Omics Approaches in Invasion Biology: Understanding Mechanisms and Impacts on Ecological Health
Abstract
1. Introduction
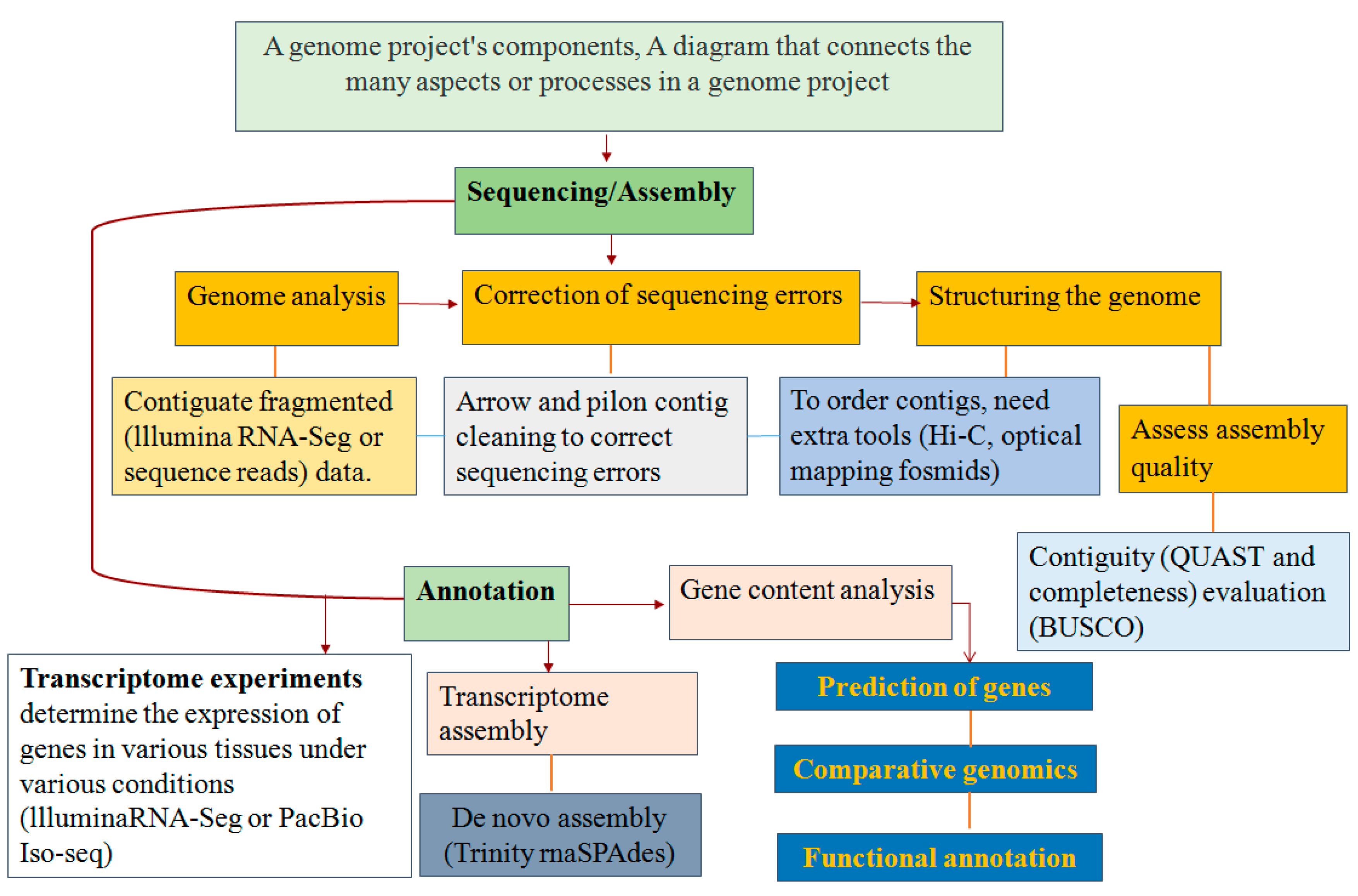
2. Genomics as a Key Tool for Understanding Plant Invasiveness
2.1. Population Genomics and Their Research Method
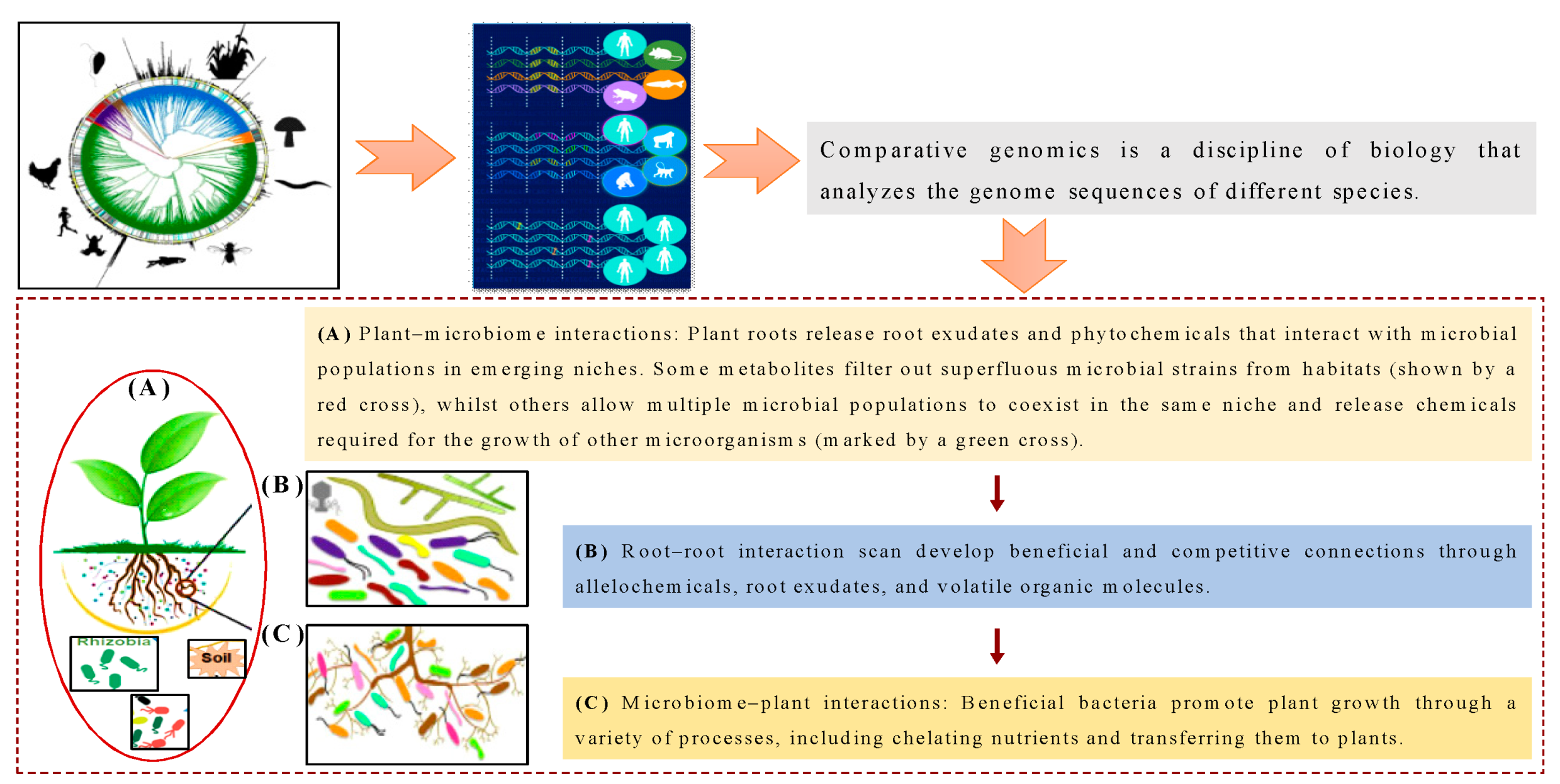
2.1.1. Comparative Genomics
2.1.2. Role of the Soil Microbiome
2.1.3. Impacts of Plant Invasion on Soil Ecological Functions
3. The Multi-Omics Methods for Plant Invasion
| Invasive Plant Species | Techniques of Omic | Findings of Study | References |
|---|---|---|---|
| Ageratina adenophora (crofton weed). | Proteomic (root exudates) | Identified proteins involved in allelopathy, which may contribute to the invasiveness of the plant. | [96,97,98] |
| Acacia saligna (golden wattle). | Proteomics (N fixing root nodules) | Identified protein elaboration in N fixation and transport; it enhances the plant’s growth and competitive ability in nutrient-poor soils. | [99] |
| Microstegium vimineum (Japanese stiltgrass). | Proteomic (invasive and native populations) | Identified differences in protein expression related to photosynthesis, stress response. | [73] |
| Cytisus scoparius (Scotch broom). | Proteomics (leaves and roots) | Identified proteins involved in plant defense, nutrient uptake. | [100] |
| S. alterniflora. | Chemico-proteomics | The function of H2S signaling in the adaptation of an invasive plant species and the important role of H2S adaptation in S. alterniflora to saline environments. | [101] |
| R. solanacearum. | Proteomics | Plant–bacterium interactions. | [102] |
| Incompatible rice/Magnaporthe grisae. | Proteomics | Plant–pathogen relationship; it is important in apoplastic protein patterns that occur during pathogen infection. | [103] |
| Potato with Ralstonia solanacearum UW551. | Proteomics | T3Es of R. solanacearum can subvert potato root immune-related proteins in a redundant manner. | [104] |
| Tomato (Solanum lycopersicum) fruit was invaded by Sclerotinia rolfsii. | Proteomics | To prioritize candidate proteins for storage organ quality improvement. | [105] |
| Aspergillus terreus invades Solanum tuberosum L. | Proteomics | During colonization, TA. terreus differently activated enzymes in potato tubers. | [106] |
| Phytophthora infestans, the pathogen responsible for potato late blight. | Proteomics | The potential magnitude of proteins encoded in the genome. | [107] |
| Expressed in Nicotiana benthamiana, R. solan. | Proteomics | Pathogens can adapt to the specific host they encounter. | [108] |
| Interactions between plants and viruses, bacteria, fungi, and nematode. | Proteomics | Interactions between plants and viruses, bacteria, fungi, and nematodes were identified and reported in proteomic studies. | [109] |
| Arabidopsis thaliana plants. | Proteomics | Providing insight into the signaling networks of a particular cell type. | [110] |
| The symbiotic interaction between Brassica napus and Piriformospora indica. | Proteomics | GO and KEGG pathway analysis revealed gene sets involved in metabolic processes. | [111] |
