Facilitated Adaptation as A Conservation Tool in the Present Climate Change Context: A Methodological Guide
Abstract
:1. Introduction
2. A Brief Overview of the Conceptual Framework
2.1. Evolutionary Adaptation
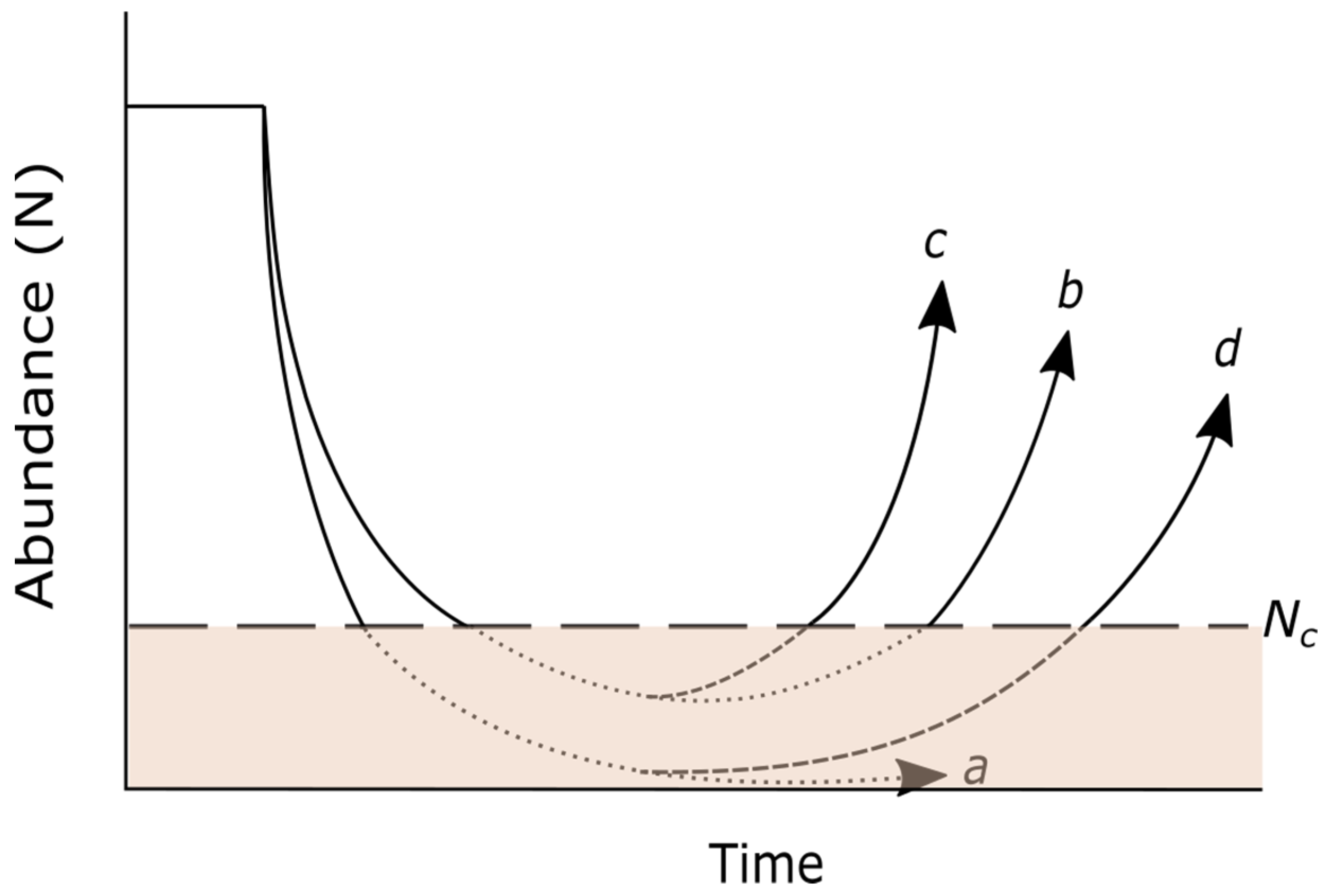
2.2. Evolutionary Rescue
2.3. Facilitated Adaptation
3. Climate-Change Adaptive Traits
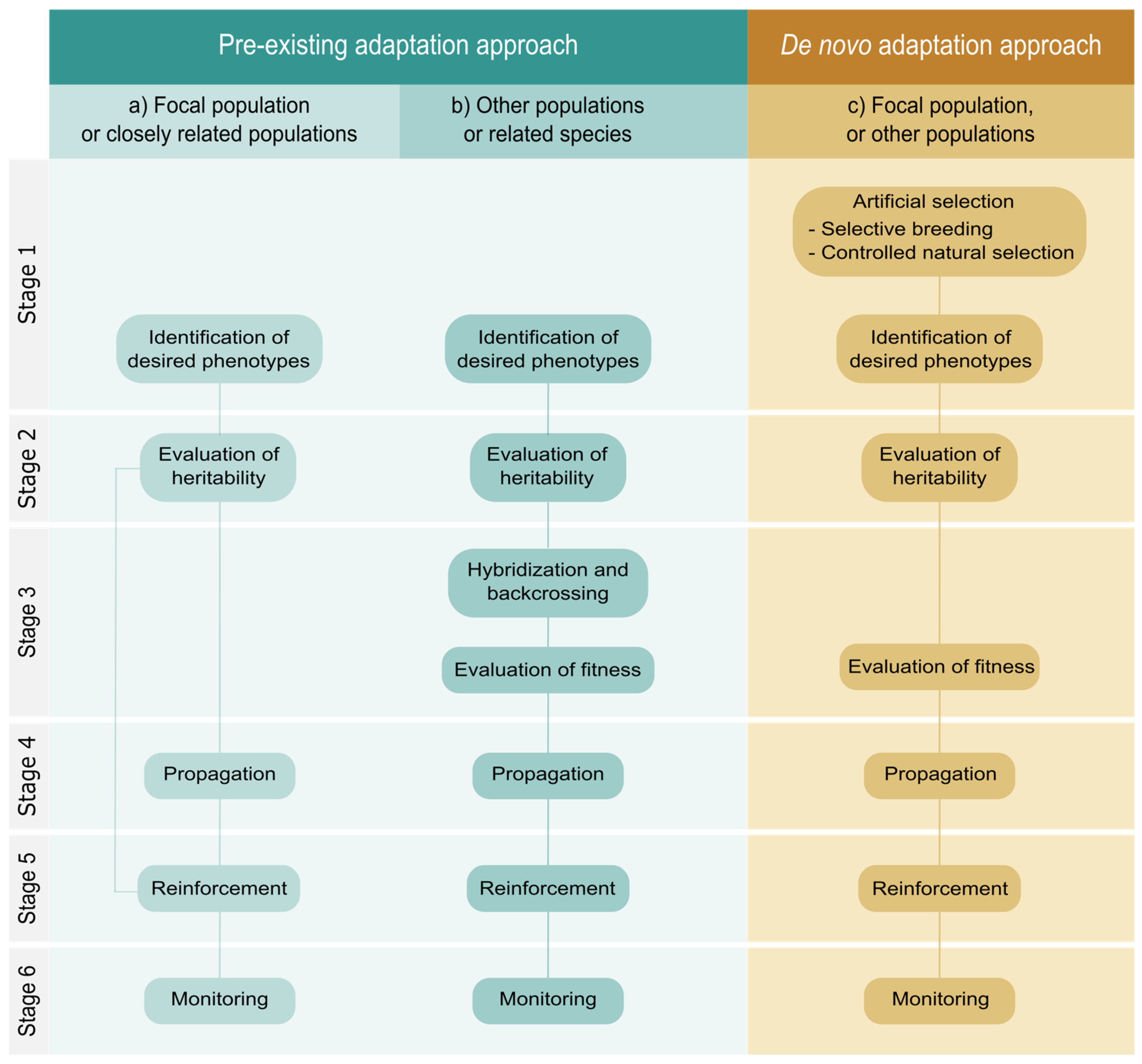
4. A Methodological Proposal for Facilitated Adaptation
4.1. Pre-Existing Adaptation Approach
4.2. De Novo Adaptation Approach
5. Risks and Difficulties of Facilitated Adaptation
6. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Scheffers, B.R.; De Meester, L.; Bridge, T.C.L.; Hoffmann, A.A.; Pandolfi, J.M.; Corlett, R.T.; Butchart, S.H.M.; Pearce-Kelly, P.; Kovacs, K.M.; Dudgeon, D.; et al. The broad footprint of climate change from genes to biomes to people. Science 2016, 354, aaf7671. [Google Scholar] [CrossRef] [PubMed]
- Wiens, J.J. Climate-related local extinctions are already widespread among plant and animal species. PLoS Biol. 2016, 14, e2001104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertin, R.I. Plant phenology and distribution in relation to recent climate change. J. Torrey Bot. Soc. 2008, 135, 126–146. [Google Scholar] [CrossRef] [Green Version]
- Chen, I.C.; Hill, J.K.; Ohlemüller, R.; Roy, D.B.; Thomas, C.D. Rapid range shifts of species associated with high levels of climate warming. Science 2011, 333, 1024–1026. [Google Scholar] [CrossRef]
- Lenoir, J.; Svenning, J.C. Latitudinal and elevational range shifts under contemporary climate change. In Encyclopedia of Biodiversity; Levin, S.A., Ed.; Elsevier: Amsterdam, The Netherlands, 2013; Volume 4, pp. 599–611. [Google Scholar]
- Freeman, B.G.; Lee-Yaw, J.A.; Sunday, J.M.; Hargreaves, A.L. Expanding, shifting and shrinking: The impact of global warming on species’ elevational distributions. Glob. Ecol. Biogeogr. 2018, 27, 1268–1276. [Google Scholar] [CrossRef]
- Cheplick, G.P. Philomatry in plants: Why do so many species have limited seed dispersal? Am. J. Bot. 2022, 109, 29–45. [Google Scholar] [CrossRef]
