Characterization of Leaf Rust Resistance in International Barley Germplasm Using Genome-Wide Association Studies
Abstract
:1. Introduction
2. Results
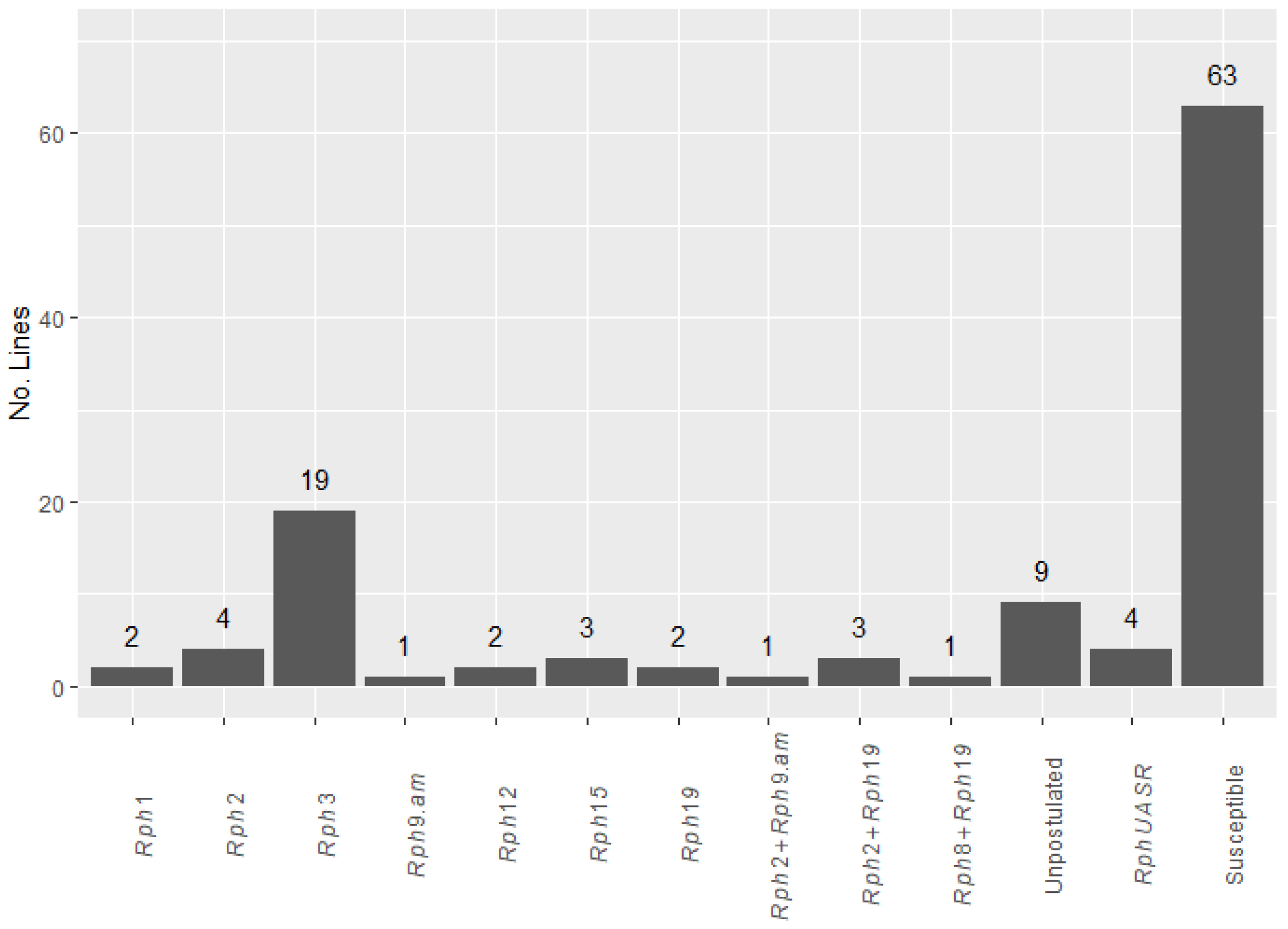
2.1. Greenhouse Seedling Tests
2.2. Variation in Field Disease Response
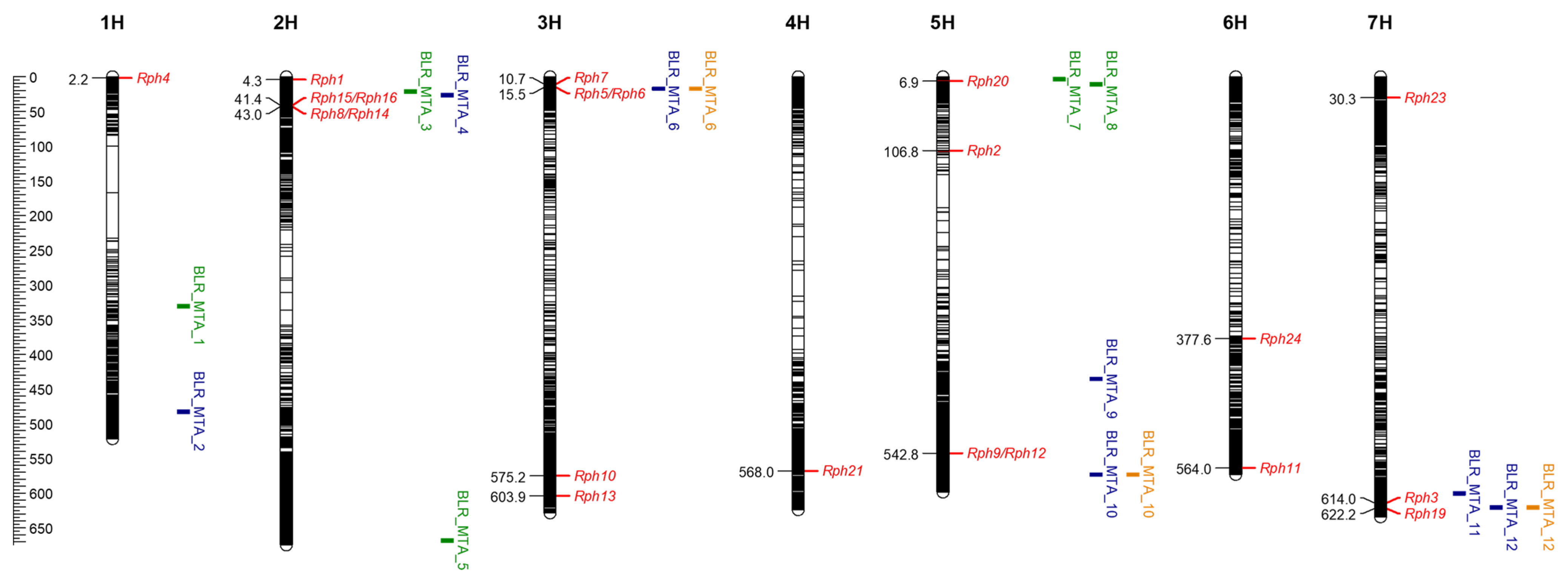
2.3. Assessment of APR in the Field and Marker Analysis
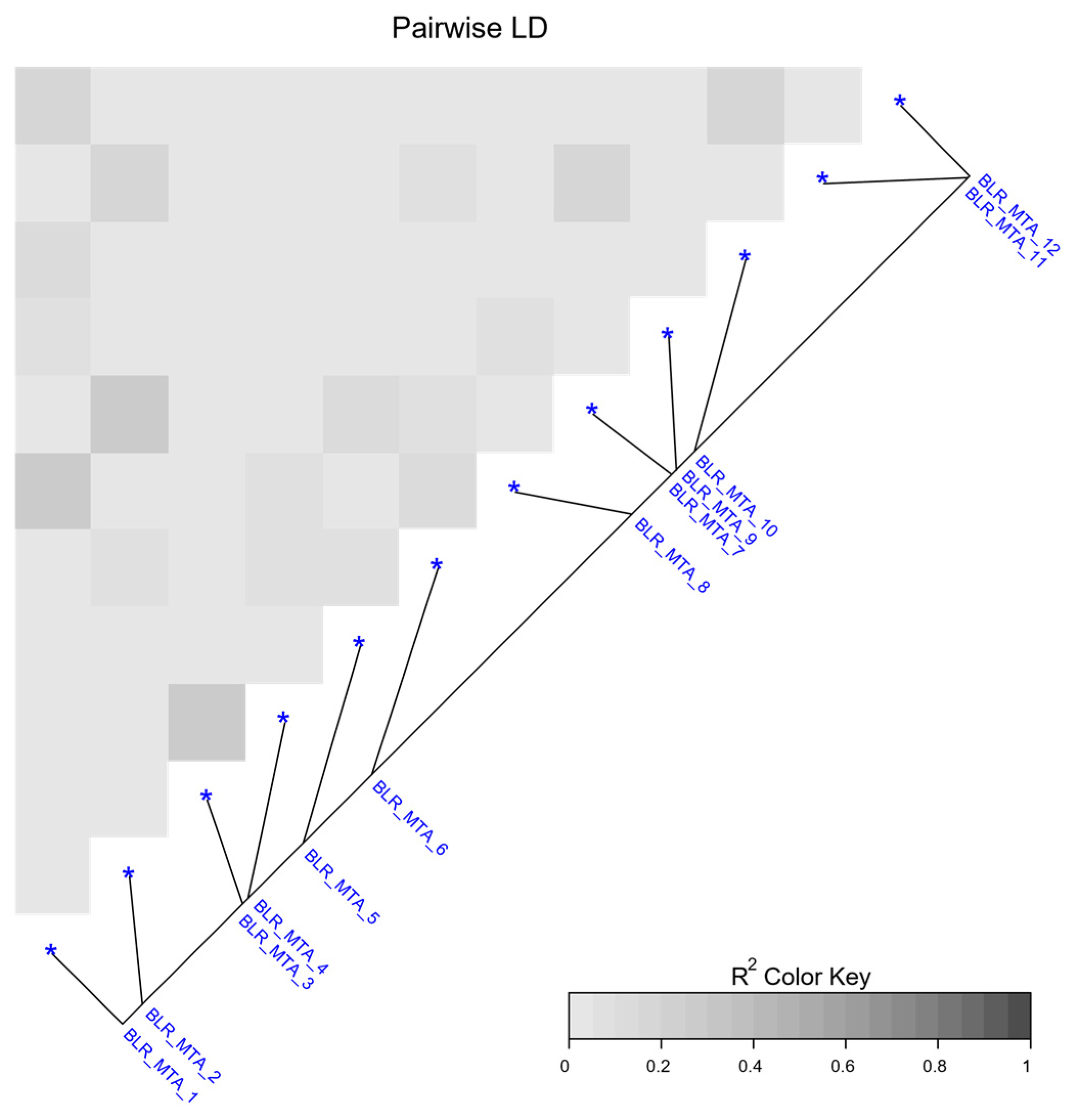
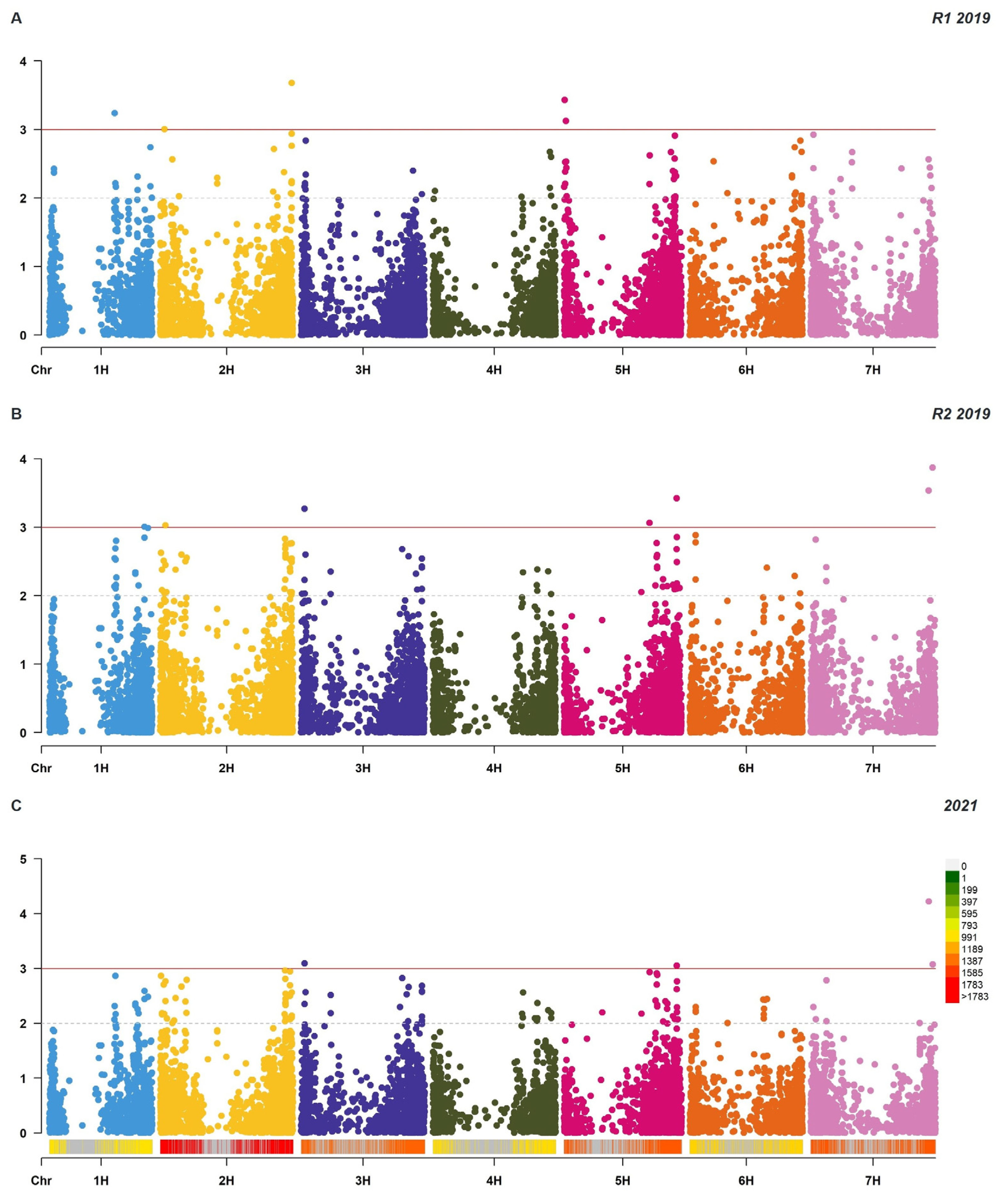
2.4. GWAS of Leaf Rust Resistance at Adult Growth Stages
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Pathogen Material
4.3. Greenhouse Screening
4.3.1. Sowing and Plant Maintenance
4.3.2. Inoculation and Disease Assessment
4.4. Field Screening
4.5. Molecular Marker Analysis for ASR and APR
4.5.1. DNA Extraction
4.5.2. Molecular Marker Screening
4.6. Phenotypic and Genotypic Data for GWAS
Filtering for Quality Markers and Genotypes
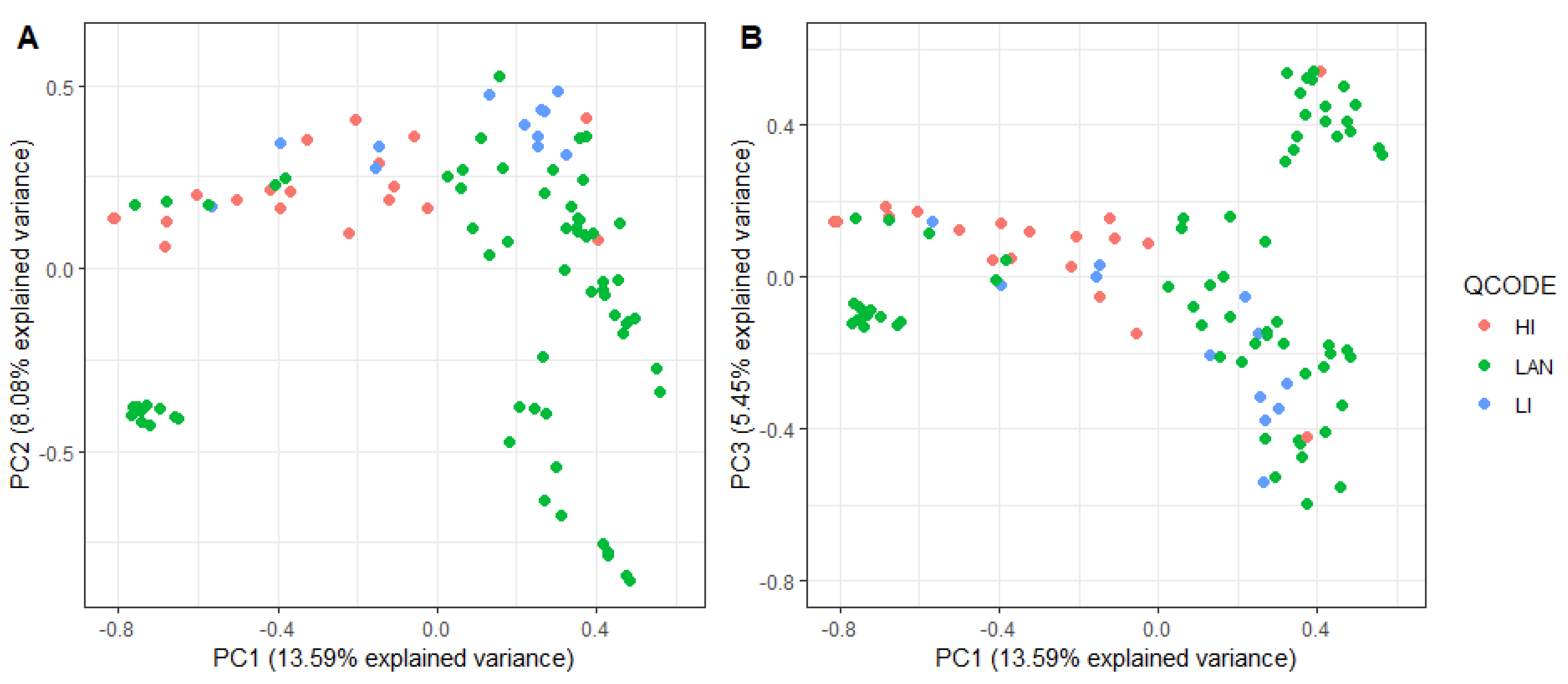
4.7. Population Structure Analysis Using PCA
4.8. Genome-Wide Association Studies of Leaf Rust Resistance
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ullrich, S.E. Barley: Production, Improvement and Uses; Wiley-Blackwell: Chichester, UK, 2011. [Google Scholar]
- Schulte, D.; Close, T.J.; Graner, A.; Langridge, P.; Matsumoto, T.; Muehlbauer, G.; Sato, K.; Schulman, A.H.; Waugh, R.; Wise, R.; et al. The international barley sequencing consortium—At the threshold of efficient access to the barley genome. Plant Physiol. 2009, 149, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.M.; Brennan, J.P. Estimating disease losses to the Australian barley industry. Australas. Plant Pathol. 2010, 39, 85–96. [Google Scholar] [CrossRef]
- Clifford, B.C. Barley leaf rust. In The Cereal Rusts Volume II: Diseases, Distribution and Control; Roelfs, A.P., Bushnell, W.R., Eds.; Academic Press: New York, NY, USA, 1985; pp. 173–205. [Google Scholar]