| Magnaporthe oryzae (M. oryzae). | Proteomics | Response to M. oryzae invasion; the iTRAQ approach was utilized to identify differentially expressed proteins (DEPs) in both the durable, resistant rice variety Gangyuan8 (GY8) and the susceptible rice variety Lijiangxintuanheigu (LTH). | [112] |
| Study interactions between plants and pathogens | Proteomics | Interactions between plants and pathogens in compatible systems. | [113] |
| Potato, a model for periderm. | Proteomics | Early tuber growth in potatoes; periderm tissue replaces the epidermis. | [114] |
| Microbial pathogens. | Proteomics | Bacterial interactions among distinct bacterial taxa, including symbiotic, pathogenic, and commensal bacteria. | [115] |
| Tomato | Proteomics | Proteome study investigation of the dynamics of various disease responses in tomato. | [116] |
| Hybrids of Solanum differing in resistance to Dickeya solani. | Proteomics | Significant differences were observed in the large-fold of various proteins between resistant and susceptible potato cultivars, and diploid clones were induced. | [117] |
| Proteomics toward the improvement of crop productivity and stress resistance. | Proteomics | The limitations of non-model organism proteomics techniques and data interpretation. | [118] |
| Plant. | Proteomics | Plant-specific issues on how proteomics can help plant systems biology. | [119] |
| Plant. | Proteomics | Plant proteomics is currently in its early stages and is subject to a significant impact on plant biology. | [120] |
| Alternanthera philoxeroides (Alligator weed). | Proteomics | The response of Alternanthera philoxeroides roots stems, and leaves to potassium deficiency stress. | [121] |
| Gibberella stalk rot in maize. | Proteomics | The defense response of corn stalks against graminearum, proteins from various immune-related pathways. | [122] |
| Rice in biotic stress. | Proteomics, metabolomics | Proteins and metabolites defense response of rice to biotic stress. | [123] |
4. Invasive Species and Environmental Change
Review of the Application of Omics to Invasion Biology and Ecology
5. Conclusions and Remarks
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ravet, K.; Patterson, E.L.; Krähmer, H.; Hamouzová, K.; Fan, L.; Jasieniuk, M.; Lawton-Rauh, A.; Malone, J.M.; McElroy, J.S.; Merotto, A., Jr. The power and potential of genomics in weed biology and management. Pest Manag. Sci. 2018, 74, 2216–2225. [Google Scholar] [CrossRef] [PubMed]
- Mable, B.K. Conservation of adaptive potential and functional diversity: Integrating old and new approaches. Conserv. Genet. 2019, 20, 89–100. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Sheflin, A.M.; Manter, D.K.; Vivanco, J.M. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 2012, 48, 489–499. [Google Scholar] [CrossRef]
- Mounger, J.; Ainouche, M.L.; Bossdorf, O.; Cavé-Radet, A.; Li, B.; Parepa, M.; Salmon, A.; Yang, J.; Richards, C.L. Epigenetics and the success of invasive plants. Philos. Trans. R. Soc. B 2021, 376, 20200117. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.R.; Ochsenkühn, M.A.; Kazlak, A.M.; Moustafa, A.; Amin, S.A. The coral microbiome: Towards an understanding of the molecular mechanisms of coral–microbiota interactions. FEMS Microbiol. Rev. 2023, 47, fuad005. [Google Scholar] [CrossRef]