- Honnay, O.; Verheyen, K.; Butaye, J.; Jacquemyn, H.; Bossuyt, B.; Hermy, M. Possible effects of habitat fragmentation and climate change on the range of forest plant species. Ecol. Lett. 2002, 5, 525–530. [Google Scholar] [CrossRef]
- Christmas, M.J.; Breed, M.F.; Lowe, A.J. Constraints to and conservation implications for climate change adaptation in plants. Conserv. Genet. 2016, 17, 305–320. [Google Scholar] [CrossRef] [Green Version]
- Anderson, J.T.; Inouye, D.W.; McKinney, A.M.; Colautti, R.I.; Mitchell-Olds, T. Phenotypic plasticity and adaptive evolution contribute to advancing flowering phenology in response to climate change. Proc. R. Soc. B Biol. Sci. 2012, 279, 3843–3852. [Google Scholar] [CrossRef] [Green Version]
- Franks, S.J.; Kane, N.C.; O’Hara, N.B.; Tittes, S.; Rest, J.S. Rapid genome-wide evolution in Brassica rapa populations following drought revealed by sequencing of ancestral and descendant gene pools. Mol. Ecol. 2016, 25, 3622–3631. [Google Scholar] [CrossRef]
- Rauschkolb, R.; Henres, L.; Lou, C.; Godefroid, S.; Dixon, L.; Durka, W.; Bossdorf, O.; Ensslin, A.; Scheepens, J.F. Historical comparisons show evolutionary changes in drought responses in European plant species after two decades of climate change. Basic Appl. Ecol. 2022, 58, 26–38. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, G.; Zeng, F.; Han, Z.; Qiu, C.-W.; Zeng, M.; Yang, Z.; Xu, F.; Wu, D.; Deng, F.; et al. Molecular evidence for adaptive evolution of drought tolerance in wild cereals. New Phytol. 2023, 237, 497–514. [Google Scholar] [CrossRef] [PubMed]
- Brunet, J.; Larson-Rabin, Z. The response of flowering time to global warming in a high-altitude plant: The impact of genetics and the environment. Botany 2012, 90, 319–326. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Sgrò, C.M.; Kristensen, T.N. Revisiting adaptive potential, population size, and conservation. Trends Ecol. Evol. 2017, 32, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.C. Accelerating extinction risk from climate change. Science 2015, 348, 571–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jump, A.S.; Peñuelas, J. Running to stand still: Adaptation and the response of plants to rapid climate change. Ecol. Lett. 2005, 8, 1010–1020. [Google Scholar] [CrossRef]
