Plant Peroxisomal Polyamine Oxidase: A Ubiquitous Enzyme Involved in Abiotic Stress Tolerance
Abstract
:1. Introduction
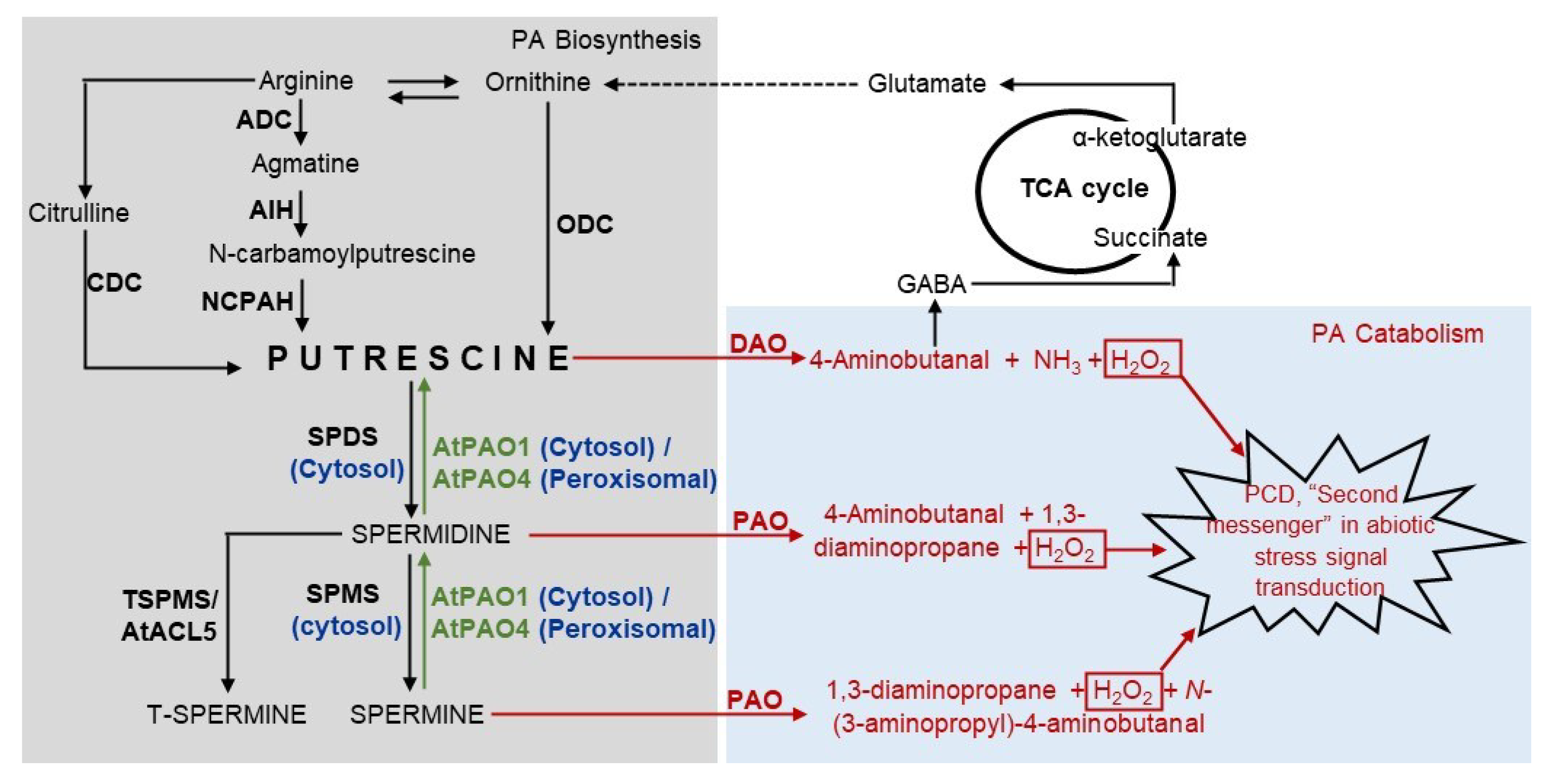
2. Polyamine Metabolism
2.1. Catabolism
2.2. Biosynthesis
3. Polyamine Oxidases
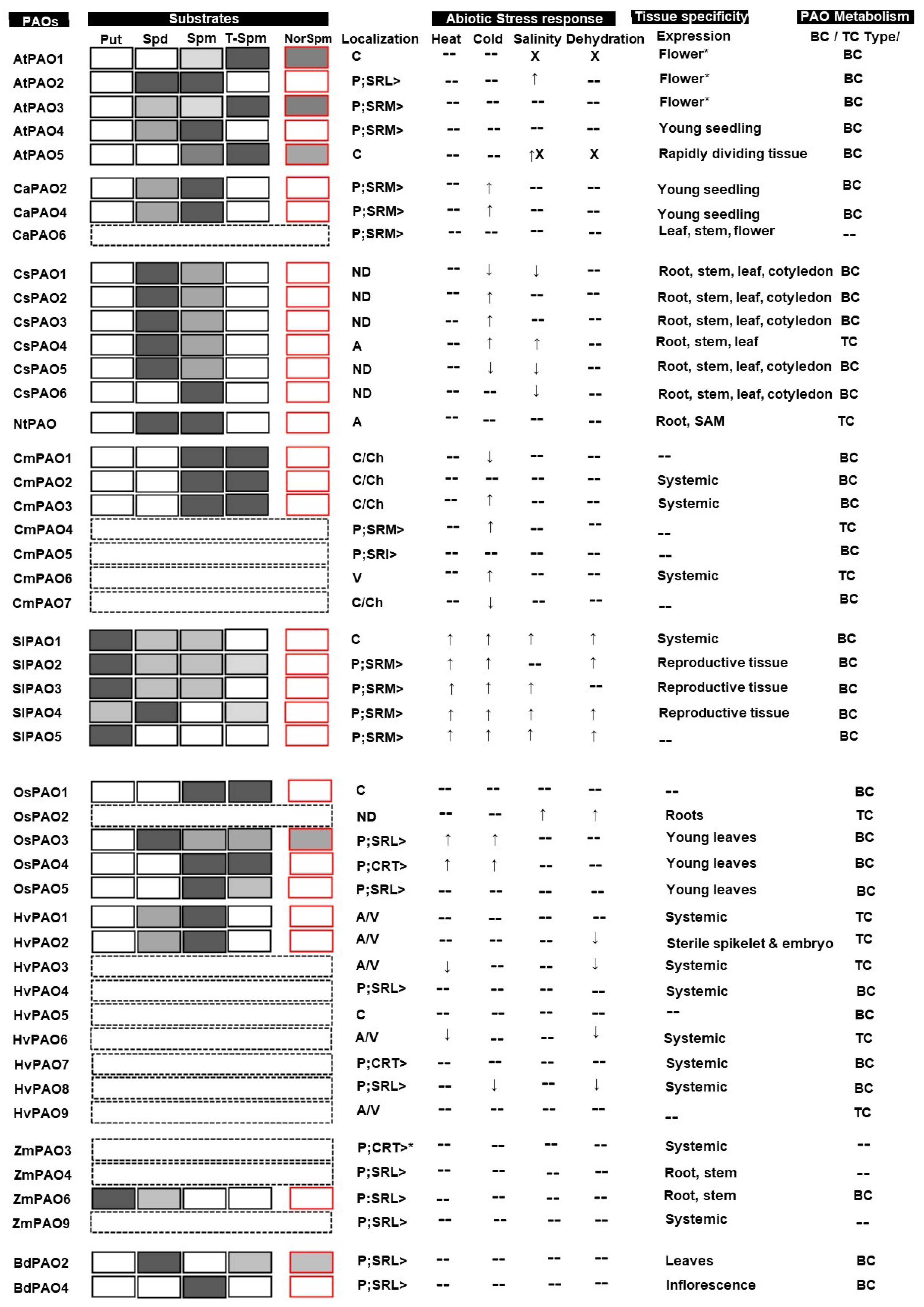
3.1. Substrate Specificity
3.1.1. Dicot PAOs
3.1.2. Monocot PAOs
3.2. Tissue Specificity of PAOs
3.2.1. Dicot PAOs
3.2.2. Monocot PAOs
3.3. Role of Polyamine Oxidases in Abiotic Stress Tolerances
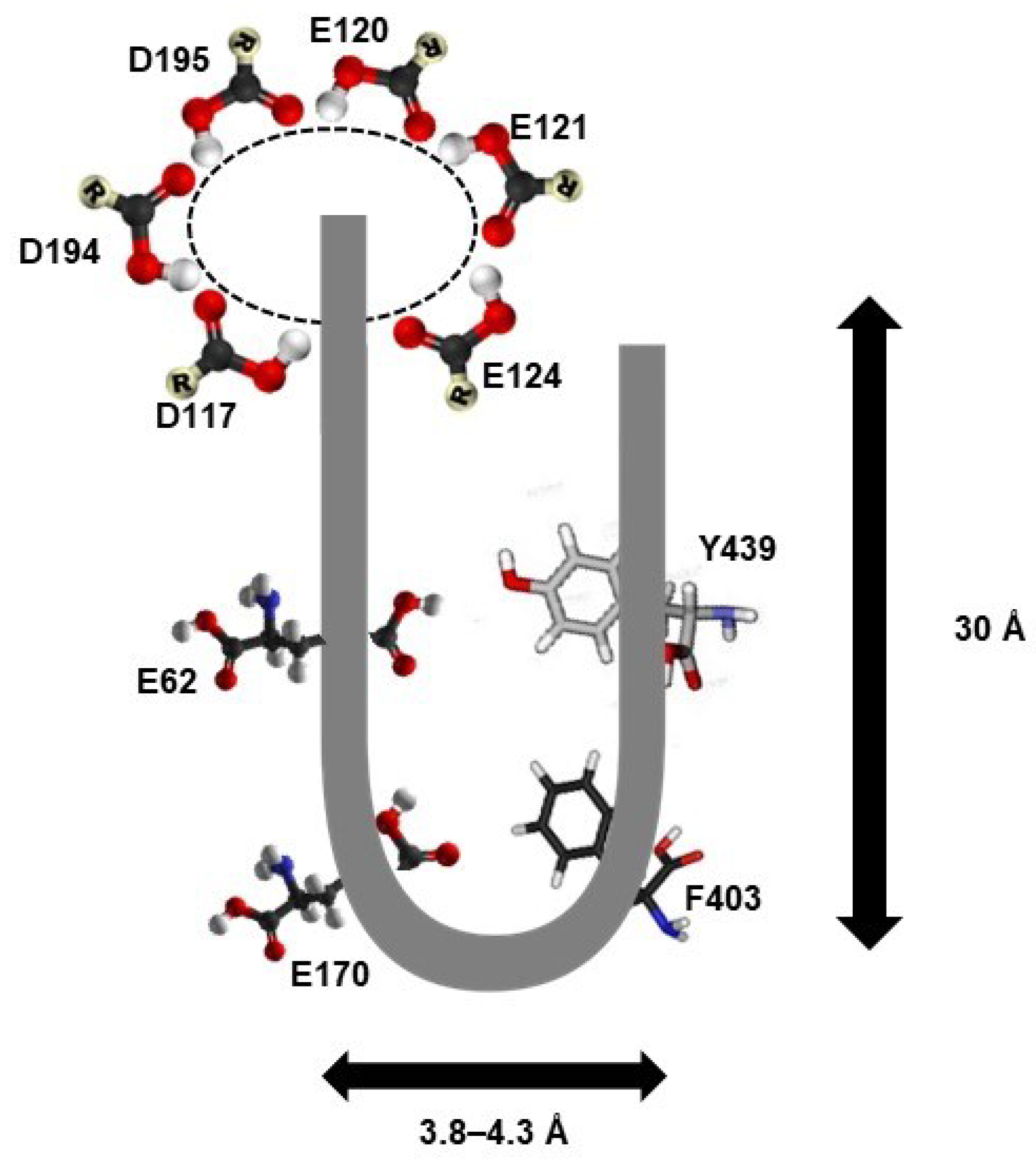
3.4. Three-Dimensional (3D) Structure of PAO
3.5. Subcellular Localization of PAOs—Peroxisome Forms the Core of Intracellular PAOs
3.6. Peroxisomal PAO: An Evolutionary Perspective
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gholami, M.; Fakhari, A.R.; Ghanati, F. Selective Regulation of Nicotine and Polyamines Biosynthesis in Tobacco Cells by Enantiomers of Ornithine. Chirality 2012, 25, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Martin-Tanguy, J. Conjugated polyamines and reproductive development: Biochemical, molecular and physiological approaches. Physiol. Plant 2010, 100, 675–688. [Google Scholar] [CrossRef]
- Kusano, T.; Berberich, T.; Tateda, C.; Takahashi, Y. Polyamines: Essential factors for growth and survival. Planta 2008, 228, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Bassard, J.-E.; Ullmann, P.; Bernier, F.; Werck-Reichhart, D. Phenolamides: Bridging polyamines to the phenolic metabolism. Phytochemistry 2010, 71, 1808–1824. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, K.; Kashiwagi, K. Modulation of protein synthesis by polyamines. IUBMB Life 2015, 67, 160–169. [Google Scholar] [CrossRef]
- Alcázar, R.; Marco, F.; Cuevas, J.C.; Patron, M.; Ferrando, A.; Carrasco, P.; Tiburcio, A.F.; Altabella, T. Involvement of polyamines in plant response to abiotic stress. Biotechnol. Lett. 2006, 28, 1867–1876. [Google Scholar] [CrossRef]
- Groppa, M.D.; Benavides, M.P. Polyamines and abiotic stress: Recent advances. Amino Acids 2007, 34, 35–45. [Google Scholar] [CrossRef]
- Sagor, G.H.M.; Inoue, M.; Kim, D.W.; Kojima, S.; Niitsu, M.; Berberich, T.; Kusano, T. The polyamine oxidase from lycophyte Selaginella lepidophylla (SelPAO5), unlike that of angiosperms, back-converts thermospermine to norspermidine. FEBS Lett. 2015, 589, 3071–3078. [Google Scholar] [CrossRef]
- Fincato, P.; Moschou, P.; Spedaletti, V.; Tavazza, R.; Angelini, R.; Federico, R.; Roubelakis-Angelakis, K.A.; Tavladoraki, P. Functional diversity inside the Arabidopsis polyamine oxidase gene family. J. Exp. Bot. 2010, 62, 1155–1168. [Google Scholar] [CrossRef]
- Michael, A.J. Polyamines in Eukaryotes, Bacteria, and Archaea. J. Biol. Chem. 2016, 291, 14896–14903. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Kim, N.W.; Niitsu, M.; Berberich, T.; Kusano, T. POLYAMINE OXIDASE 1 from rice (Oryza sativa) is a functional ortholog of Arabidopsis POLYAMINE OXIDASE 5. Plant Signal. Behav. 2014, 9, e29773. [Google Scholar] [CrossRef]
- Fincato, P.; Moschou, P.; Ahou, A.; Angelini, R.; Roubelakis-Angelakis, K.A.; Federico, R.; Tavladoraki, P. The members of Arabidopsis thaliana PAO gene family exhibit distinct tissue- and organ-specific expression pattern during seedling growth and flower development. Amino Acids 2011, 42, 831–841. [Google Scholar] [CrossRef]
- Knott, J.M.; Romer, P.; Sumper, M. Putative spermine synthases from Thalassiosira pseudonana and Arabidopsis thaliana synthesize thermospermine rather than spermine. FEBS Lett. 2007, 581, 3081–3086. [Google Scholar] [CrossRef]
- Takano, A.; Kakehi, J.-I.; Takahashi, T. Thermospermine is Not a Minor Polyamine in the Plant Kingdom. Plant Cell Physiol. 2012, 53, 606–616. [Google Scholar] [CrossRef]
- Gamarnik, A.; Frydman, R.B. Cadaverine, an Essential Diamine for the Normal Root Development of Germinating Soybean (Glycine max) Seeds. Plant Physiol. 1991, 97, 778–785. [Google Scholar] [CrossRef]
- Takahashi, Y.; Cong, R.; Sagor, G.H.M.; Niitsu, M.; Berberich, T.; Kusano, T. Characterization of five polyamine oxidase isoforms in Arabidopsis thaliana. Plant Cell Rep. 2010, 29, 955–965. [Google Scholar] [CrossRef]
