Leaf Chlorophyll a, b and Carotenoid Contents and Their Ratios Using Hyperspectral Reflectance. Remote Sens. 2020, 12, 3265. [Google Scholar] [CrossRef]
- Hengl, T.; Leenaars, J.G.B.; Shepherd, K.D.; Walsh, M.G.; Heuvelink, G.B.M.; Mamo, T.; Tilahun, H.; Berkhout, E.; Cooper, M.; Fegraus, E.; et al. Soil nutrient maps of Sub-Saharan Africa: Assessment of soil nutrient content at 250 m spatial resolution using machine learning. Nutr. Cycl. Agroecosyst. 2017, 109, 77–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taghizadeh-Mehrjardi, R.; Schmidt, K.; Chakan, A.A.; Rentschler, T.; Scholten, T. Improving the Spatial Prediction of Soil Organic Carbon Content in Two Contrasting Climatic Regions by Stacking Machine Learning Models and Rescanning Covariate Space. Remote Sens. 2020, 12, 1095. [Google Scholar] [CrossRef]





| Data Source | Name of the Sensor | Type of Spectra or Image Data | Methods | Study Area | Object of Study | Regression Statistics | Research Contents | Reference | Year of References |
|---|---|---|---|---|---|---|---|---|---|
| Airborne | Compact Airborne Spectrographic Imager (CASI) | Hyperspectral remote sensing imagery (HIS) | Lookup-table (LUT)-based inversion | Ten black spruce stands near Sudbury, Ontario | Ten black spruce | R2 = 0.47, RMSE = 4.34 μg/cm2 | Estimated LCC from the CASI imagery by combining the geometrical-optical model 4-Scale and the modified leaf optical model PROSPECT | [30] | 2008 |
| Ground-based | ASD FieldSpec spectrometer | Hyperspectral remote sensing reflectance data | Narrowband vegetation indices (VIs) | The Naeba Mountains, Japan | Beech leaves | CI (R2 =0.73, WAIC = 2241.5, RPD = 1.76) D2 (R2 = 0.71, WAIC = 582.4, RPD = 1.94) | Evaluated the performances of hyperspectral indices for both leaf types within beech canopies, developed a new index for estimating LCC in both sunlit and sun-shaded areas. | [5] | 2017 |
| Ground-based | Gaia hyperspectral imaging system | HSI | Linear extrapolation method | The experimental greenhouse of China Agricultural University | Potato | R2 = 0.8682 | Inverted the LCC of potato by using the selected optimal red edge position | [31] | 2018 |
| Ground-based | Hyperspectral lidar (HSL) system | HSL | PROSPECT-4 model, support vector regression (SVR) | Junchuan County, Suizhou, China | Rice | PROSPECT-4 model inversion (R2 = 0.55) | Investigated the possibility of estimating foliar Chl through the PROSPECT-4 model using the HSL system. | [32] | 2018 |