- Skelly, D.K.; Joseph, L.N.; Possingham, H.P.; Freidenburg, L.K.; Farrugia, T.J.; Kinnison, M.T.; Hendry, A.P. Evolutionary responses to climate change. Conserv. Biol. 2007, 21, 1353–1355. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Sgrò, C. Climate change and evolutionary adaptation. Nature 2011, 470, 479–485. [Google Scholar] [CrossRef]
- Keeley, A.T.H.; Ackerly, D.D.; Cameron, D.R.; Heller, N.E.; Huber, P.R.; Schloss, C.A.; Thorne, J.H.; Merenlender, A.M. New concepts, models, and assessments of climate-wise connectivity. Environ. Res. Lett. 2018, 13, 73002. [Google Scholar] [CrossRef]
- Hällfors, M.H.; Vaara, E.M.; Hyvärinen, M.; Oksanen, M.; Schulman, L.E.; Siipi, H.; Lehvävirta, S. Coming to terms with the concept of moving species threatened by climate change—A systematic review of the terminology and definitions. PLoS ONE 2014, 9, e102979. [Google Scholar] [CrossRef]
- Hunter, M.L. Climate change and moving species: Furthering the debate on assisted colonization. Conserv. Biol. 2007, 21, 1356–1358. [Google Scholar] [CrossRef] [PubMed]
- McLachlan, J.S.; Hellmann, J.J.; Schwartz, M.W. A framework for debate of assisted migration in an era of climate change. Conserv. Biol. 2007, 21, 297–302. [Google Scholar] [CrossRef]
- Ricciardi, A.; Simberloff, D. Assisted colonization is not a viable conservation strategy. Trends Ecol. Evol. 2009, 24, 248–253. [Google Scholar] [CrossRef]
- Carrete, M.; Tella, J.L. Is assisted colonization feasible? Lessons from past introductions. Front. Ecol. Environ. 2012, 10, 12–13. [Google Scholar] [CrossRef]
- Dukes, J.S.; Mooney, H.A. Disruption of ecosystem processes in western North America by invasive species. Rev. Chil. Hist. Nat. 2004, 77, 411–437. [Google Scholar] [CrossRef]
- Scarela, R.; Genovesi, P.; Essl, F.; Rabitsch, W. The Impacts of Invasive Alien Species in Europe; European Environment Agency: Copenhagen, Denmark, 2012. [Google Scholar]
- Williams, M.C.; Wardle, G.M. Pinus radiata invasion in Australia: Identifying key knowledge gaps and research directions. Austral Ecol. 2007, 32, 721–739. [Google Scholar] [CrossRef]
- Kew, R. The State of the World’s Plants Report—2016; Royal Botanic Gardens: Kew, UK, 2016.