- Xu, L.; Pierroz, G.; Wipf, H.M.-L.; Gao, C.; Taylor, J.W.; Lemaux, P.G.; Coleman-Derr, D. Holo-omics for deciphering plant-microbiome interactions. Microbiome 2021, 9, 69. [Google Scholar] [CrossRef]
- Prakash, O.; Shouche, Y.; Jangid, K.; Kostka, J.E. Microbial cultivation and the role of microbial resource centers in the omics era. Appl. Microbiol. Biotechnol. 2013, 97, 51–62. [Google Scholar] [CrossRef]
- Beale, D.J.; Karpe, A.V.; Ahmed, W. Beyond metabolomics: A review of multi-omics-based approaches. In Microbial Metabolomics: Applications in Clinical, Environmental, and Industrial Microbiology; Springer: Berlin/Heidelberg, Germany, 2016; pp. 289–312. [Google Scholar]
- Bell, T.H.; Joly, S.; Pitre, F.E.; Yergeau, E. Increasing phytoremediation efficiency and reliability using novel omics approaches. Trends Biotechnol. 2014, 32, 271–280. [Google Scholar] [CrossRef]
- Crandall, S.G.; Gold, K.M.; Jiménez-Gasco, M.d.M.; Filgueiras, C.C.; Willett, D.S. A multi-omics approach to solving problems in plant disease ecology. PLoS ONE 2020, 15, e0237975. [Google Scholar] [CrossRef]
- Kaul, S.; Sharma, T.K.; Dhar, M. “Omics” tools for better understanding the plant–endophyte interactions. Front. Plant Sci. 2016, 7, 955. [Google Scholar] [CrossRef]
- Oberbeckmann, S.; Osborn, A.M.; Duhaime, M.B. Microbes on a bottle: Substrate, season and geography influence community composition of microbes colonizing marine plastic debris. PLoS ONE 2016, 11, e0159289. [Google Scholar] [CrossRef]
- Apweiler, R.; Aslanidis, C.; Deufel, T.; Gerstner, A.; Hansen, J.; Hochstrasser, D.; Kellner, R.; Kubicek, M.; Lottspeich, F.; Maser, E. Approaching clinical proteomics: Current state and future fields of application in fluid proteomics. Clin. Chem. Lab. Med. 2009, 47, 724–744. [Google Scholar] [CrossRef]
- Kumar, M.; Kuzhiumparambil, U.; Pernice, M.; Jiang, Z.; Ralph, P.J. Metabolomics: An emerging frontier of systems biology in marine macrophytes. Algal Res. 2016, 16, 76–92. [Google Scholar] [CrossRef]
- Cranford, S.W.; De Boer, J.; Van Blitterswijk, C.; Buehler, M.J. Materiomics: An-omics approach to biomaterials research. Adv. Mater. 2013, 25, 802–824. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, L.; Liu, Z.; Zhao, Z.; Zhao, J.; Wang, Z.; Zhou, G.; Liu, P.; Liu, M. Transcriptome and metabolome profiling unveil the mechanisms of Ziziphus jujuba Mill. peel coloration. Food Chem. 2020, 312, 125903. [Google Scholar] [CrossRef]
- Chatterjee, A.; Shankar, A.; Singh, S.; Kesari, V.; Rai, R.; Patel, A.K.; Rai, L. Beneficial microorganisms and abiotic stress tolerance in plants. In Approaches for Enhancing Abiotic Stress Tolerance in Plants; CRC Press: Boca Raton, FL, USA, 2019; pp. 473–502. [Google Scholar]
- Mamta, B.; Rajam, M. RNAi technology: A new platform for crop pest control. Physiol. Mol. Biol. Plants 2017, 23, 487–501. [Google Scholar] [CrossRef]
- Yurkovich, J.T.; Tian, Q.; Price, N.D.; Hood, L. A systems approach to clinical oncology uses deep phenotyping to deliver personalized care. Nat. Rev. Clin. Oncol. 2020, 17, 183–194. [Google Scholar] [CrossRef]