| Unmanned Aerial Vehicle (UAV) | Parrot sequoia multi-spectral sensor | Multi-spectral images (MSIs) | Machine learning regression algorithms (MLRAs) | ICAR research complex for NEH region at Umiam, Meghalaya | Maize | R2 = 0.904, RMSE = 0.057 mg/gm | Estimated the LCC of a standing maize plant from multi-spectral UAV images by using machine learning algorithms. | [33] | 2019 |
| UAV | Cubert S185 hyperspectral sensor | HSI | LR (linear regression), SVR (support vector regression) | Luozhuang village, Zhangziying Town, Daxing District, Beijing, China | Soybean and maize | MCARI1 for soybean (MAE = 1.617) MCARI/OSAVI for maize (MAE = 2.422); | Retrieved canopy SPAD values of maize and soybean by using the 16 VIs at different observation angles and their combinations. | [34] | 2020 |
| Satellite | Landsat-8 Operational Land Imager (OLI) | MSI | VIs (vegetation indices), MLRAs (machine learning regression algorithms), LUT (lookup-table)-based inversion, and hybrid regression approaches | Shunyi District, Beijing, China | Winter wheat | MTVI2 (RMSE = 5.99 µg/cm2, RRMSE = 10.49%) GPR (RMSE = 5.50 µg/cm2, RRMSE = 9.62%) LUT (RMSE = 8.08 µg/cm2, RRMSE = 14.14%) AL-GPR (RMSE = 12.43 µg/cm2, RRMSE = 21.77%) | Evaluated capabilities and potentials of Landsat-8 (OLI) imagery using four different retrieval methods for LCC modeling | [35] | 2020 |
| UAV | Cubert S185 hyperspectral sensor | HSI | MLR (multi-variable linear regression), RF (random forest), BPNN (backpropagation neural network), and SVM (support vector machine) | Yucheng Comprehensive Experiment Station (YCES) of the Chinese Academy of Sciences | Maize and wheat | SVM for maize (R2 = 0.83, RMSE = 5.80, MRE = 0.12); SVM for wheat (R2 = 0.78, RMSE = 2.80, MRE = 0.11) | Examined the effects of spectral information and spatial scale of unmanned drone images, as well as phenological types and phenology, on LCC estimation of maize and wheat. | [36] | 2020 |
| Satellite | Sentinel-2 | MSI | PROSPECT-5 leaf optical model | The Borden Forest Research Station | Mixed temperate forest | R2 = 0.849, RMSE = 0.304 μg/cm2 | Estimated LCC from Sentinel-2 (MSI) data via a physically based, two-step inversion approach | [37] | 2021 |