- Park, R.F.; Golegaonkar, P.G.; Derevnina, L.; Sandhu, K.S.; Karaoglu, H.; Elmansour, H.M.; Dracatos, P.M.; Singh, D. Leaf rust of cultivated barley: Pathology and Control. Annu. Rev. Phytopathol. 2015, 53, 565–589. [Google Scholar] [CrossRef] [PubMed]
- Park, R.F.; Karakousis, A. Characterization and mapping of gene Rph19 conferring resistance to Puccinia hordei in the cultivar ‘Reka 1’ and several Australian barleys. Plant Breed. 2002, 121, 232–236. [Google Scholar] [CrossRef]
- Cotterill, P.J.; Rees, R.G.; Platz, G.J.; Dillmacky, R. Effects of leaf rust on selected Australian barleys. Aust. J. Exp. Agric. 1992, 32, 747–751. [Google Scholar] [CrossRef]
- Griffey, C.A.; Das, M.K.; Baldwin, R.E.; Waldenmaier, C.M. Yield losses in winter barley resulting from a new race of Puccinia hordei in North America. Plant Dis. 1994, 78, 256–260. [Google Scholar] [CrossRef]
- Park, R.F. Pathogenic specialisation and pathotype distribution of Puccinia hordei Otth. in Australia, 1992–2001. Plant Disease 2003, 87, 1311–1316. [Google Scholar] [CrossRef] [PubMed]
- Park, R.F. Breeding cereals for rust resistance in Australia. Plant Pathol. 2008, 57, 591–602. [Google Scholar] [CrossRef]
- Singh, D.; Dracatos, P.; Derevnina, L.; Zhou, M.; Park, R.F. Rph23: A new designated additive adult plant resistance gene to leaf rust in barley on chromosome 7H. Plant Breed. 2014, 134, 62–69. [Google Scholar] [CrossRef]
- Derevnina, L.; Singh, D.; Park, R.F. Identification and characterization of seedling and adult plant resistance to Puccinia hordei in Chinese barley germplasm. Plant Breed. 2013, 132, 571–579. [Google Scholar] [CrossRef]