- Yoda, H.; Hiroi, Y.; Sano, H. Polyamine Oxidase Is One of the Key Elements for Oxidative Burst to Induce Programmed Cell Death in Tobacco Cultured Cells. Plant Physiol. 2006, 142, 193–206. [Google Scholar] [CrossRef]
- Kakehi, J.-I.; Kuwashiro, Y.; Niitsu, M.; Takahashi, T. Thermospermine is Required for Stem Elongation in Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 1342–1349. [Google Scholar] [CrossRef]
- Ono, Y.; Kim, D.W.; Watanabe, K.; Sasaki, A.; Niitsu, M.; Berberich, T.; Kusano, T.; Takahashi, Y. Constitutively and highly expressed Oryza sativa polyamine oxidases localize in peroxisomes and catalyze polyamine back conversion. Amino Acids 2011, 42, 867–876. [Google Scholar] [CrossRef]
- Jaana, V.; Riina, M.; Komlan, A.; Marko, S.; Johanna, K.; Esa, L.; Hely, H.; Outi, S.; Tytti, S. Thermospermine Synthase (ACL5) and Diamine Oxidase (DAO) Expression Is Needed for Zygotic Embryogenesis and Vascular Development in Scots Pine. Front. Plant Sci. 2019, 10, 1600. [Google Scholar]
- Takahashi, Y.; Ono, K.; Akamine, Y.; Asano, T.; Ezaki, M.; Mouri, I. Highly-expressed polyamine oxidases catalyze polyamine back conversion in Brachypodium distachyon. J. Plant Res. 2017, 131, 341–348. [Google Scholar] [CrossRef]
- Hao, Y.; Huang, B.; Jia, D.; Mann, T.; Jiang, X.; Qiu, Y.; Niitsu, M.; Berberich, T.; Kusano, T.; Liu, T. Identification of seven polyamine oxidase genes in tomato (Solanum lycopersicum L.) and their expression profiles under physiological and various stress conditions. J. Plant Physiol. 2018, 228, 1–11. [Google Scholar] [CrossRef]
- Cervelli, M.; Cona, A.; Angelini, R.; Polticelli, F.; Federico, R.; Mariottini, P. A barley polyamine oxidase isoform with distinct structural features and subcellular localization. JBIC J. Biol. Inorg. Chem. 2001, 268, 3816–3830. [Google Scholar] [CrossRef]
- Cervelli, M.; Di Caro, O.; Di Penta, A.; Angelini, R.; Federico, R.; Vitale, A.; Mariottini, P. A novel C-terminal sequence from barley polyamine oxidase is a vacuolar sorting signal. Plant J. 2004, 40, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Cervelli, M.; Bianchi, M.; Cona, A.; Crosatti, C.; Stanca, M.; Angelini, R.; Federico, R.; Mariottini, P. Barley polyamine oxidase isoforms 1 and 2, a peculiar case of gene duplication. FEBS J. 2006, 273, 3990–4002. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Kim, D.W.; Niitsu, M.; Berberich, T.; Kusano, T. Oryza sativa polyamine oxidase 1 back-converts tetraamines, spermine and thermospermine, to spermidine. Plant Cell Rep. 2013, 33, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Freitas, V.S.; Miranda, R.D.S.; Costa, J.H.; De Oliveira, D.F.; Paula, S.D.O.; Miguel, E.D.C.; Freire, R.S.; Prisco, J.T.; Gomes-Filho, E. Ethylene triggers salt tolerance in maize genotypes by modulating polyamine catabolism enzymes associated with H2O2 production. Environ. Exp. Bot. 2018, 145, 75–86. [Google Scholar] [CrossRef]
- Mellidou, I.; Karamanoli, K.; Beris, D.; Haralampidis, K.; Constantinidou, H.-I.A.; Roubelakis-Angelakis, K.A. Underexpression of apoplastic polyamine oxidase improves thermotolerance in Nicotiana tabacum. J. Plant Physiol. 2017, 218, 171–174. [Google Scholar] [CrossRef]
- Ni Tun, N.; Santa-Catarina, C.; Begum, T.; Silveira, V.; Handro, W.; Floh, E.I.S.; Scherer, G.F.E. Polyamines Induce Rapid Biosynthesis of Nitric Oxide (NO) in Arabidopsis thaliana Seedlings. Plant Cell Physiol. 2006, 47, 346–354. [Google Scholar] [CrossRef]
- An, Z.F.; Jing, W.; Liu, Y.L.; Zhang, W.H. Hydrogen peroxide generated by copper amine oxidase is involved in abscisic acid-induced stomatal closure in Vicia faba. J. Exp. Bot. 2008, 59, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Pál, M.; Szalai, G.; Janda, T. Speculation: Polyamines are important in abiotic stress signaling. Plant Sci. 2015, 237, 16–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agurla, S.; Gayatri, G.; Raghavendra, A.S. Polyamines increase nitric oxide and reactive oxygen species in guard cells of Arabidopsis thaliana during stomatal closure. Protoplasma 2018, 255, 153–162. [Google Scholar] [CrossRef]
- Mattoo, A.K.; Minocha, S.C.; Minocha, R.; Handa, A.K. Polyamines and cellular metabolism in plants: Transgenic approaches reveal different responses to diamine putrescine versus higher polyamines spermidine and spermine. Amino Acids 2009, 38, 405–413. [Google Scholar] [CrossRef]