- Sgrò, C.M.; Lowe, A.J.; Hoffmann, A.A. Building evolutionary resilience for conserving biodiversity under climate change. Evol. Appl. 2011, 4, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Aitken, S.N.; Whitlock, M.C. Assisted gene flow to facilitate local adaptation to climate change. Annu. Rev. Ecol. Evol. Syst. 2013, 44, 367–388. [Google Scholar] [CrossRef]
- Thomas, M.A.; Roemer, G.W.; Donlan, C.J.; Dickson, B.G.; Matocq, M.; Malaney, J. Gene tweaking for conservation. Nature 2013, 501, 485–486. [Google Scholar] [CrossRef] [Green Version]
- Ahuja, I.; de Vos, R.C.H.; Bones, A.M.; Hall, R.D. Plant molecular stress responses face climate change. Trends Plant Sci. 2010, 15, 664–674. [Google Scholar] [CrossRef]
- Franks, S.J.; Hoffmann, A.A. Genetics of climate change adaptation. Annu. Rev. Genet. 2012, 46, 185–208. [Google Scholar] [CrossRef] [PubMed]
- Vanwallendael, A.; Soltani, A.; Emery, N.C.; Peixoto, M.M.; Olsen, J.; Lowry, D.B. A molecular view of plant local adaptation: Incorporating stress-response networks. Annu. Rev. Plant Biol. 2019, 70, 559–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlson, S.M.; Cunningham, C.J.; Westley, P.A.H. Evolutionary rescue in a changing world. Trends Ecol. Evol. 2014, 29, 521–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hufbauer, R.A.; Szűcs, M.; Kasyon, E.; Youngberg, C.; Koontz, M.J.; Richards, C.; Tuff, T.; Melbourne, B.A. Three types of rescue can avert extinction in a changing environment. Proc. Natl. Acad. Sci. USA 2015, 112, 10557–10562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, G.S.; Morris, M.R.; Genis, A.B.; Szűcs, M.; Melbourne, B.A.; Tavener, S.J.; Hufbauer, R.A. The power of evolutionary rescue is constrained by genetic load. Evol. Appl. 2017, 10, 731–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson-Manning, C.F.; Wagner, M.R.; Mitchell-Olds, T. Adaptive evolution: Evaluating empirical support for theoretical predictions. Nat. Rev. Genet. 2012, 13, 867–877. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, J.K.; Pickrell, J.K.; Coop, G. The genetics of human adaptation: Hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 2010, 20, R208–R215. [Google Scholar] [CrossRef] [Green Version]
- Panigrahi, J.; Mishra, R.R.; Sahu, A.R.; Rath, S.C.; Kole, C.R. Marker-assisted breeding for stress resistence in crop plants. In Molecular Stress Physiology of Plants; Rout, G.R., Das, A.B., Eds.; Springer: New Delhi, India, 2013; pp. 387–426. [Google Scholar]
- Pritchard, J.K.; Di Rienzo, A. Adaptation—not by sweeps alone. Nat. Publ. Gr. 2010, 11, 665–667. [Google Scholar] [CrossRef] [Green Version]
- Csilléry, K.; Rodríguez-Verdugo, A.; Rellstab, C.; Guillaume, F. Detecting the genomic signal of polygenic adaptation and the role of epistasis in evolution. Mol. Ecol. 2018, 27, 606–612. [Google Scholar] [CrossRef] [Green Version]
- Fagny, M.; Austerlitz, F. Polygenic adaptation: Integrating population genetics and gene regulatory networks. Trends Genet. 2021, 37, 631–638. [Google Scholar] [CrossRef]
- Pavličev, M.; Cheverud, J.M. Constraints evolve: Context dependency of gene effects allows evolution of pleiotropy. Annu. Rev. Ecol. Evol. Syst. 2015, 46, 413–434. [Google Scholar] [CrossRef] [Green Version]
- Pepper, J.W. The evolution of evolvability in genetic linkage patterns. BioSystems 2003, 69, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Fox, R.J.; Donelson, J.M.; Schunter, C.; Ravasi, T.; Gaitán-Espitia, J.D. Beyond buying time: The role of plasticity in phenotypic adaptation to rapid environmental change. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180174. [Google Scholar] [CrossRef] [PubMed]
- Thiebaut, F.; Hemerly, A.S.; Ferreira, P.C.G. A role for epigenetic regulation in the adaptation and stress responses of non-model plants. Front. Plant Sci. 2019, 10, 246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stajic, D.; Jansen, L.E.T. Empirical evidence for epigenetic inheritance driving evolutionary adaptation. Philos. Trans. R. Soc. B Biol. Sci. 2021, 376, 20200121. [Google Scholar] [CrossRef] [PubMed]
- Gomulkiewicz, R.; Shaw, R.G. Evolutionary rescue beyond the models. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2013, 368, 20120093. [Google Scholar] [CrossRef] [PubMed]
- Bell, G. Evolutionary rescue and the limits of adaptation. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120080. [Google Scholar] [CrossRef] [PubMed]
- Bell, G.; Gonzalez, A. Evolutionary rescue can prevent extinction following environmental change. Ecol. Lett. 2009, 12, 942–948. [Google Scholar] [CrossRef]
- Bell, G.; Gonzalez, A. Adaptation and evolutionary rescue in metapopulations experiencing environmental deterioration. Science 2011, 332, 1327–1330. [Google Scholar] [CrossRef]
- Schiffers, K.; Bourne, E.C.; Lavergne, S.; Thuiller, W.; Travis, J.M.J. Limited evolutionary rescue of locally adapted populations facing climate change. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2013, 368, 20120083. [Google Scholar] [CrossRef] [Green Version]