- Mehnaz, M.; Dracatos, P.M.; Park, R.F.; Singh, D. Mining Middle Eastern and Central Asian Barley Germplasm to Understand Diversity for Resistance to Puccinia hordei, Causal Agent of Leaf Rust. Agronomy 2021, 11, 2146. [Google Scholar] [CrossRef]
- Singh, D.; Ziems, L.A.; Dracatos, P.M.; Pourkheirandish, M.; Tshewang, S.; Czembor, P.; German, S.; Fowler, R.A.; Snyman, L.; Platz, G.J.; et al. Genome-wide association studies provide insights on genetic architecture of resistance to leaf rust in a worldwide barley collection. Mol. Breed. 2018, 38, 43. [Google Scholar] [CrossRef]
- Hickey, L.T.; Lawson, W.; Platz, G.J.; Dieters, M.; Arief, V.N.; German, S.; Fletcher, S.; Park, R.F.; Singh, D.; Pereyra, S.; et al. Mapping Rph20: A gene conferring adult plant resistance to Puccinia hordei in barley. Theor. Appl. Genet. 2011, 123, 55–68. [Google Scholar] [CrossRef]
- Ziems, L.A.; Hickey, L.T.; Platz, G.J.; Franckowiak, J.D.; Dracatos, P.M.; Singh, D.; Park, R.F. Characterisation of Rph24: A gene conferring adult plant resistance to Puccinia hordei in barley. Phytopathology 2017, 107, 834–841. [Google Scholar] [CrossRef]
- Pathan, A.K.; Park, R.F. Evaluation of seedling and adult plant resistance to leaf rust in European wheat cultivars. Euphytica 2006, 149, 327–342. [Google Scholar] [CrossRef]
- Golegaonkar, P.G.; Singh, D.; Park, R.F. Evaluation of seedling and adult plant resistance to Puccinia hordei in barley. Euphytica 2008, 166, 183–197. [Google Scholar] [CrossRef]
- Dracatos, P.M.; Park, R.F.; Singh, D. Validating molecular markers for barley leaf rust resistance genes Rph20 and Rph24. Plant Dis. 2021, 105, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Dracatos, P.M.; (The University of Sydney, School of Life and Environmental Sciences, Faculty of Science, Sydney, Australia). Makers Linked to Rph7. Personal communication, 2020. [Google Scholar]