- Ding, A.; Zhang, R.; Ngo, H.H.; He, X.; Ma, J.; Nan, J.; Li, G. Life cycle assessment of sewage sludge treatment and disposal based on nutrient and energy recovery: A review. Sci. Total Environ. 2021, 769, 144451. [Google Scholar] [CrossRef]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; De Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Prim. 2021, 1, 59. [Google Scholar] [CrossRef]
- Li, F.; Li, C.; Chen, Y.; Liu, J.; Zhang, C.; Irving, B.; Fitzsimmons, C.; Plastow, G.; Guan, L.L. Host genetics influence the rumen microbiota and heritable rumen microbial features associate with feed efficiency in cattle. Microbiome 2019, 7, 92. [Google Scholar] [CrossRef]
- Benson, A.K.; Kelly, S.A.; Legge, R.; Ma, F.; Low, S.J.; Kim, J.; Zhang, M.; Oh, P.L.; Nehrenberg, D.; Hua, K. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18933–18938. [Google Scholar] [CrossRef] [PubMed]
- Ferone, M.; Gowen, A.; Fanning, S.; Scannell, A.G. Microbial detection and identification methods: Bench top assays to omics approaches. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3106–3129. [Google Scholar] [CrossRef] [PubMed]
- Van Emon, J.M. The omics revolution in agricultural research. J. Agric. Food Chem. 2016, 64, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Kouselya, M.; Muhilan, B.; Chattopadhyay, I. 1 Omics approaches for characterization of environmental microorganisms. In Environmental Microbiology: Emerging Technologies; De Gruyter: Vienna, Austria, 2022. [Google Scholar]
- Gaertner, M.; Holmes, P.M.; Richardson, D.M. Biological invasions, resilience and restoration. In Restoration Ecology: The New Frontier; Wiley Online Library: Hoboken, NJ, USA, 2012; pp. 265–280. [Google Scholar]
- Gladman, N.; Goodwin, S.; Chougule, K.; McCombie, W.R.; Ware, D. Era of gapless plant genomes: Innovations in sequencing and mapping technologies revolutionize genomics and breeding. Curr. Opin. Biotechnol. 2023, 79, 102886. [Google Scholar] [CrossRef]
- Gioria, M.; Hulme, P.E.; Richardson, D.M.; Pyšek, P. Why Are Invasive Plants Successful? Annu. Rev. Plant Biol. 2022, 47, 777–780. [Google Scholar] [CrossRef]
- Moravcová, L.; Pyšek, P.; Jarošík, V.; Pergl, J. Getting the right traits: Reproductive and dispersal characteristics predict the invasiveness of herbaceous plant species. PLoS ONE 2015, 10, e0123634. [Google Scholar] [CrossRef]
- Hodkinson, T.R.; Perdereau, A.; Klaas, M.; Cormican, P.; Barth, S. Genotyping by sequencing and plastome analysis finds high genetic variability and geographical structure in Dactylis glomerata L. in Northwest Europe despite lack of ploidy variation. Agronomy 2019, 9, 342. [Google Scholar] [CrossRef]
- North, H.L.; McGaughran, A.; Jiggins, C.D. Insights into invasive species from whole-genome resequencing. Mol. Ecol. 2021, 30, 6289–6308. [Google Scholar] [CrossRef]
- Prentis, P.J.; Wilson, J.R.; Dormontt, E.E.; Richardson, D.M.; Lowe, A.J. Adaptive evolution in invasive species. Trends Plant Sci. 2008, 13, 288–294. [Google Scholar] [CrossRef]
- Le Roux, J.; Wieczorek, A. Molecular systematics and population genetics of biological invasions: Towards a better understanding of invasive species management. Ann. Appl. Biol. 2009, 154, 1–17. [Google Scholar] [CrossRef]