- Czuppon, P.; Blanquart, F.; Uecker, H.; Débarre, F. The effect of habitat choice on evolutionary rescue in subdivided populations. Am. Nat. 2021, 197, 625–643. [Google Scholar] [CrossRef] [PubMed]
- Tomasini, M.; Peischl, S. When does gene flow facilitate evolutionary rescue? Evolution 2020, 74, 1640–1653. [Google Scholar] [CrossRef] [PubMed]
- Gomulkiewicz, R.; Holt, R.D. When does evolution by natural selection prevent extinction? Evolution 1995, 49, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Phelps, M.P.; Seeb, L.W.; Seeb, J.E. Transforming ecology and conservation biology through genome editing. Conserv. Biol. 2020, 34, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.Y.; Hoffmann, A.A.; van Oppen, M.J.H. Hybridization as a conservation management tool. Conserv. Lett. 2019, 12, e12652. [Google Scholar] [CrossRef] [Green Version]
- Lind, M.I.; Spagopoulou, F. Evolutionary consequences of epigenetic inheritance. Heredity 2018, 121, 205–209. [Google Scholar] [CrossRef] [Green Version]
- Rey, O.; Eizaguirre, C.; Angers, B.; Baltazar-Soares, M.; Sagonas, K.; Prunier, J.G.; Blanchet, S. Linking epigenetics and biological conservation: Towards a conservation epigenetics perspective. Funct. Ecol. 2020, 34, 414–427. [Google Scholar] [CrossRef] [Green Version]
- van Oppen, M.J.H.; Oliver, J.K.; Putnam, H.M.; Gates, R.D. Building coral reef resilience through assisted evolution. Proc. Natl. Acad. Sci. USA 2015, 112, 2307–2313. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, J.A.; Miller, J.M. Adaptive introgression as a resource for management and genetic conservation in a changing climate. Conserv. Biol. 2016, 30, 33–41. [Google Scholar] [CrossRef]
- Weeks, A.R.; Sgro, C.M.; Young, A.G.; Frankham, R.; Mitchell, N.J.; Miller, K.A.; Byrne, M.; Coates, D.J.; Eldridge, M.D.B.; Sunnucks, P.; et al. Assessing the benefits and risks of translocations in changing environments: A genetic perspective. Evol. Appl. 2011, 4, 709–725. [Google Scholar] [CrossRef] [Green Version]
- Kelly, E.; Phillips, B.L. Targeted gene flow for conservation. Conserv. Biol. 2016, 30, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Becklin, K.M.; Anderson, J.T.; Gerhart, L.M.; Wadgymar, S.M.; Wessinger, C.A.; Ward, J.K. Examining plant physiological responses to climate change through an evolutionary lens. Plant Physiol. 2016, 172, 635–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamann, E.; Denney, D.; Day, S.; Lombardi, E.; Jameel, M.I.; MacTavish, R.; Anderson, J.T. Plant eco-evolutionary responses to climate change: Emerging directions. Plant Sci. 2021, 304, 110737. [Google Scholar] [CrossRef] [PubMed]
- Fitter, A.H.; Fitter, R.S.R. Rapid changes in flowering time in British plants. Science 2002, 296, 1689–1691. [Google Scholar] [CrossRef] [PubMed]
- Amano, T.; Smithers, R.J.; Sparks, T.H.; Sutherland, W.J. A 250-year index of first flowering dates and its response to temperature changes. Proc. R. Soc. B Biol. Sci. 2010, 277, 2451–2457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellwood, E.R.; Temple, S.A.; Primack, R.B.; Bradley, N.L.; Davis, C.C. Record-breaking early flowering in the eastern United States. PLoS ONE 2013, 8, e53788. [Google Scholar] [CrossRef] [PubMed]
- Parmesan, C.; Hanley, M.E. Plants and climate change: Complexities and surprises. Ann. Bot. 2015, 116, 849–864. [Google Scholar] [CrossRef]
- Qian, C.; Yan, X.; Shi, Y.; Yin, H.; Chang, Y.; Chen, J.; Ingvarsson, P.K.; Nevo, E.; Ma, X.F. Adaptive signals of flowering time pathways in wild barley from Israel over 28 generations. Heredity 2020, 124, 62–76. [Google Scholar] [CrossRef] [Green Version]
- Kooyers, N.J. The evolution of drought escape and avoidance in natural herbaceous populations. Plant Sci. 2015, 234, 155–162. [Google Scholar] [CrossRef]
- Hu, X.; Zhou, W.; Sun, S. Responses of plant reproductive phenology to winter-biased warming in an alpine meadow. Front. Plant Sci. 2020, 11, 534703. [Google Scholar] [CrossRef]
- Hopkins, R.; Schmitt, J.; Stinchcombe, J.R. A latitudinal cline and response to vernalization in leaf angle and morphology in Arabidopsis thaliana (Brassicaceae). New Phytol. 2008, 179, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Tonsor, S.J.; Scott, C.; Boumaza, I.; Liss, T.R.; Brodsky, J.L.; Vierling, E. Heat shock protein 101 effects in A. thaliana: Genetic variation, fitness and pleiotropy in controlled temperature conditions. Mol. Ecol. 2008, 17, 1614–1626. [Google Scholar] [CrossRef] [Green Version]
- Hong, S.W.; Vierling, E. Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc. Natl. Acad. Sci. USA 2000, 97, 4392–4397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Queitsch, C.; Hong, S.-W.; Vierling, E.; Lindquist, S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 2000, 12, 479–492. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.; Khungar, L.; Shimphrui, R.; Tiwari, L.D.; Tripathi, G.; Sarkar, N.K.; Agarwal, S.K.; Agarwal, M.; Grover, A. AtHsp101 research sets course of action for the genetic improvement of crops against heat stress. J. Plant Biochem. Biotechnol. 2020, 29, 715–732. [Google Scholar] [CrossRef]