- Jost, M.; Singh, D.; Evans, L.; Park, R.F.; Dracatos, P. Fine mapping of leaf rust resistance gene Rph13 from wild barley. Theor. Appl. Genet. 2020, 133, 1887–1895. [Google Scholar] [CrossRef] [PubMed]
- Dinh, H.X.; Park, R.F.; Singh, D.; Pourkheirandish, M. Fine mapping and identification of leaf rust resistance gene, Rph3 in barley. In Proceedings of the Plant and Animal Conference XXVII, San Diego, CA, USA, 12–16 January 2019. [Google Scholar]
- Dracatos, P.M.; (The University of Sydney, School of Life and Environmental Sciences, Faculty of Science, Sydney, Australia). Makers Linked to Rph15. Personal communication, 2020. [Google Scholar]
- Prins, R.; Dreisigacker, S.; Pretorius, Z.; van Schalkwyk, H.; Wessels, E.; Smit, C.; Bender, C.; Singh, D.; Boyd, L.A. Stem rust resistance in a geographically diverse collection of spring wheat lines collected from across Africa. Front. Plant Sci. 2016, 7, 973. [Google Scholar] [CrossRef]
- Ziems, L.A.; Hickey, L.T.; Hunt, C.H.; Mace, E.S.; Platz, G.J.; Franckowiak, J.D.; Jordan, D.R. Association mapping of resistance to Puccinia hordei in Australian barley breeding germplasm. Theor. Appl. Genet. 2014, 127, 1199–1212. [Google Scholar] [CrossRef]
- Turuspekov, Y.; Ormanbekova, D.; Rsaliev, A.; Abugalieva, S. Genome-wide association study on stem rust resistance in Ka-zakh spring barley lines. BMC Plant Biol. 2016, 16, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, L.; Germán, S.; Pereyra, S.; Hayes, P.M.; Pérez, C.A.; Capettini, F.; Locatelli, A.; Berberian, N.M.; Falconi, E.E.; Estrada, R.; et al. Multi-environment multi-QTL association mapping identifies disease resistance QTL in barley germplasm from Latin America. Theor. Appl. Genet. 2014, 128, 501–516. [Google Scholar] [CrossRef]