| UAV | Pika L hyperspectral imaging system | HSI | Narrowband vegetation indices (VIs) Multiple linear regression (MLR) | A commercial wine estate at the eastern base of Helan Mountain in Ningxia Province, China | Wine grapes | (D735 − D573)/(D735 + D573) (R2 = 0.50) | Investigated the SPAD changes of grape leaves at different growth stages, and explored a new method for predicting these parameters using hyperspectral imaging. | [38] | 2021 |
| Ground-based | ASD FieldSpec spectrometer | Hyperspectral reflectance data | VIs, PROSPECT, PLSR, SVR | Changchun, China | Different plant species (trees, bushes, and lianas) | Modified difference ratio index (MDRI), R2 = 0.92, RMSE = 5.65 μg/cm2 | Developed a new algorithm for estimating the LCC of different plant species by combining SIs (spectral indices) with multi-angular hyperspectral reflectance of leaves. | [10] | 2022 |
| Orders | Tb | Pb | Nb | rmax | Corresponding Bands/nm |
|---|---|---|---|---|---|
| 0 | 1946 | 1720 | 226 | 0.8542 | 709 |
| 0.2 | 1953 | 1756 | 197 | 0.8722 | 704 |
| 0.4 | 1778 | 1609 | 169 | 0.8835 | 698 |
| 0.6 | 1631 | 1426 | 205 | 0.8884 | 694 |
| 0.8 | 1535 | 1235 | 300 | 0.9179 | 756 |
| 1 | 1340 | 931 | 409 | 0.8929 | 756 |
| 1.2 | 1264 | 515 | 649 | 0.9015 | 742 |
| 1.4 | 1086 | 406 | 680 | 0.8994 | 726 |
| 1.6 | 963 | 375 | 588 | 0.9018 | 723 |
| 1.8 | 701 | 290 | 411 | 0.9020 | 720 |
| 2 | 429 | 197 | 232 | 0.9001 | 718 |
| Orders | Nvar | Nlvs | RMSEC | RMSECV | RMSEP |
|---|---|---|---|---|---|
| 0 | 5 | 9 | 3.41 | 3.61 | 5.74 |
| 0.2 | 54 | 10 | 3.35 | 3.59 | 5.42 |
| 0.4 | 16 | 9 | 3.38 | 3.58 | 5.54 |
| 0.6 | 18 | 10 | 3.33 | 3.55 | 5.41 |
| 0.8 | 14 | 10 | 3.16 | 3.33 | 5.04 |
| 1 | 27 | 8 | 3.05 | 3.34 | 5.61 |
| 1.2 | 19 | 8 | 2.81 | 3.04 | 5.51 |
| 1.4 | 11 | 9 | 2.82 | 3.14 | 5.24 |
| 1.6 | 32 | 9 | 2.59 | 3.12 | 5.86 |
| 1.8 | 15 | 5 | 2.82 | 3.15 | 5.25 |
| 2 | 43 | 5 | 2.59 | 3.02 | 5.39 |
| Orders | Algorithm | Training Set | Testing Set | Validation Set | RPIQ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 | MAE | RMSE | R2 | MAE | RMSE | R2 | MAE | RMSE | |||
| 0 | RR | 0.82 | 3.56 | 4.61 | 0.82 | 4.08 | 5.10 | 0.83 | 3.94 | 5.04 | 2.77 |
| RF | 0.85 | 3.31 | 4.30 | 0.66 | 5.45 | 6.94 | 0.66 | 6.02 | 7.20 | 1.67 | |
| XGBoost | 0.99 | 0.53 | 0.71 | 0.67 | 5.46 | 6.82 | 0.72 | 5.34 | 6.51 | 1.99 | |
| SVR | 0.85 | 3.14 | 4.26 | 0.84 | 3.72 | 4.76 | 0.84 | 3.93 | 4.92 | 3.07 | |
| GPR | 0.85 | 3.29 | 4.30 | 0.83 | 3.79 | 4.87 | 0.85 | 3.88 | 4.84 | 3.13 | |
| 0.2 | RR | 0.86 | 3.17 | 4.06 | 0.84 | 3.75 | 4.71 | 0.88 | 3.40 | 4.23 | 3.59 |
| RF | 0.91 | 2.51 | 3.29 | 0.77 | 4.30 | 5.74 | 0.80 | 4.28 | 5.47 | 2.57 | |