- Barua, D.; Heckathorn, S.A.; Coleman, J.S. Variation in heat-shock proteins and photosynthetic thermotolerance among natural populations of Chenopodium album L. from contrasting thermal environments: Implications for plant responses to global warming. J. Integr. Plant Biol. 2008, 50, 1440–1451. [Google Scholar] [CrossRef]
- Sastry, A.; Barua, D. Leaf thermotolerance in tropical trees from a seasonally dry climate varies along the slow-fast resource acquisition spectrum. Sci. Rep. 2017, 7, 11246. [Google Scholar] [CrossRef] [Green Version]
- Sentinella, A.T.; Warton, D.I.; Sherwin, W.B.; Offord, C.A.; Moles, A.T. Tropical plants do not have narrower temperature tolerances, but are more at risk from warming because they are close to their upper thermal limits. Glob. Ecol. Biogeogr. 2020, 29, 1387–1398. [Google Scholar] [CrossRef]
- Yeaman, S.; Hodgins, K.A.; Lotterhos, K.E.; Suren, H.; Nadeau, S.; Degner, J.C.; Nurkowski, K.A.; Smets, P.; Wang, T.; Gray, L.K.; et al. Convergent local adaptation to climate in distantly related conifers. Science 2016, 353, 23–26. [Google Scholar] [CrossRef] [Green Version]
- Sacristán-Bajo, S.; García-Fernández, A.; Lara-Romero, C.; Prieto-Benítez, S.; Tabarés, P.; Morente-López, J.; Rubio Teso, M.L.; Alameda-Martín, A.; Torres, E.; Iriondo, J.M. Population origin determines the adaptive potential for the advancement of flowering onset in Lupinus angustifolius L. (Fabaceae). Evol. Appl. 2023, 16, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Reed, P.B.; Peterson, M.L.; Pfeifer-Meister, L.E.; Morris, W.F.; Doak, D.F.; Roy, B.A.; Johnson, B.R.; Bailes, G.T.; Nelson, A.A.; Bridgham, S.D. Climate manipulations differentially affect plant population dynamics within versus beyond northern range limits. J. Ecol. 2021, 109, 664–675. [Google Scholar] [CrossRef]
- Akçakaya, H.R.; Burgman, M.A.; Kindvall, O.; Wood, C.C.; Sjögren-Gulve, P.; Hatfield, J.S.; McCarthy, M.A. Species Conservation and Management: Case Studies; Oxford University Press: Oxford, UK, 2004. [Google Scholar]
- Giménez-Benavides, L.; Albert, M.J.; Iriondo, J.M.; Escudero, A. Demographic processes of upward range contraction in a long-lived Mediterranean high mountain plant. Ecography 2011, 34, 85–93. [Google Scholar] [CrossRef]
- Iriondo, J.M.; Albert, M.J.; Escudero, A. Structural equation modelling: An alternative for assessing causal relationships in threatened plant populations. Biol. Conserv. 2003, 113, 367–377. [Google Scholar] [CrossRef]
- Bramer, I.; Anderson, B.J.; Bennie, J.; Bladon, A.J.; De Frenne, P.; Hemming, D.; Hill, R.A.; Kearney, M.R.; Körner, C.; Korstjens, A.H.; et al. Advances in monitoring and modelling climate at ecologically relevant scales. In Advances in Ecological Research; Bohan, D.A., Dumbrell, A.J., Woodward, G., Jackson, M., Eds.; Academic Press: London, UK, 2018. [Google Scholar]
- Kozak, K.H.; Graham, C.H.; Wiens, J.J. Integrating GIS-based environmental data into evolutionary biology. Trends Ecol. Evol. 2008, 23, 141–148. [Google Scholar] [CrossRef]
- Thormann, I.; Parra-Quijano, M.; Endresen, D.T.F.; Rubio-Teso, M.L.; Iriondo, M.J.; Maxted, N. Predictive Characterization of Crop Wild Relatives and Landraces; Bioversity International: Rome, Italy, 2014. [Google Scholar]
- Storfer, A.; Patton, A.; Fraik, A.K. Navigating the interface between landscape genetics and landscape genomics. Front. Genet. 2018, 9, 68. [Google Scholar] [CrossRef] [Green Version]
- Browne, L.; Wright, J.W.; Fitz-Gibbon, S.; Gugger, P.F.; Sork, V.L. Adaptational lag to temperature in valley oak (Quercus lobata) can be mitigated by genome-informed assisted gene flow. Proc. Natl. Acad. Sci. USA 2019, 116, 25179–25185. [Google Scholar] [CrossRef] [Green Version]
- Exposito-Alonso, M.; Gómez Rodríguez, R.; Barragán, C.; Capovilla, G.; Chae, E.; Devos, J.; Dogan, E.S.; Friedemann, C.; Gross, C.; Lang, P.; et al. Natural selection on the Arabidopsis thaliana genome in present and future climates. Nature 2019, 573, 126–129. [Google Scholar] [CrossRef]
- Forester, B.R.; Landguth, E.L.; Hand, B.K.; Balkenhol, N. Landscape genomics for wildlife research. In Population Genomics: Wildlife; Hohenlohe, P.A., Rajora, O.P., Eds.; Springer: Cham, Switzerland, 2021; pp. 145–184. [Google Scholar]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics; Pearson: Harlow, UK, 1996. [Google Scholar]
- Postma, E. Four decades of estimating heritabilities in wild vertebrate populations: Improved methods, more data, better estimates? In Quantitative Genetics in the Wild; Charmantier, A., Garant, D., Kruuk, L.E.B., Eds.; Oxford University Press: Oxford, UK, 2014. [Google Scholar]
- Kruuk, L.E.B. Estimating genetic parameters in natural populations using the “animal model”. Philos. Trans. R. Soc. B Biol. Sci. 2004, 359, 873–890. [Google Scholar] [CrossRef] [Green Version]