- Wu, N.; Zhang, S.; Li, X.; Cao, Y.; Liu, X.; Wang, Q.; Liu, Q.; Liu, H.; Hu, X.; Zhou, X.J. Fall webworm genomes yield insights into rapid adaptation of invasive species. Nat. Ecol. Evol. 2019, 3, 105–115. [Google Scholar] [CrossRef]
- Ekblom, R.; Galindo, J. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 2011, 107, 1–15. [Google Scholar] [CrossRef]
- MariaKatherineMejia-Guerra, D.; Sheehan, M.J. Genomic Resources for Breeding in Alfalfa: Availability, Utility, and Adoption. In The Alfalfa Genome; Springer International Publishing: Cham, Switzerland, 2021; p. 177. [Google Scholar]
- Hohenlohe, P.A.; Funk, W.C.; Rajora, O.P. Population genomics for wildlife conservation and management. Mol. Ecol. 2021, 30, 62–82. [Google Scholar] [CrossRef]
- Zhou, Q.; Xin, Z.; Wang, Y.; Miao, R.; Liu, Z.; Zong, L.; Li, X.; Ma, Q.; Liang, W.; Yu, H. The Adaptive Capacity of Alien and Rare Species in China. Forests 2022, 13, 2005. [Google Scholar] [CrossRef]
- Paz-Kagan, T.; Silver, M.; Panov, N.; Karnieli, A. Multispectral approach for identifying invasive plant species based on flowering phenology characteristics. Remote Sens. 2019, 11, 953. [Google Scholar] [CrossRef]
- Pfennig, K.; Pfennig, D. Character displacement: Ecological and reproductive responses to a common evolutionary problem. Q. Rev. Biol. 2009, 84, 253–276. [Google Scholar] [CrossRef]
- De Vries, R.P.; Riley, R.; Wiebenga, A.; Aguilar-Osorio, G.; Amillis, S.; Uchima, C.A.; Anderluh, G.; Asadollahi, M.; Askin, M.; Barry, K. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017, 18, 28. [Google Scholar] [CrossRef]
- Ogura, A.; Akizuki, Y.; Imoda, H.; Mineta, K.; Gojobori, T.; Nagai, S. Comparative genome and transcriptome analysis of diatom, Skeletonema costatum, reveals evolution of genes for harmful algal bloom. BMC Genom. 2018, 19, 765. [Google Scholar] [CrossRef]
- Bennetzen, J.L.; Wang, H. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu. Rev. Plant Biol. 2014, 65, 505–530. [Google Scholar] [CrossRef]
- Jian-Hui, H.; Xing-Guo, H.; Qin-Er, Y.; Yong-Fei, B. Fundamentals of invasive species biology and ecology. Biodivers. Sci. 2003, 11, 240. [Google Scholar]
- Blommaert, J. Genome size evolution: Towards new model systems for old questions. Proc. R. Soc. B 2020, 287, 20201441. [Google Scholar] [CrossRef] [PubMed]
- Kraaijeveld, K. Genome size and species diversification. Evol. Biol. 2010, 37, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Bock, D.G.; Caseys, C.; Cousens, R.D.; Hahn, M.A.; Heredia, S.M.; Hübner, S.; Turner, K.G.; Whitney, K.D.; Rieseberg, L.H. What we still don’t know about invasion genetics. In Invasion Genetics: The Baker and Stebbins Legacy; Wiley Online Library: Hoboken, NJ, USA, 2016; pp. 346–370. [Google Scholar]
- Großkinsky, D.K.; Syaifullah, S.J.; Roitsch, T. Integration of multi-omics techniques and physiological phenotyping within a holistic phenomics approach to study senescence in model and crop plants. J. Exp. Bot. 2018, 69, 825–844. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-S.; Qian, Z.-H.; Shi, T.; Li, Z.-Z.; Chen, J.-M. Chromosome-level genome assembly of the aquatic plant Nymphoides indica reveals transposable element bursts and NBS-LRR gene family expansion shedding light on its invasiveness. DNA Res. 2022, 29, dsac022. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef]