| XGBoost | 0.98 | 1.21 | 1.70 | 0.83 | 3.84 | 4.96 | 0.85 | 3.90 | 4.84 | 2.67 | |
| SVR | 0.88 | 2.69 | 4.74 | 0.84 | 3.51 | 4.74 | 0.81 | 3.94 | 5.37 | 3.09 | |
| GPR | 0.87 | 3.03 | 3.89 | 0.85 | 3.67 | 4.62 | 0.88 | 3.55 | 4.29 | 3.86 | |
| 0.4 | RR | 0.84 | 3.27 | 4.43 | 0.84 | 3.71 | 4.83 | 0.82 | 4.11 | 5.21 | 2.88 |
| RF | 0.90 | 2.64 | 3.48 | 0.73 | 4.74 | 6.17 | 0.71 | 5.46 | 6.60 | 1.99 | |
| XGBoost | 0.94 | 2.06 | 2.75 | 0.76 | 4.38 | 5.86 | 0.73 | 5.18 | 6.37 | 2.47 | |
| SVR | 0.86 | 2.95 | 4.15 | 0.82 | 3.82 | 5.08 | 0.21 | 5.40 | 10.92 | 1.29 | |
| GPR | 0.86 | 3.18 | 4.09 | 0.86 | 3.51 | 4.43 | 0.84 | 3.99 | 4.88 | 2.89 | |
| 0.6 | RR | 0.87 | 3.06 | 4.01 | 0.85 | 3.72 | 4.68 | 0.86 | 3.75 | 4.65 | 3.44 |
| RF | 0.90 | 2.69 | 3.54 | 0.81 | 2.69 | 3.54 | 0.84 | 3.91 | 4.95 | 3.08 | |
| XGBoost | 0.91 | 2.56 | 3.36 | 0.80 | 4.03 | 5.27 | 0.84 | 3.98 | 4.87 | 3.13 | |
| SVR | 0.86 | 2.87 | 4.06 | 0.84 | 2.53 | 4.75 | 0.83 | 3.96 | 5.14 | 3.42 | |
| GPR | 0.88 | 2.99 | 3.83 | 0.85 | 3.70 | 4.68 | 0.87 | 3.75 | 4.52 | 3.89 | |
| 0.8 | RR | 0.86 | 3.10 | 4.04 | 0.84 | 3.81 | 4.74 | 0.86 | 3.76 | 4.68 | 3.38 |
| RF | 0.91 | 2.55 | 3.27 | 0.81 | 4.17 | 5.20 | 0.85 | 3.78 | 4.84 | 2.81 | |
| XGBoost | 0.92 | 2.41 | 3.10 | 0.80 | 4.21 | 5.36 | 0.82 | 4.09 | 5.27 | 2.41 | |
| SVR | 0.86 | 3.03 | 4.11 | 0.82 | 3.91 | 5.05 | 0.86 | 3.55 | 4.58 | 3.36 | |
| GPR | 0.87 | 2.98 | 3.88 | 0.85 | 3.74 | 4.69 | 0.85 | 3.86 | 4.77 | 3.23 | |
| 1 | RR | 0.89 | 3.02 | 3.71 | 0.85 | 3.68 | 4.67 | 0.86 | 3.60 | 4.63 | 3.34 |
| RF | 0.89 | 2.78 | 3.60 | 0.84 | 3.63 | 4.72 | 0.85 | 3.79 | 4.75 | 2.48 | |
| XGBoost | 0.91 | 2.53 | 3.25 | 0.84 | 3.82 | 4.71 | 0.84 | 3.91 | 4.98 | 2.59 | |
| SVR | 0.84 | 3.23 | 4.45 | 0.82 | 3.95 | 5.04 | 0.83 | 3.90 | 5.06 | 3.05 | |
| GPR | 0.89 | 3.02 | 3.71 | 0.85 | 3.68 | 4.67 | 0.86 | 3.60 | 4.63 | 3.34 | |
| 1.2 | RR | 0.87 | 3.10 | 4.00 | 0.85 | 3.77 | 4.63 | 0.83 | 4.03 | 5.10 | 2.86 |
| RF | 0.90 | 2.58 | 3.49 | 0.83 | 3.73 | 4.91 | 0.83 | 3.78 | 5.10 | 2.50 | |
| XGBoost | 0.93 | 2.27 | 2.96 | 0.83 | 3.72 | 4.99 | 0.82 | 3.96 | 5.18 | 2.82 | |
| SVR | 0.88 | 2.58 | 3.84 | 0.85 | 3.61 | 4.68 | 0.84 | 4.06 | 4.96 | 2.94 | |
| GPR | 0.87 | 3.09 | 3.99 | 0.85 | 3.77 | 4.64 | 0.83 | 4.05 | 5.13 | 2.85 | |
| 1.4 | RR | 0.84 | 3.32 | 4.38 | 0.84 | 3.72 | 4.79 | 0.82 | 4.15 | 5.20 | 2.99 |
| RF | 0.88 | 2.90 | 3.78 | 0.81 | 3.76 | 5.19 | 0.81 | 4.24 | 5.37 | 2.62 | |