- Park, R.F.; McIntosh, R.A. Adult plant resistances to Puccinia recondita f. sp. tritici in wheat. N. Z. J. Crop Hort. Sci. 1994, 22, 151–158. [Google Scholar] [CrossRef]
- Chen, C.; Jost, M.; Clark, B.; Martin, M.; Matny, O.; Steffenson, B.J.; Franckowiak, J.D.; Mascher, M.; Singh, D.; Perovic, D.; et al. BED-domain-containing immune receptor from wild barley confers widely effective resistance to leaf rust. Plant Biotech J. 2021, 19, 1206–1215. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.J.; Chicaiza, O.; Caffarel, J.C.; Sallam, A.H.; Druka, A.; Waugh, R.; Ordon, F.; Kopahnke, D.; Keilwagen, J.; Perovic, D.; et al. Development of barley introgression lines carrying the leaf rust resistance genes Rph1 to Rph15. Crop Sci. 2020, 60, 282–302. [Google Scholar] [CrossRef]
- Weerasena, J.S.; Steffenson, B.J.; Falk, A.B. Conversion of an amplified fragment length polymorphism marker into a co-dominant marker in the mapping of the Rph15 gene conferring resistance to barley leaf rust, Puccinia hordei Otth. Theor. Appl. Genet. 2003, 108, 712–719. [Google Scholar] [CrossRef]
- Rothwell, C.T.; Singh, D.; van Ogtrop, F.; Sørensen, C.; Fowler, R.; Germán, S.; Park, R.F.; Dracatos, P.M. Rapid phenotyping of adult plant resistance in barley (Hordeum vulgare) to leaf rust under controlled conditions. Plant Breed. 2019, 138, 51–61. [Google Scholar] [CrossRef]
- Castro, A.J.; Gamba, F.; German, S.; Gonzalez, S.; Hayes, P.M.; Pereyra, S.; Perez, C. Quantitative trait locus analysis of spot blotch and leaf rust resistance in the BCD47 × Baronesse barley mapping population. Plant Breed. 2012, 131, 258–266. [Google Scholar] [CrossRef]
- Ziems, L.A.; Franckowiak, J.D.; Platz, G.J.; Mace, E.S.; Park, R.F.; Singh, D.; Jordan, D.R.; Hickey, L.T. Investigating successive Australian barley breeding populations for stable resistance to leaf rust. Theor. Appl. Genet. 2017, 130, 2463–2477. [Google Scholar] [CrossRef]