- Wuddineh, W.; Minocha, R.; Minocha, S.C. Polyamines in the Context of Metabolic Networks. Polyam. Methods Mol. Biol. 2017, 1694, 1–23. [Google Scholar] [CrossRef]
- Docimo, T.; Reichelt, M.; Schneider, B.; Kai, M.; Kunert, G.; Gershenzon, J.; D’Auria, J.C. The first step in the biosynthesis of cocaine in Erythroxylum coca: The characterization of arginine and ornithine decarboxylases. Plant Mol. Biol. 2012, 78, 599–615. [Google Scholar] [CrossRef]
- Pegg, A.E. Functions of Polyamines in Mammals. J. Biol. Chem. 2016, 291, 14904–14912. [Google Scholar] [CrossRef]
- Han, L. Studies on Mechanism of Low-Temperature Storage and Polyamine Impact in Cut Flowers of Herbaceous Peony Postharvest Senescence; Shandong Agricultural University: Shandong, China, 2016. [Google Scholar]
- Ouyang, J.; Song, C.; Chen, D. Research progress on heat-tolerance mechanism and transports of polyamines in plant. Mol. Plant Breed. 2017, 15, 3286–3294. [Google Scholar]
- De Oliveira, L.F.; Navarro, B.V.; Cerruti, G.V.; Elbl, P.; Minocha, R.; Minocha, S.C.; Dos Santos, A.L.W.; Floh, E.I.S. Polyamine- and Amino Acid-Related Metabolism: The Roles of Arginine and Ornithine are Associated with the Embryogenic Potential. Plant Cell Physiol. 2018, 59, 1084–1098. [Google Scholar] [CrossRef]
- Chen, D.; Shao, Q.; Yin, L.; Younis, A.; Zheng, B. Polyamine function in plants: Metabolism, regulation on development, and roles in abiotic stress responses. Front. Plant Sci. 2019, 9, 1945. [Google Scholar] [CrossRef]
- Hanfrey, C.; Sommer, S.; Mayer, M.J.; Al, E. Arabidopsis polyamine biosynthesis: Absence of ornithine decarboxylase and the mechanism of arginine decarboxylase activity. Plant J. 2010, 27, 551–560. [Google Scholar] [CrossRef]
- Sindhu, R.K.; Cohen, S.S. Subcellular Localization of Spermidine Synthase in the Protoplasts of Chinese Cabbage Leaves. Plant Physiol. 1984, 76, 219–223. [Google Scholar] [CrossRef]
- Hanzawa, Y.; Takahashi, T.; Michael, A.J.; Burtin, D.; Long, D.; Piñeiro, M.; Coupland, G.; Komeda, Y. ACAULIS5, an Arabidopsis gene required for stem elongation, encodes a spermine synthase. EMBO J. 2000, 19, 4248–4256. [Google Scholar] [CrossRef] [Green Version]
- Tavladoraki, P.; Rossi, M.N.; Saccuti, G.; Perez-Amador, M.A.; Polticelli, F.; Angelini, R.; Federico, R. Heterologous Expression and Biochemical Characterization of a Polyamine Oxidase from Arabidopsis Involved in Polyamine Back Conversion. Plant Physiol. 2006, 141, 1519–1532. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Watanabe, K.; Murayama, C.; Izawa, S.; Niitsu, M.; Michael, A.J.; Berberich, T.; Kusano, T. Polyamine Oxidase5 Regulates Arabidopsis Growth through Thermospermine Oxidase Activity. Plant Physiol. 2014, 165, 1575–1590. [Google Scholar] [CrossRef] [PubMed]
- Šebela, M.; Radová, A.; Angelini, R.; Tavladoraki, P.; Frébort, I.; Peč, P. FAD-containing polyamine oxidases: A timely challenge for researchers in biochemistry and physiology of plants. Plant Sci. 2001, 160, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Moschou, P.N.; Paschalidis, K.A.; Roubelakis-Angelakis, K.A. Plant polyamine catabolism. Plant Signal. Behav. 2008, 3, 1061–1066. [Google Scholar] [CrossRef]
- Xiao, H.-J.; Liu, K.-K.; Li, D.-W.; Arisha, M.H.; Chai, W.-G.; Gong, Z.-H. Cloning and characterization of the pepper CaPAO gene for defense responses to salt-induced leaf senescence. BMC Biotechnol. 2015, 15, 100. [Google Scholar] [CrossRef]
- Wang, W.; Liu, J.-H. CsPAO4 of Citrus sinensis functions in polyamine terminal catabolism and inhibits plant growth under salt stress. Sci. Rep. 2016, 6, 31384. [Google Scholar] [CrossRef]
- Li, M.; Lu, J.; Tao, M.; Li, M.; Yang, H.; Xia, E.-H.; Chen, Q.; Wan, X. Genome-Wide Identification of Seven Polyamine Oxidase Genes in Camellia sinensis (L.) and Their Expression Patterns Under Various Abiotic Stresses. Front. Plant Sci. 2020, 11, 544933. [Google Scholar] [CrossRef]
- Clay, N.K.; Nelson, T. Arabidopsis thick vein mutation affects vein thickness and organ vascularization, and resides in a provascular cell specific spermine synthase involved in vein definition and in polar auxin transport. Plant Physiol. 2005, 138, 767–777. [Google Scholar] [CrossRef]