- Gao, G.F.; Li, H.; Shi, Y.; Yang, T.; Gao, C.H.; Fan, K.; Zhang, Y.; Zhu, Y.G.; Delgado-Baquerizo, M.; Zheng, H.L. Continental-scale plant invasions reshuffle the soil microbiome of blue carbon ecosystems. Glob. Change Biol. 2022, 28, 4423–4438. [Google Scholar] [CrossRef]
- Yin, L.; Liu, B.; Wang, H.; Zhang, Y.; Wang, S.; Jiang, F.; Ren, Y.; Liu, H.; Liu, C.; Wan, F. The rhizosphere microbiome of Mikania micrantha provides insight into adaptation and invasion. Front. Microbiol. 2020, 11, 1462. [Google Scholar] [CrossRef]
- Coats, V.C.; Rumpho, M.E. The rhizosphere microbiota of plant invaders: An overview of recent advances in the microbiomics of invasive plants. Front. Microbiol. 2014, 5, 368. [Google Scholar] [CrossRef]
- Trognitz, F.; Hackl, E.; Widhalm, S.; Sessitsch, A. The role of plant–microbiome interactions in weed establishment and control. FEMS Microbiol. Ecol. 2016, 92, fiw138. [Google Scholar] [CrossRef]
- Weidenhamer, J.D.; Callaway, R.M. Direct and indirect effects of invasive plants on soil chemistry and ecosystem function. J. Chem. Ecol. 2010, 36, 59–69. [Google Scholar] [CrossRef]
- Yang, W.; Zhao, H.; Leng, X.; Cheng, X.; An, S. Soil organic carbon and nitrogen dynamics following Spartina alterniflora invasion in a coastal wetland of eastern China. Catena 2017, 156, 281–289. [Google Scholar] [CrossRef]
- Sapsford, S.J.; Brandt, A.J.; Davis, K.T.; Peralta, G.; Dickie, I.A.; Gibson, R.D.; Green, J.L.; Hulme, P.E.; Nuñez, M.A.; Orwin, K.H. Towards a framework for understanding the context dependence of impacts of non-native tree species. Funct. Ecol. 2020, 34, 944–955. [Google Scholar] [CrossRef]
- Willis, C.G.; Ruhfel, B.R.; Primack, R.B.; Miller-Rushing, A.J.; Losos, J.B.; Davis, C.C. Favorable climate change response explains non-native species’ success in Thoreau’s woods. PLoS ONE 2010, 5, e8878. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Krauss, G.-J.; Sole, M.; Krauss, G.; Schlosser, D.; Wesenberg, D.; Baerlocher, F. Fungi in freshwaters: Ecology, physiology and biochemical potential. FEMS Microbiol. Rev. 2011, 35, 620–651. [Google Scholar] [CrossRef]
- Diwan, D.; Rashid, M.; Vaishnav, A. Current understanding of plant-microbe interaction through the lenses of multi-omics approaches and their benefits in sustainable agriculture. Microbiol. Res. 2022, 265, 127180. [Google Scholar] [CrossRef]
- Mishra, A.K.; Sudalaimuthuasari, N.; Hazzouri, K.M.; Saeed, E.E.; Shah, I.; Amiri, K.M. Tapping into Plant–Microbiome Interactions through the Lens of Multi-Omics Techniques. Cells 2022, 11, 3254. [Google Scholar] [CrossRef]
- Zavahir, J.S.; Wijepala, P.C.; Seneviratne, G. Role of Microbial Communities in Plant–Microbe Interactions, Metabolic Cooperation, and Self-Sufficiency Leading to Sustainable Agriculture. Role Microb. Communities Sustain. 2021, 29, 1–35. [Google Scholar]