| XGBoost | 0.89 | 2.67 | 3.63 | 0.79 | 4.15 | 5.46 | 0.78 | 4.69 | 5.74 | 2.58 | |
| SVR | 0.86 | 2.72 | 4.09 | 0.81 | 4.09 | 5.24 | 0.82 | 4.27 | 5.29 | 2.88 | |
| GPR | 0.84 | 3.32 | 4.26 | 0.84 | 3.68 | 4.76 | 0.82 | 4.16 | 5.21 | 2.92 | |
| 1.6 | RR | 0.86 | 3.11 | 4.07 | 0.85 | 3.74 | 4.59 | 0.86 | 3.56 | 4.68 | 2.98 |
| RF | 0.92 | 2.42 | 3.19 | 0.81 | 3.82 | 5.24 | 0.85 | 3.52 | 4.75 | 2.79 | |
| XGBoost | 0.99 | 0.88 | 1.24 | 0.80 | 3.80 | 5.35 | 0.82 | 4.04 | 5.28 | 2.71 | |
| SVR | 0.86 | 2.94 | 4.16 | 0.84 | 3.70 | 4.77 | 0.83 | 3.96 | 5.09 | 2.85 | |
| GPR | 0.84 | 3.31 | 4.35 | 0.85 | 3.70 | 4.66 | 0.85 | 3.67 | 4.75 | 3.06 | |
| 1.8 | RR | 0.84 | 3.41 | 4.37 | 0.85 | 3.59 | 4.67 | 0.84 | 3.78 | 4.96 | 2.89 |
| RF | 0.91 | 2.47 | 3.25 | 0.81 | 3.76 | 5.14 | 0.86 | 3.42 | 4.63 | 2.80 | |
| XGBoost | 0.97 | 1.36 | 1.80 | 0.81 | 3.81 | 5.14 | 0.84 | 3.78 | 4.95 | 2.88 | |
| SVR | 0.83 | 3.27 | 4.47 | 0.83 | 3.74 | 4.95 | 0.81 | 3.97 | 5.30 | 2.70 | |
| GPR | 0.83 | 3.47 | 4.46 | 0.84 | 3.68 | 4.78 | 0.84 | 3.73 | 4.91 | 2.91 | |
| 2.0 | RR | 0.86 | 3.21 | 4.09 | 0.83 | 3.93 | 4.87 | 0.82 | 3.93 | 4.87 | 2.66 |
| RF | 0.87 | 3.06 | 3.89 | 0.82 | 3.83 | 5.10 | 0.84 | 3.71 | 4.94 | 2.67 | |
| XGBoost | 0.93 | 2.23 | 2.86 | 0.81 | 3.87 | 5.24 | 0.85 | 3.34 | 4.73 | 2.69 | |
| SVR | 0.87 | 2.60 | 3.88 | 0.81 | 3.99 | 5.22 | 0.77 | 4.59 | 5.93 | 2.26 | |
| GPR | 0.85 | 3.31 | 4.27 | 0.84 | 3.76 | 4.83 | 0.83 | 3.97 | 5.10 | 2.75 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasan, U.; Jia, K.; Wang, L.; Wang, C.; Shen, Z.; Yu, W.; Sun, Y.; Jiang, H.; Zhang, Z.; Guo, J.; et al. Retrieval of Leaf Chlorophyll Contents (LCCs) in Litchi Based on Fractional Order Derivatives and VCPA-GA-ML Algorithms. Plants 2023, 12, 501. https://doi.org/10.3390/plants12030501
Hasan U, Jia K, Wang L, Wang C, Shen Z, Yu W, Sun Y, Jiang H, Zhang Z, Guo J, et al. Retrieval of Leaf Chlorophyll Contents (LCCs) in Litchi Based on Fractional Order Derivatives and VCPA-GA-ML Algorithms. Plants. 2023; 12(3):501. https://doi.org/10.3390/plants12030501
Chicago/Turabian StyleHasan, Umut, Kai Jia, Li Wang, Chongyang Wang, Ziqi Shen, Wenjie Yu, Yishan Sun, Hao Jiang, Zhicong Zhang, Jinfeng Guo, and et al. 2023. "Retrieval of Leaf Chlorophyll Contents (LCCs) in Litchi Based on Fractional Order Derivatives and VCPA-GA-ML Algorithms" Plants 12, no. 3: 501. https://doi.org/10.3390/plants12030501