- Muñiz, L.; Minguet, E.G.; Singh, S.K.; Pesquet, E.; Vera-Sirera, F.; Moreau-Courtois, C.L.; Carbonell, J.; Blázquez, M.A.; Tuominen, H. ACAULIS5 controls Arabidopsis xylem specification through the prevention of premature cell death. Development 2008, 135, 2573–2582. [Google Scholar] [CrossRef]
- Vera-Sirera, F.; Minguet, E.G.; Singh, S.K.; Ljung, K.; Tuominen, H.; Blázquez, M.A.; Carbonell, J. Role of polyamines in plant vascular development. Plant Physiol. Biochem. 2010, 48, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Polticelli, F.; Basran, J.; Faso, C.; Cona, A.; Minervini, G.; Angelini, R.; Federico, R.; Scrutton, N.S.; Tavladoraki, P. Lys300 Plays a Major Role in the Catalytic Mechanism of Maize Polyamine Oxidase. Biochemistry 2005, 44, 16108–16120. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Jiang, W.; Tian, L.; Fu, Y.; Tan, L.; Zhu, Z.; Sun, C.; Liu, F. Polyamine oxidase 3 is involved in salt tolerance at the germination stage in rice. J. Genet. Genom. 2022, 49, 458–468. [Google Scholar] [CrossRef]
- Gholizadeh, F.; Mirzaghaderi, G. Genome-wide analysis of the polyamine oxidase gene family in wheat (Triticum aestivum L.) reveals involvement in temperature stress response. PLoS ONE 2020, 15, e0236226. [Google Scholar] [CrossRef]
- Xi, Y.; Hu, W.; Zhou, Y.; Liu, X.; Qian, Y. Genome-Wide Identification and Functional Analysis of Polyamine Oxidase Genes in Maize Reveal Essential Roles in Abiotic Stress Tolerance. Front. Plant Sci. 2022, 13, 950064. [Google Scholar] [CrossRef]
- Liu, J.-H.; Wang, W.; Wu, H.; Gong, X.; Moriguchi, T. Polyamines function in stress tolerance: From synthesis to regulation. Front. Plant Sci. 2015, 6, 827. [Google Scholar] [CrossRef]
- Zarza, X.; Atanasov, K.E.; Marco, F.; Arbona, V.; Carrasco, P.; Kopka, J.; Fotopoulos, V.; Munnik, T.; Gómez-Cadenas, A.; Tiburcio, A.F.; et al. Polyamine oxidase 5 loss-of-function mutations in Arabidopsis thaliana trigger metabolic and transcriptional reprogramming and promote salt stress tolerance. Plant, Cell Environ. 2016, 40, 527–542. [Google Scholar] [CrossRef] [PubMed]
- Sagor, G.H.M.; Zhang, S.; Kojima, S.; Simm, S.; Berberich, T.; Kusano, T. Reducing Cytoplasmic Polyamine Oxidase Activity in Arabidopsis Increases Salt and Drought Tolerance by Reducing Reactive Oxygen Species Production and Increasing Defense Gene Expression. Front. Plant Sci. 2016, 7, 214. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.S.; Guo, Y.; Quintero, F.J.; Pardo, J.M.; Schumaker, K.S.; Zhu, J.K. Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the Salt-Overly-Sensitive (SOS) pathway. J. Biol. Chem. 2004, 279, 207–215. [Google Scholar] [CrossRef]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liang, L.; Xiao, J.; Xie, Y.; Zhu, L.; Xue, X.; Xu, L.; Zhou, P.; Ran, J.; Huang, Z.; et al. Genome-Wide Identification of Polyamine Oxidase (PAO) Family Genes: Roles of CaPAO2 and CaPAO4 in the Cold Tolerance of Pepper (Capsicum annuum L.). Int. J. Mol. Sci. 2022, 23, 9999. [Google Scholar] [CrossRef] [PubMed]
- Sagor, G.H.M.; Inoue, M.; Kusano, T.; Berberich, T. Expression profile of seven polyamine oxidase genes in rice (Oryza sativa) in response to abiotic stresses, phytohormones and polyamines. Physiol. Mol. Biol. Plants 2021, 27, 1353–1359. [Google Scholar] [CrossRef] [PubMed]
- Tanwar, U.K.; Stolarska, E.; Paluch-Lubawa, E.; Mattoo, A.K.; Arasimowicz-Jelonek, M.; Sobieszczuk-Nowicka, E. Unraveling the genetics of polyamine metabolism in barley for senescence-related crop improvement. Int. J. Biol. Macromol. 2022, 221, 585–603. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Takahashi, Y.; Berberich, T.; Imai, A.; Miyazaki, A.; Takahashi, T.; Michael, A.; Kusano, T. The polyamine spermine protects against high salt stress in Arabidopsis thaliana. FEBS Lett. 2006, 580, 6783–6788. [Google Scholar] [CrossRef]
- Pottosin, I.; Shabala, S. Polyamines control of cation transport across plant membranes: Implications for ion homeostasis and abiotic stress signaling. Front. Plant Sci. 2014, 5, 154. [Google Scholar] [CrossRef]
- Pottosin, I.; Velarde-Buendía, A.M.; Bose, J.; Fuglsang, A.T.; Shabala, S. Polyamines cause plasma membrane depolarization, activate Ca-, and modulate H-ATPase pump activity in pea roots. J. Exp. Bot. 2014, 65, 2463–2472. [Google Scholar] [CrossRef]