- Frankham, R.; Ballou, J.D.; Eldridge, M.D.B.; Lacy, R.C.; Ralls, K.; Dudash, M.R.; Fenster, C.B. Predicting the probability of outbreeding depression. Conserv. Biol. 2011, 25, 465–475. [Google Scholar] [CrossRef]
- IUCN/SSC. Guidelines for Reintroductions and Other Conservation Translocations. Version 1.0; IUCN Species Survival Commission: Gland, Switzerland, 2013. [Google Scholar]
- Maschinski, J.; Haskins, K.E. Plant Reintroduction in a Changing Climate. Promises and Peril; Island Press: Washington, DC, USA, 2012. [Google Scholar]
- van Tienderen, P.H. Elasticities and the link between demographic and evolutionary dynamics. Ecology 2000, 81, 666–679. [Google Scholar] [CrossRef]
- Walsh, B.; Lynch, M. Evolution and Selection of Quantitative Traits; Oxford University Press: Oxford, UK, 2018. [Google Scholar]
- Kelly, E.; Phillips, B. How many and when? Optimising targeted gene flow for a step change in the environment. Ecol. Lett. 2019, 22, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Morris, W.F.; Doak, D.F. Quantitative Conservation Biology: Theory and Practice of Population Viability Analysis; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- van Rossum, F.; Hardy, O.J. Guidelines for genetic monitoring of translocated plant populations. Conserv. Biol. 2022, 36, e13670. [Google Scholar] [CrossRef]
- Acquaah, G. Conventional plant breeding principles and techniques. In Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer: Cham, Switzerland, 2015; pp. 115–158. [Google Scholar]
- Griffiths, A.J.F.; Miller, J.H.; Suzuki, D.T.; Lewontin, R.C.; Gelbart, W.M. An Introduction to Genetic Analysis, 7th ed.; W.H. Freeman: New York, NY, USA, 2000. [Google Scholar]
- Chakravarti, L.J.; Beltran, V.H.; van Oppen, M.J.H. Rapid thermal adaptation in photosymbionts of reef-building corals. Glob. Chang. Biol. 2017, 23, 4675–4688. [Google Scholar] [CrossRef] [PubMed]
- Acquaah, G. Principles of Plant Genetics and Breeding; Wiley-Blackwell: Chichester, UK, 2012. [Google Scholar]
- Orton, T.J. Horticultural Plant Breeding; Elsevier: London, UK, 2020. [Google Scholar]
- Chen, Y.; Lübberstedt, T. Molecular basis of trait correlations. Trends Plant Sci. 2010, 15, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Galloway, L.F.; Burgess, K.S. Artificial selection on flowering time: Influence on reproductive phenology across natural light environments. J. Ecol. 2012, 100, 852–861. [Google Scholar] [CrossRef]
- Satake, A.; Kawagoe, T.; Saburi, Y.; Chiba, Y.; Sakurai, G.; Kudoh, H. Forecasting flowering phenology under climate warming by modelling the regulatory dynamics of flowering-time genes. Nat. Commun. 2013, 4, 2303. [Google Scholar] [CrossRef] [Green Version]
- Frankham, R.; Ballou, J.D.; Ralls, K.; Eldridge, M.D.B.; Dudash, M.R.; Fenster, C.B.; Lacy, R.C.; Sunnucks, P. Genetic Managment of Fragmented Animal and Plant Populations; Oxford University Press: Oxford, UK, 2017. [Google Scholar]
- Ellstrand, N.C.; Rieseberg, L.H. When gene flow really matters: Gene flow in applied evolutionary biology. Evol. Appl. 2016, 9, 833–836. [Google Scholar] [CrossRef] [Green Version]
- Gómez, J.M.; González-Megías, A.; Lorite, J.; Abdelaziz, M.; Perfectti, F. The silent extinction: Climate change and the potential hybridization-mediated extinction of endemic high-mountain plants. Biodivers. Conserv. 2015, 24, 1843–1857. [Google Scholar] [CrossRef]
- Inoue, K.; Araki, T.; Endo, M. Circadian clock during plant development. J. Plant Res. 2018, 131, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Züst, T.; Heichinger, C.; Grossniklaus, U.; Harrington, R.; Kliebenstein, D.J.; Turnbull, L.A. Natural enemies drive geographic variation in plant defenses. Science 2012, 338, 116–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tigano, A.; Friesen, V.L. Genomics of local adaptation with gene flow. Mol. Ecol. 2016, 25, 2144–2164. [Google Scholar] [CrossRef] [PubMed]
- Ensslin, A.; Tschöpe, O.; Burkart, M.; Joshi, J. Fitness decline and adaptation to novel environments in ex situ plant collections: Current knowledge and future perspectives. Biol. Conserv. 2015, 192, 394–401. [Google Scholar] [CrossRef]
- Sun, Q.; Lai, L.; Zhou, J.; Yi, S.; Liu, X.; Guo, J.; Zheng, Y. Differences in ecological traits between plants grown in situ and ex situ and implications for conservation. Sustainability 2022, 14, 5199. [Google Scholar] [CrossRef]
- Havens, K.; Guerrant, E.O., Jr.; Maunder, M.; Vitt, P. Guidelines for ex situ conservation collection management: Minimizing risks. In Ex Situ Plant Conservation: Supporting Species Survival in the Wild; Guerrant, E.O., Jr., Havens, K., Maunder, M., Eds.; Island Press: Washington, DC, USA, 2004; pp. 454–473. [Google Scholar]
- Husband, B.C.; Campbell, L.G. Population responses to novel environments: Implications for ex situ plant conservation. In Ex Situ Plant Conservation: Supporting Species Survival in the Wild; Guerrant, E.O., Jr., Havens, K., Maunder, M., Eds.; Island Press: Washington, DC, USA, 2004; pp. 231–266. [Google Scholar]