- Qi, X.; Niks, R.E.; Stam, P.; Lindhout, P. Identification of QTLs for partial resistance to leaf rust (Puccinia hordei) in barley. Theor. Appl. Genet. 1998, 96, 1205–1215. [Google Scholar] [CrossRef]
- Von Korff, M.; Wang, H.; Léon, J.; Pillen, K. AB-QTL analysis in spring barley. I. Detection of resistance genes against powdery mildew, leaf rust and scald introgressed from wild barley. Theor. Appl. Genet. 2005, 111, 583–590. [Google Scholar] [CrossRef]
- Marcel, T.C.; Varshney, R.K.; Barbieri, M.; Jafary, H.; De Kock, M.J.D.; Graner, A.; Niks, R.E. A high-density consensus map of barley to compare the distribution of QTLs for partial resistance to Puccinia hordei and of defence gene homologues. Theor. Appl. Genet. 2006, 114, 487–500. [Google Scholar] [CrossRef]
- Vatter, T.; Maurer, A.; Perovic, D.; Kopahnke, D.; Pillen, K.; Ordon, F. Identification of QTL conferring resistance to stripe rust (Puccinia striiformis f. sp. hordei) and leaf rust (Puccinia hordei) in barley using nested association mapping (NAM). PLoS ONE 2018, 13, e0191666. [Google Scholar] [CrossRef]
- Park, R.F.; (The University of Sydney, School of Life and Environmental Sciences, Faculty of Science, Sydney, Australia). Pathotypes of P. hordei (pts) to Postulate ASR Rph Genes. Personal communication, 2020. [Google Scholar]
- Gilmour, J. Octal Notation for Designating Physiologic Races of Plant Pathogens. Nature. 1973, 242, 620. [Google Scholar] [CrossRef]
- Peterson, R.F.; Campbell, A.B.; Hannah, A.E. A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can. J. Res. 1948, 26, 496–500. [Google Scholar] [CrossRef]
- Visual Crossing Corporation. Visual Crossing Weather. 2019–2021. Available online: www.visualcrossing.com (accessed on 9 February 2023).