- Saha, J.; Brauer, E.K.; Sengupta, A.; Popescu, S.C.; Gupta, K.; Gupta, B. Polyamines as redox homeostasis regulators during salt stress in plants. Front. Environ. Sci. 2015, 3, 21. [Google Scholar] [CrossRef]
- Binda, C.; Coda, A.; Angelini, R.; Federico, R.; Ascenzi, P.; Mattevi, A. A 30 Å long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase. Structure 1999, 7, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Kamada-Nobusada, T.; Hayashi, M.; Fukazawa, M.; Sakakibara, H.; Nishimura, M. A Putative Peroxisomal Polyamine Oxidase, AtPAO4, is Involved in Polyamine Catabolism in Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 1272–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neuberger, G.; Maurer-Stroh, S.; Eisenhaber, B.; Hartig, A.; Eisenhaber, F. Prediction of Peroxisomal Targeting Signal 1 Containing Proteins from Amino Acid Sequence. J. Mol. Biol. 2003, 328, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Brocard, C.; Hartig, A. Peroxisome targeting signal 1: Is it really a simple tripeptide? Biochim. Biophys. Acta Mol. Cell Res. 2006, 1763, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Lingner, T.; Kataya, A.R.; Antonicelli, G.E.; Benichou, A.; Nilssen, K.; Chen, X.-Y.; Siemsen, T.; Morgenstern, B.; Meinicke, P.; Reumann, S. Identification of Novel Plant Peroxisomal Targeting Signals by a Combination of Machine Learning Methods and in Vivo Subcellular Targeting Analyses. Plant Cell 2011, 23, 1556–1572. [Google Scholar] [CrossRef]
- Lametschwandtner, G.; Brocard, C.; Fransen, M.; Van Veldhoven, P.; Berger, J.; Hartig, A. The Difference in Recognition of Terminal Tripeptides as Peroxisomal Targeting Signal 1 between Yeast and Human Is Due to Different Affinities of Their Receptor Pex5p to the Cognate Signal and to Residues Adjacent to It. J. Biol. Chem. 1998, 273, 33635–33643. [Google Scholar] [CrossRef] [PubMed]
- Reumann, S.; Chowdhary, G.; Lingner, T. Characterization, prediction and evolution of plant peroxisomal targeting signals type 1 (PTS1s). Biochim. Biophys. Acta BBA Mol. Cell Res. 2016, 1863, 790–803. [Google Scholar] [CrossRef]
- Fodor, K.; Wolf, J.; Erdmann, R.; Schliebs, W.; Wilmanns, M. Molecular Requirements for Peroxisomal Targeting of Alanine-Glyoxylate Aminotransferase as an Essential Determinant in Primary Hyperoxaluria Type 1. PLoS Biol. 2012, 10, e1001309. [Google Scholar] [CrossRef]
- Sacksteder, K.A.; Gould, S.J. The Genetics of Peroxisome Biogenesis. Annu. Rev. Genet. 2000, 34, 623–652. [Google Scholar] [CrossRef]
- Fodor, K.; Wolf, J.; Reglinski, K.; Passon, D.M.; Lou, Y.; Schliebs, W.; Erdmann, R.; Wilmanns, M. Ligand-Induced Compaction of the PEX5 Receptor-Binding Cavity Impacts Protein Import Efficiency into Peroxisomes. Traffic 2014, 16, 85–98. [Google Scholar] [CrossRef]
- Skoulding, N.S.; Chowdhary, G.; Deus, M.J.; Baker, A.; Reumann, S.; Warriner, S.L. Experimental Validation of Plant Peroxisomal Targeting Prediction Algorithms by Systematic Comparison of In Vivo Import Efficiency and In Vitro PTS1 Binding Affinity. J. Mol. Biol. 2015, 427, 1085–1101. [Google Scholar] [CrossRef] [PubMed]
- Glover, J.R.; Andrews, D.W.; Subramani, S.; Rachubinski, R.A. Mutagenesis of the amino targeting signal of Saccharomyces cerevisiae 3- ketoacyl-CoA thiolase reveals conserved amino acids required for import into peroxisomes in vivo. J. Biol. Chem. 1994, 269, 7558–7563. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Mullen, R.T.; Trelease, R.N. Oilseed isocitrate lyases lacking their essential type 1 peroxisomal targeting signal are piggybacked to glyoxysomes. Plant Cell 1997, 9, 185–197. [Google Scholar]
- Kataya, A.R.; Heidari, B.; Hagen, L.; Kommedal, R.; Slupphaug, G.; Lillo, C. Protein Phosphatase 2A Holoenzyme Is Targeted to Peroxisomes by Piggybacking and Positively Affects Peroxisomal β-Oxidation. Plant Physiol. 2014, 167, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Purdue, P.E.; Lazarow, P.B. Eci1p uses a PTS1 to enter peroxisomes: Either its own or that of a partner, Dci1p. Eur. J. Cell Biol. 2001, 80, 126–138. [Google Scholar] [CrossRef]