- Constán-Nava, S.; Soliveres, S.; Torices, R.; Serra, L.; Bonet, A. Direct and indirect effects of invasion by the alien tree Ailanthus altissima on riparian plant communities and ecosystem multifunctionality. Biol. Invasions 2015, 17, 1095–1108. [Google Scholar] [CrossRef]
- Rasool, F.; Khan, M.R.; Schneider, M.; Uzair, M.; Aqeel, M.; Ajmal, W.; Léon, J.; Naz, A.A. Transcriptome unveiled the gene expression patterns of root architecture in drought-tolerant and sensitive wheat genotypes. Plant Physiol. Biochem. 2022, 178, 20–30. [Google Scholar] [CrossRef]
- Lockwood, B.L.; Somero, G.N. Transcriptomic responses to salinity stress in invasive and native blue mussels (Genus mytilus). Mol. Ecol. 2011, 20, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Walker, W.B.; Roy, A.; Anderson, P.; Schlyter, F.; Hansson, B.S.; Larsson, M.C. Transcriptome analysis of gene families involved in chemosensory function in Spodoptera littoralis (Lepidoptera: Noctuidae). BMC Genom. 2019, 20, 428. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.-S.; Manoharan, B.; Dhandapani, V.; Jegadeesan, S.; Rutherford, S.; Wan, J.S.; Huang, P.; Dai, Z.-C.; Du, D.-L. Pathogen resistance in Sphagneticola trilobata (Singapore daisy): Molecular associations and differentially expressed genes in response to disease from a widespread fungus. Genetica 2022, 150, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Rutherford, S.; Qi, S.; Huang, P.; Dai, Z.; Du, D. Transcriptome profiling of Arabidopsis thaliana roots in response to allelopathic effects of Conyza canadensis. Ecotoxicology 2022, 31, 53–63. [Google Scholar] [CrossRef]
- Luo, L.; Kong, X.; Gao, Z.; Zheng, Y.; Yang, Y.; Li, X.; Yang, D.; Geng, Y.; Yang, Y. Comparative transcriptome analysis reveals ecological adaption of cold tolerance in northward invasion of Alternanthera philoxeroides. BMC Genom. 2020, 21, 532. [Google Scholar] [CrossRef]
- Saminathan, T.; Malkaram, S.A.; Patel, D.; Taylor, K.; Hass, A.; Nimmakayala, P.; Huber, D.H.; Reddy, U.K. Transcriptome analysis of invasive plants in response to mineral toxicity of reclaimed coal-mine soil in the appalachian region. Environ. Sci. Technol. 2015, 49, 10320–10329. [Google Scholar] [CrossRef]
- Allison, S.D.; Gartner, T.B.; Holland, K.; Weintraub, M.; Sinsabaugh, R.L. Soil enzymes: Linking proteomics and ecological processes. In Manual of Environmental Microbiology; Wiley Online Library: Hoboken, NJ, USA, 2007; pp. 704–711. [Google Scholar]
- Ye, W.; Wang, T.; Wei, W.; Lou, S.; Lan, F.; Zhu, S.; Li, Q.; Ji, G.; Lin, C.; Wu, X. The full-length transcriptome of Spartina alterniflora reveals the complexity of high salt tolerance in monocotyledonous halophyte. Plant Cell Physiol. 2020, 61, 882–896. [Google Scholar] [CrossRef]
- Elsheikh, E.A.; El-Keblawy, A.; Mosa, K.A.; Okoh, A.I.; Saadoun, I. Role of endophytes and rhizosphere microbes in promoting the invasion of exotic plants in arid and semi-arid areas: A review. Sustainability 2021, 13, 13081. [Google Scholar] [CrossRef]
- Immel, F.; Renaut, J.; Masfaraud, J.-F. Physiological response and differential leaf proteome pattern in the European invasive Asteraceae Solidago canadensis colonizing a former cokery soil. J. Proteom. 2012, 75, 1129–1143. [Google Scholar] [CrossRef]