- Monat, C.; Padmarasu, S.; Lux, T.; Wicker, T.; Gundlach, H.; Himmelbach, A.; Ens, J.; Li, C.; Muehlbauer, G.J.; Schulman, A.H.; et al. TRITEX: Chromo-some-scale sequence assembly of Triticeae genomes with open-source tools. Genome Biol. 2019, 20, 284. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Wimmer, V.; Albrecht, T.; Auinger, H.-J.; Schön, C.-C. Synbreed: A framework for the analysis of genomic prediction data using R. Bioinformatics 2012, 28, 2086–2087. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Endelman, J.B. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 2011, 4, 255–258. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Leung, H.K. Chopsticks: The ‘snp.matrix’ and ‘X.snp.matrix’ Classes, R Package Version 1.56.0; Bioconductor: Seattle, WA, USA, 2016.
- Shin, J.H.; Blay, S.; McNeney, B.; Graham, J. LDheatmap: An R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J. Stat. Softw. 2006, 16, 1–9. [Google Scholar] [CrossRef] [Green Version]
| MTA Name | Marker ID | Chr 1 | Position (Mbp) 2 | Co-Located Rph Gene 3 | R1_2019 4 | R2_2019 4 | 2021 4 | SNP Position in Marker Sequence | Allele Resistant | Allele Susceptible |
|---|---|---|---|---|---|---|---|---|---|---|
| MTA_1 | 3258774 | 1H | 331.01 | 34.16 *** | 33.57 ** | 31.92 ** | 8 | G | A | |
| MTA_2 | 3262665 | 1H | 482.85 | NS 5 | 37.12 *** | 34.01 ** | 7 | G | A | |
| MTA_3 | 3929195 | 2H | 21.47 | Rph15/16/8/14 | 29.41 *** | 28.37 ** | 25.18 * | 33 | C | T |
| MTA_4 | 3256022 | 2H | 26.34 | Rph15/16/8/14 | NS 5 | 31.70 *** | 30.10 ** | 8 | T | C |
| MTA_5 | 3261067 | 2H | 668.5 | 32.93 *** | 29.26 ** | 26.47 ** | 67 | A | G | |
| MTA_6 | 3266799 | 3H | 17.35 | Rph7/5/6 | 19.86 ** | 28.74 *** | 28.02 *** | 9 | G | C |
| MTA_7 | 3434139 | 5H | 4.05 | Rph20 | 32.05 *** | NS 5 | NS 5 | 61 | C | T |
| MTA_8 | 5249701 | 5H | 11.35 | Rph20 | 31.10 *** | NS 5 | NS 5 | 49 | T | C |
| MTA_9 | 3396786 | 5H | 436.05 | 21.49 ** | 28.48 *** | 27.61 ** | 55 | T | G | |
| MTA_10 | 3256468 | 5H | 573.69 | NS 5 | 39.35 *** | 37.17 *** | 67 | C | G | |
| MTA_11 | 3260244 | 7H | 600.48 | Rph3 | 29.31 ** | 38.98 *** | 43.01 **** | 61 | A | G |
| MTA_12 | 4792921 | 7H | 620.61 | Rph3/19 | 28.24 * | 49.06 *** | 43.47 *** | 15 | A | C |
| Differential Genotype | Rph Gene | 200 P− | 220 P+ +Rph13 | 253 P− | 4610 P+ | 5610 P+ | 5453 P+ | 5656 P+ | 5457 P− | 5457 P+ |
|---|---|---|---|---|---|---|---|---|---|---|
| Sudan | Rph1 | −CN | 0 | 3+ | N | −N | 3+ | 1−CN | 3+ | 3+ |
| Peruvian | Rph2 | N | N | 3− | 1+CN | 1+CN | 3+ | 3+ | 3+C | 3+C |
| Estate | Rph3 | C | 0 | 0 | N | -CN | 1− | 3+ | 3+C | 3+C |
| Gold | Rph4 | 1- | 33+ | 3+ | 33+ | 3C | 3+ | 3+ | 33+C | 33+C |
| Magnif 104 | Rph5 | 1−N | 33+ | N | N | -N | N | 1−N | N | N |
| Bolivia | Rph2+Rph6 | N | N | 3+ | CN | CN | 33+ | 12- | 3+ | 3+ |