- Gabay-Maskit, S.; Cruz-Zaragoza, L.D.; Shai, N.; Eisenstein, M.; Bibi, C.; Cohen, N.; Hansen, T.; Yifrach, E.; Harpaz, N.; Belostotsky, R.; et al. A piggybacking mechanism enables peroxisomal localization of the glyoxylate cycle enzyme Mdh2 in yeast. J. Cell Sci. 2020, 133, jcs244376. [Google Scholar] [CrossRef]
- Scandalios, J.G. Oxygen Stress and Superoxide Dismutases. Plant Physiol. 1993, 101, 7–12. [Google Scholar] [CrossRef]
- Noctor, G.; Foyer, C.H. Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef]
- Langebartels, C.; Kerner, K.; Leonardi, S.; Schraudner, M.; Trost, M.; Heller, W. Biochemical-plant responses to ozone: 1. differential induction of polyamine and ethylene biosynthesis in tobacco. Plant Physiol. 1991, 95, 882–889. [Google Scholar] [CrossRef]
- Kubis, J. The effect of exogenous spermidine on superoxide dismutase activity, H2O2 and superoxide radical level in barley leaves under water deficit conditions. Acta Physiol. Plant. 2005, 27, 289–295. [Google Scholar] [CrossRef]
- Kuznetsov, V.V.; Shevyakova, N.I. Polyamines and stress tolerance of plants. Plant Stress 2007, 1, 50–71. [Google Scholar]
- Bordenave, C.D.; Mendoza, C.G.; Bremont, J.F.J.; Gárriz, A.; Rodríguez, A. Defining novel plant polyamine oxidase subfamilies through molecular modeling and sequence analysis. BMC Evol. Biol. 2019, 19, 28. [Google Scholar] [CrossRef] [PubMed]
- Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Byng, J.W.; Judd, W.S.; Soltis, D.E.; Mabberley, D.J.; Sennikov, A.N.; Soltis, P.S.; Stevens, P.F. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. 2016, 181, 1–20. [Google Scholar]
- Salvi, D.; Tavladoraki, P. The tree of life of polyamine oxidases. Sci. Rep. 2020, 10, 17858. [Google Scholar] [CrossRef] [PubMed]

| Sl. No. | The Scientific Name of the Plant | Common Name of the Plant | Accession No. of the Sequence | Family of Plant | C-Ter Tripeptide |
|---|---|---|---|---|---|
| Dicotyledons | |||||
| 1 | Amborella trichopoda | Amborella | >XP_006836334.1 | Amborellaceae pichon | SRM |
| 2 | Actinidia chinensis | Golden kiwifruit | >PSS16430.1 | Actinidiaceae | SRM |
| 3 | Artemisia annua | Sweet wormwood | >PWA74707.1 >PWA66356.1 | Asteraceae | SRF SRM |
| 4 | Cynara cardunculus | Cardoon | >XP_024988081.1 >KVH97550.1 | Asteraceae | SRI |
| 5 | Lactuca sativa | Lettuce | >XP_023736501.1 | Asteraceae | SRI |
| 6 | Helianthus annus | Common Sunflower | >XP_021986264.1 | Asteraceae | SRI |
| 7 | Beta vulgaris | Beet | >XP_010688837.1 | Amaranthaceae | SRM |
| 8 | Chenopodium quinoa | Quinoa | >XP_021724622.1 | Amaranthaceae | SRM |
| 9 | Spinacia oleracea | Spinach | >XP_021845331.1 | Amaranthaceae | SRM |
| 10 | Daucus corota | Queen Anne’s lace | >XP_017237286.1 | Apiaceae | SRI |
| 11 | Arabis alpine | Alpine rock cress | >KFK37143.1 >KFK35146.1 | Brassicaceae | SRL SRI |
| 12 | Arabidopsis thaliana | Mouse ear cress | >At2g43020 >NP_191464.1 | Brassicaceae | SRL SRM |
| 13 | Arabidopsis lyrata subsp.lyrata | Lyre-leaved rock cress | >XP_020884243.1 >XP_020880523.1 | Brassicaceae | SRL SRM |
| 14 | Brassica rapa | Field mustard | >XP_009133564.1 >XP_009104181.1 | Brassicaceae | SRL SRM |
| 15 | Brassica napus | Rapeseed | >XP_013687858.1 >XP_022560874.1 | Brassicaceae | SRL SRM |
| 16 | Brassica oleracea var.oleracea | Wild cabbage | >XP_013630886.1 >XP_013588398.1 | Brassicaceae | SRL SRM |
| 17 | Capsella rubella | Pink shepherd’s purse | >XP_006294097.1 | Brassicaceae | SRL |
| 18 | Camelina sativa | False flax | >XP_019101215.1 >XP_010516573.1 | Brassicaceae | SRL SRM |
| 19 | Eutrema salsugineum | Saltwater cress | >ESQ44181.1 | Brassicaceae | SRM |
| 20 | Raphanus sativus | Radish | >XP_018485151.1 >XP_018460415.1 | Brassicaceae | SRL SRM |
| 21 | Handroanthus impetiginosus | Pink Tumpet Tree | >PIN20378.1 | Bignoniaceae | SRM |
| 22 | Cucurbita moschata | Crookneck pumpkin | >XP_022936044.1 >XP_022931432.1 | Cucurbitaceae | SRM SRL |
| 23 | Cucurbita maxima | Winter Squash | >XP_022976762.1 >XP_022984772.1 | Cucurbitaceae | SRL SRM |
| 24 | Cucumis melo | Muskmelon | >XP_008464648.1 >XP_008451845.1 | Cucurbitaceae | SRL SRM |
| 25 | Cucurbita pepo | Field pumpkin | >XP_023535748.1 >XP_023553553.1 | Cucurbitaceae | SRL SRM |
| 26 | Momordica charantia | Bitter Squash | >ALO20334.1 | Cucurbitaceae | SPL |
| 27 | Ipomea nil | Blue Morning glory | >XP_019193306.1 | Convolvulaceae | SRM |