- Crutsinger, G.M.; Collins, M.D.; Fordyce, J.A.; Gompert, Z.; Nice, C.C.; Sanders, N.J. Plant genotypic diversity predicts community structure and governs an ecosystem process. Science 2006, 313, 966–968. [Google Scholar] [CrossRef] [Green Version]
- Johnson, M.T.J.; Agrawal, A.A. Plant genotype and environment interact to shape a diverse arthropod community on evening primrose (Oenothera biennis). Ecology 2005, 86, 874–885. [Google Scholar] [CrossRef] [Green Version]
- Crawford, K.M.; Rudgers, J.A. Genetic diversity within a dominant plant outweighs plant species diversity in structuring an arthropod community. Ecology 2013, 94, 1025–1035. [Google Scholar] [CrossRef]
- Catford, J.A.; Jansson, R.; Nilsson, C. Reducing redundancy in invasion ecology by integrating hypotheses into a single theoretical framework. Divers. Distrib. 2009, 15, 22–40. [Google Scholar] [CrossRef] [Green Version]
- Wadgymar, S.M.; Weis, A.E. Phenological mismatch and the effectiveness of assisted gene flow. Conserv. Biol. 2017, 31, 547–558. [Google Scholar] [CrossRef]
- Prieto-Benítez, S.; Morente-López, J.; Rubio Teso, M.L.; Lara-Romero, C.; García-Fernández, A.; Torres, E.; Iriondo, J.M. Evaluating assisted gene flow in marginal populations of a high mountain species. Front. Ecol. Evol. 2021, 9, 638837. [Google Scholar] [CrossRef]
- Andersson, D.I.; Jerlström-Hultqvist, J.; Näsvall, J. Evolution of new functions de novo and from preexisting genes. Cold Spring Harb. Perspect. Biol. 2015, 7, a017996. [Google Scholar] [CrossRef] [PubMed]
- Kronholm, I.; Collins, S. Epigenetic mutations can both help and hinder adaptive evolution. Mol. Ecol. 2016, 25, 1856–1868. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.; Soltis, P.S.; Soltis, D.E.; Yang, B. Considerations in adapting CRISPR/Cas9 in nongenetic model plant systems. Appl. Plant Sci. 2020, 8, e11314. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.A.; Monaco, T.A.; Larson, S.R.; Hamerlynck, E.P.; Crain, J.L. Using genomic selection to develop performance-based restoration plant materials. Int. J. Mol. Sci. 2022, 23, 8275. [Google Scholar] [CrossRef] [PubMed]
- Shamshad, M.; Sharma, A. The usage of genomic selection strategy in plant breeding. In Next Generation Plant Breeding; Çiftçi, Y.Ö., Ed.; IntechOpen: London, UK, 2018; pp. 93–108. [Google Scholar]
- Willi, Y.; Van Buskirk, J.; Hoffmann, A.A. Limits to the adaptive potential of small populations. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 433–458. [Google Scholar] [CrossRef]
- Huang, Y.; Tran, I.; Agrawal, A.F. Does genetic variation maintained by environmental heterogeneity facilitate adaptation to novel selection? Am. Nat. 2016, 188, 27–37. [Google Scholar] [CrossRef] [Green Version]
| Term | Definition | Reference |
|---|---|---|
| Adaptive introgression | Movement of genetic material from the genome of one species into the genome of another through repeated interbreeding for increasing species’ ability to respond to a changing climate. | [63] |
| Assisted evolution | Any approach that accelerates the rate of naturally occurring evolutionary processes. | [62] |
| Assisted gene flow | The managed movement of individuals or gametes between populations within species ranges to mitigate local maladaptation in the short and long term. | [31] |
| Facilitated adaptation | Intervention to rescue a target population or species by endowing it with adaptive alleles, or gene variants, using genetic engineering. | [32] |
| Genetic adaptation | Genetic translocation to introduce new alleles for traits important for environmental change. | [64] 1 |
| Targeted gene flow | Individuals that are pre-adapted to future conditions are translocated to increase the adaptive capacity of another population. | [65] 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres, E.; García-Fernández, A.; Iñigo, D.; Lara-Romero, C.; Morente-López, J.; Prieto-Benítez, S.; Rubio Teso, M.L.; Iriondo, J.M. Facilitated Adaptation as A Conservation Tool in the Present Climate Change Context: A Methodological Guide. Plants 2023, 12, 1258. https://doi.org/10.3390/plants12061258
Torres E, García-Fernández A, Iñigo D, Lara-Romero C, Morente-López J, Prieto-Benítez S, Rubio Teso ML, Iriondo JM. Facilitated Adaptation as A Conservation Tool in the Present Climate Change Context: A Methodological Guide. Plants. 2023; 12(6):1258. https://doi.org/10.3390/plants12061258
Chicago/Turabian StyleTorres, Elena, Alfredo García-Fernández, Diana Iñigo, Carlos Lara-Romero, Javier Morente-López, Samuel Prieto-Benítez, María Luisa Rubio Teso, and José M. Iriondo. 2023. "Facilitated Adaptation as A Conservation Tool in the Present Climate Change Context: A Methodological Guide" Plants 12, no. 6: 1258. https://doi.org/10.3390/plants12061258
APA StyleTorres, E., García-Fernández, A., Iñigo, D., Lara-Romero, C., Morente-López, J., Prieto-Benítez, S., Rubio Teso, M. L., & Iriondo, J. M. (2023). Facilitated Adaptation as A Conservation Tool in the Present Climate Change Context: A Methodological Guide. Plants, 12(6), 1258. https://doi.org/10.3390/plants12061258