| Cebada Capa | Rph7 | N | 0 | N | CN | -N | +N | 1−CN | +CN | +N |
| Egypt 4 | Rph8 | 33+ | 33+ | 33+ | 3+ | 33+ | 3+ | 12-CN | 3+ | 3+ |
| Abyssinian | Rph9 | 1N | N | 3+ | 1CN | 1+CN | 33+ | 33+ | 3+ | 3+ |
| Clipper BC8 | Rph10 | 3+ | 33+ | 1+CN | 1+CN | 3+ | 3+ | 33+ | 33+ | 3+ |
| Clipper BC67 | Rph11 | 12+ | 1CN | 12CN | 2+C | 2++C | 3+ | 12−CN | 33+ | 33+ |
| Triumph | Rph12 | +N | +N | 1CN | 33+C | 3 | 3+ | 33+ | 3+ | 3+ |
| PI 531849 | Rph13 | 0 | 3+ | −N | N | -N | CN | 12−CN | +CN | +CN |
| PI 584760 | Rph14 | 12+ | 1CN | 1+CN | 12C | 12+CN | 1+CN | 33+ | 1CN | 1+CN |
| Prior | Rph19 | 1 | 33+ | 1−CN | 3+ | 3+ | 3+ | 33+ | 12− | 3+ |
| Cantala | Rph9.am | 3+ | 3+ | 12−C | 3+ | 3+ | 3+ | 3+ | 3+C | 3+ |
| Pathotype (Culture No.) | Virulence 1 |
|---|---|
| 200 P− (518) 2 | Rph8 |
| 220 P+ +Rph13 (577) 2 | Rph5, Rph8, Rph13 |
| 253 P− (490) | Rph1, Rph2, Rph4, Rph6, Rph8 |
| 4610 P+ (491) 2 | Rph4, Rph8, Rph9, Rph12 |
| 5610 P+ (520) 2 | Rph4, Rph8, Rph9, Rph10, Rph12 |
| 5453 P+ (584) | Rph1, Rph2, Rph4, Rph6, Rph9, Rph10, Rph12, Rph19 |
| 5656 P+ (623) | Rph2, Rph3, Rph4, Rph6, Rph8, Rph9, Rph12, Rph19 |
| 5457 P− (626) | Rph1, Rph2, Rph3, Rph4, Rph6, Rph9, Rph10, Rph12 |
| 5457 P+ (612) | Rph1, Rph2, Rph3, Rph4, Rph6, Rph9, Rph10, Rph12, Rph19 |
| Gene | Marker | Chr 1 | Forward Primer Sequence, 5′-3′ | Reverse primer Sequence, 5′-3′ | Reference |
|---|---|---|---|---|---|
| Rph7 | Unknown | 3H | GAGATAAAAGCATTACCAAAGGCTCAT | GCGCGCGCAACAGCAAACGGC | [20] |
| Rph15 | Unknown | 2H | TGAAGAAGCTGGAAGGTCACC | AGCCAAAAACCCTTCTGGCT | [23] |
| Rph20 | bPb0837 | 5H | GACACTTCGTGCCAGTTTG | CCTCCCTCCCTCTTCTCAAC | [15] |
| Rph23 | Ebmac0603 | 7H | ACCGAAACTAAATGAACTACTTCG | TGCAAACTGTGCTATTAAGGG | [11] |
| Rph24 | Sun43-44 | 6H | CTAGACACCACCACCACACC | ATACCAGAGTTTGCGTCCGG | [19] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ziems, L.A.; Singh, L.; Dracatos, P.M.; Dieters, M.J.; Sanchez-Garcia, M.; Amri, A.; Verma, R.P.S.; Park, R.F.; Singh, D. Characterization of Leaf Rust Resistance in International Barley Germplasm Using Genome-Wide Association Studies. Plants 2023, 12, 862. https://doi.org/10.3390/plants12040862
Ziems LA, Singh L, Dracatos PM, Dieters MJ, Sanchez-Garcia M, Amri A, Verma RPS, Park RF, Singh D. Characterization of Leaf Rust Resistance in International Barley Germplasm Using Genome-Wide Association Studies. Plants. 2023; 12(4):862. https://doi.org/10.3390/plants12040862
Chicago/Turabian StyleZiems, Laura A., Lovepreet Singh, Peter M. Dracatos, Mark J. Dieters, Miguel Sanchez-Garcia, Ahmed Amri, Ramesh Pal Singh Verma, Robert F. Park, and Davinder Singh. 2023. "Characterization of Leaf Rust Resistance in International Barley Germplasm Using Genome-Wide Association Studies" Plants 12, no. 4: 862. https://doi.org/10.3390/plants12040862