| 28 | Tarenaya hassleriana | Spider flower | >XP_010525644.1 >XP_010550699.1 | Cleomaceae | SRL SRM |
| 29 | Carica papaya | Papaya | >XP_021898383.1 | Caricaceae | SRM |
| 30 | Cephalotus follicularis | Western Australian Pitcher plant | >GAV59997.1 | Cephalotaceae | SRM |
| 31 | Hevea brasiliensis | Pará rubber tree | >XP_021665846.1 | Euphorbiaceae | SRM |
| 32 | Jatropha curcas | Physic nut | >XP_012072709.1 | Euphorbiaceae | SRM |
| 33 | Manihot esculenta | Cassava | >XP_021603628.1 | Euphorbiaceae | SRM |
| 34 | Ricinus communis | Castor bean | >XP_002521588.1 | Euphorbiaceae | SRM |
| 35 | Arachis duranensis | Wild herb | >XP_015973279.1 | Fabaceae | SRL |
| 36 | Arachis hypogea | Peanut | >XP_025669047.1 | Fabaceae | SRL |
| 37 | Cajanus cajan | Pigeon pea | >XP_020204978.1 >XP_020210206.1 | Fabaceae | SRM SRL |
| 38 | Cicer arietinum | Chickpea | >XP_004491274.1 >XP_004499541.1 | Fabaceae | SRF SRM |
| 39 | Glycine max | Soybean | >XP_003551948.1 | Fabaceae | SRL |
| 40 | Glycine soja | Wild Soybean | >KHN12003.1 | Fabaceae | SRL |
| 41 | Lupinus augustifolius | Blue Lupin, Narrowleaved Lupin | >XP_019455951.1 | Fabaceae | SRL |
| 42 | Mucuna pruriens | Velvet beans | >RDX68841.1 | Fabaceae | SRL |
| 43 | Phaseolus vulgaris | Common bean | >XP_007146297.1 >XP_007141453.1 | Fabaceae | SRM SRL |
| 44 | Medicago trancatula | Strong-spined medlick | >XP_003617318.1 >XP_013459605.1 | Fabaceae | SRI SRM |
| 45 | Trifolium subterraneum | Subterranean clover | >GAU12612.1 >GAU22182.1 | Fabaceae | SRM SRI |
| 46 | Trifolium pratense | Red clover | >PNY04428.1 | Fabaceae | SRI |
| 47 | Trifolium repens | White clover | >AQQ81875.1 | Fabaceae | SRI |
| 48 | Vigna angularis | Adzuki bean | >XP_017436636.1 >XP_017430881.1 | Fabaceae | SRM SRL |
| 49 | Vigna radiata | Mung bean | >XP_014490314.1 >XP_014505168.1 | Fabaceae | SRM SRL |
| 50 | Quercus suber | Cork oak | >RLW29351.1 | Fagaceae | SRM |
| 51 | Dorcoceras hygrometricum | --- | >KZV25408.1 | Gesneriaceae | SRI |
| 52 | Juglans regia | Common Walnut | >XP_018824097.1 | Juglandaceae | SRM |
| 53 | Genlisea aurea | Corkscrew Plant | >EPS67202.1 | Lentibulariceae | SRM |
| 54 | Punica granatum | Pomegranate | >OWM73258.1 | Lythraceae | SRL |
| 55 | Corchorus olitorius | Jute mallow | >OMO68085.1 | Malvaceae | SRM |
| 56 | Corchorus capsularis | White jute | >OMO49622.1 | Malvaceae | SRM |
| 57 | Durio zibethinus | Duria | >XP_022739756.1 | Malvaceae | SRL |
| 58 | Gossypium Raimondi | Cotton Plant Species | >XP_012437381.1 | Malvaceae | TRL |
| 59 | Gossypium arboretum | Tree cotton | >XP_017642334.1 | Malvaceae | TRL |
| 60 | Gossypium hirsutum | Upland Cotton, Mexican Cotton | >XP_016712212.1 | Malvaceae | TRL |
| 61 | Herrania umbratica | Monkey Cacao | >XP_021274234.1 | Malvaceae | SRM |
| 62 | Theobroma cacao | Cacao tree | >XP_007048902.2 | Malvaceae | SRM |
| 63 | Eucalyptus grandis | Flooeded gum, rose gum | >XP_010054154.1 | Myrtaceae | SRM |
| 64 | Morus notabilis | Mulberry Tree | >XP_024027830.1 | Moraceae | SRM |
| 65 | Nelumbo nucifera | Water Lily | >XP_010244717.1 >XP_010275888.1 | Nelumbonaceae | SRM SRL |
| 66 | Olea europea var. sylvestris | Olive | >XP_022871877.1 | Oleaceae | SRM |
| 67 | Sesamum indicum | Sesame | >XP_011085441.1 | Pedaliaceae | SRM |
| 68 | Papaver somniferum | Opium poppy | >XP_026393108.1 >XP_026391831.1 | Papaveraceae | SRL SRM |
| 69 | Macleaya cordata | Five-Seeded Plume Poppy | >OVA19352.1 | Papaveraceae | SRL |
| 70 | Erythranthe guttata | Seep monkeyflower | >XP_012830906.1 | Phrymaceae | SRM |
| 71 | Aquilegia caerulea | Colorado blue columbine | >PIA45277.1 >PIA30210.1 | Ranunculaceae | SRV SRL |
| 72 | Citrus sinensis | Sweet orange | >XP_006485009.1 | Rutaceae | SRL |
| 73 | Citrus trifoliata | Trifoliate orange | >AJP16790.1 | Rutaceae | SRL |
| 74 | Ziziphus jujuba | Jujube | >XP_015880626.1 | Rhamnaceae | SRL |
| 75 | Coffea canephora | Robusta coffee | >CDP16058.1 | Rubiaceae | SRM |
| 76 | Fragaria vesca | Wild Strawberry | >XP_004303904.1 | Rosaceae | SRL |
| 77 | Malus domestica | Apple | >ANJ77639.1 | Rosaceae | SRL |
| 78 | Prunus yedoensis | Yoshino cherry | >XP_011032740.1 | Rosaceae | IPL |
| 79 | Prunus persica | Peach | >XP_007215363.2 | Rosaceae | SRI |
| 80 | Prunus avium | Sweet cherry | >XP_021824861.1 | Rosaceae | SRI |
| 81 | Rosa chinensis | China rose | >XP_024180697.1 | Rosaceae | SRL |
| 82 | Prunus trichocarpa | Black cottonwood | >PNT47987.1 >XP_002306765.2 | Salicaceae | SRM SRI |
| 83 | Populus euphratica | Desert poplar | >XP_011032740.1 | Salicaceae | SRM |
| 84 | Capsicum annuum | Sweet and chili pepper | >XP_016541238.1 | Solanaceae | SRM |
| 85 | Capsicum baccatum | Pepper | >PHT42735.1 | Solanaceae | SRM |
| 86 | Capsicum chinense | Habanero type pepper | >PHU11700.1 | Solanaceae | SRM |
| 87 | Citrus clementina | Clementine | >XP_006437065.1 | SRL | |
| 88 | Nicotiana sylvestris | Flowering Tobacco | >XP_009777198.1 >XP_009757614.1 | Solanaceae | SRL SRM |
| 89 | Nicotiana attenuata | Coyote Tobacco | >XP_019249249.1 >XP_019262812.1 | Solanaceae | SRM SRL |
| 90 | Nicotiana tomentosiformis | Tobacco (wild species) | >XP_009588592.1 >XP_009602218.1 | Solanaceae | SRL SRM |
| 91 | Nicotiana tabacum | Common Tobacco | >XP_016451254.1 >XP_016478455.1 | Solanaceae | SRL SRM |
| 92 | Solanum tuberosum | Potato | >XP_006357889.1 | Solanaceae | SRM |
| 93 | Solanum pennelli | Wild tomato | >XP_015082560.1 | Solanaceae | SRM |
| 94 | Solanum lycopersicum | Tomato | >XP_004243630.1 | Solanaceae | SRM |
| 95 | Populus trichocarpa | >XP_002306765.2 | Salicaceae | SRI | |
| 96 | Camelia sinensis | Tea | >QPO25410.1 >QPO25411.1 | Theaceae | SRM SRI |
| 97 | Vitis vinifera | Common grape vine | >XP_002282970.1 | Vitaceae | SRM |
| Monocotyledons | |||||
| 98 | Asparagus officinalis | Sparrow grass | >XP_020270229.1 | Asparagaceae | SRM |
| 99 | Elaeis guineensis | African Oil Palm | >XP_010909649.1 | Arecaceae | SRM |
| 100 | Phoenix dactylifera | Date palm | >XP_008787574.1 | Arecaceae | SRM |
| 101 | Ananas comosus | Pineapple | >XP_020102904.1 | Bromeliaceae | SRI |
| 102 | Musa acuminate | Cavendish Banana | >XP_009399229.1 | Musaceae | SRM |
| 103 | Dendrobium catenatum | The chained dendrobium | >XP_020682153.1 | Orchidaceae | SRI |
| 104 | Phalaenopsis equestris | Moth orchids | XP_020579254.1 | Orchidaceae | SRI |
| 105 | Oryza sativa | Asian Rice | >BAM17621.1 | Poaceae | SRL |
| 106 | Oryza brachyantha | African Rice | >XP_006652847.1 | Poaceae | SRL |
| 107 | Oryza meyeriana | South-Asian Wild Rice | >KAF0892522.1 | Poaceae | SRL |
| 108 | Triticum aestivum | Bread Wheat | >SPT20037.1 | Poaceae | SRL |
| 109 | Aegilops tauschii | Rough-spike hard grass | >XP_020174159.1 | Poaceae | SRL |
| 110 | Hordeum vulgare | Barley | >KAE8785488.1 >KAE8772463.1 | Poaceae | SRL CRT |
| 111 | Brachypodium distachyon | Purple false brome | >XP_010240449.1 | Poaceae | SRL |
| 112 | Panicum hallii | Hall’s panicgrass | >PUZ48969.1 | Poaceae | SRL |
| 113 | Panicum miliaceum | Proso millet | >RLM75211.1 | Poaceae | SRL |
| 114 | Zea mays | Corn | >XP_020400822.1 | Poaceae | SRL |
| 115 | Setaria italic | Foxtail millet | >XP_004976853.1 | Poaceae | SRL |
| 116 | Eragrostis curvula | Lovegrass | >TVU14770.1 | Poaceae | SRL |
| 117 | Sorghum bicolor | Great millet | >XP_002448555.1 | Poaceae | SRL |
| 118 | Dichanthelium oligosanthes | Heller’s rosette grass | >OEL27565.1 | Poaceae | SRL |
| Lower plants | |||||
| 119 | Selaginella moellendorffi | Spikemoss | >XP_002965599.1 | Pteridophytes | SRL |
| 120 | Physcomitrella patens | Spreading earthmoss | >XP_001756864.1 | Bryophytes | SRM |
| 121 | Volvox carteri | --- | >XP_002954733.1 | Green Algae | SKL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samanta, I.; Roy, P.C.; Das, E.; Mishra, S.; Chowdhary, G. Plant Peroxisomal Polyamine Oxidase: A Ubiquitous Enzyme Involved in Abiotic Stress Tolerance. Plants 2023, 12, 652. https://doi.org/10.3390/plants12030652
Samanta I, Roy PC, Das E, Mishra S, Chowdhary G. Plant Peroxisomal Polyamine Oxidase: A Ubiquitous Enzyme Involved in Abiotic Stress Tolerance. Plants. 2023; 12(3):652. https://doi.org/10.3390/plants12030652
Chicago/Turabian StyleSamanta, Ishita, Pamela Chanda Roy, Eshani Das, Sasmita Mishra, and Gopal Chowdhary. 2023. "Plant Peroxisomal Polyamine Oxidase: A Ubiquitous Enzyme Involved in Abiotic Stress Tolerance" Plants 12, no. 3: 652. https://doi.org/10.3390/plants12030652