- Tugizimana, F.; Mhlongo, M.I.; Piater, L.A.; Dubery, I.A. Metabolomics in plant priming research: The way forward? Int. J. Mol. Sci. 2018, 19, 1759. [Google Scholar] [CrossRef]
- Nephali, L.; Piater, L.A.; Dubery, I.A.; Patterson, V.; Huyser, J.; Burgess, K.; Tugizimana, F. Biostimulants for plant growth and mitigation of abiotic stresses: A metabolomics perspective. Metabolites 2020, 10, 505. [Google Scholar] [CrossRef]
- Kalu, C.M.; Ogola, H.J.; Selvarajan, R.; Tekere, M.; Ntushelo, K. Correlations between root metabolomics and bacterial community structures in the phragmites australis under acid mine drainage-Polluted wetland ecosystem. Curr. Microbiol. 2022, 79, 34. [Google Scholar] [CrossRef]
- Ramalingam, A.; Kudapa, H.; Pazhamala, L.T.; Weckwerth, W.; Varshney, R.K. Proteomics and metabolomics: Two emerging areas for legume improvement. Front. Plant Sci. 2015, 6, 1116. [Google Scholar] [CrossRef]
- Skubel, S.A.; Su, X.; Poulev, A.; Foxcroft, L.C.; Dushenkov, V.; Raskin, I. Metabolomic differences between invasive alien plants from native and invaded habitats. Sci. Rep. 2020, 10, 9749. [Google Scholar] [CrossRef]
- Xiong, R.; Gunter, C.; Fleming, E.; Vernon, S.D.; Bateman, L.; Unutmaz, D.; Oh, J. Multi-‘omics of gut microbiome-host interactions in short-and long-term myalgic encephalomyelitis/chronic fatigue syndrome patients. Cell Host Microbe 2023, 31, 273–287.e275. [Google Scholar] [CrossRef]
- Van Der Heijden, M.G.; Bardgett, R.D.; Van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 2018, 9, 112. [Google Scholar] [CrossRef]
- Kamutando, C.N.; Vikram, S.; Kamgan-Nkuekam, G.; Makhalanyane, T.P.; Greve, M.; Le Roux, J.J.; Richardson, D.M.; Cowan, D.A.; Valverde, A. The functional potential of the rhizospheric microbiome of an invasive tree species, Acacia dealbata. Microb. Ecol. 2019, 77, 191–200. [Google Scholar] [CrossRef]
- Kowalski, K.P.; Bacon, C.; Bickford, W.; Braun, H.; Clay, K.; Leduc-Lapierre, M.; Lillard, E.; McCormick, M.K.; Nelson, E.; Torres, M. Advancing the science of microbial symbiosis to support invasive species management: A case study on Phragmites in the Great Lakes. Front. Microbiol. 2015, 6, 95. [Google Scholar] [CrossRef]
- Zhang, H.-Y.; Goncalves, P.; Copeland, E.; Qi, S.-S.; Dai, Z.-C.; Li, G.-L.; Wang, C.-Y.; Du, D.-L.; Thomas, T. Invasion by the weed Conyza canadensis alters soil nutrient supply and shifts microbiota structure. Soil Biol. Biochem. 2020, 143, 107739. [Google Scholar] [CrossRef]
- Mei, Y.-H.; Li, X.; Zhou, J.-Y.; Kong, F.-L.; Qi, S.-S.; Zhu, B.; Naz, M.; Dai, Z.-C.; Du, D.-L. Both Adaptability and Endophytic Bacteria Are Linked to the Functional Traits in the Invasive Clonal Plant Wedelia trilobata. Plants 2022, 11, 3369. [Google Scholar] [CrossRef] [PubMed]
- Tosi, M.; Mitter, E.K.; Gaiero, J.; Dunfield, K. It takes three to tango: The importance of microbes, host plant, and soil management to elucidate manipulation strategies for the plant microbiome. Can. J. Microbiol. 2020, 66, 413–433. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Wu, P.; Ling, H.; Xu, G.; Xu, F.; Zhang, Q. Plant nutriomics in China: An overview. Ann. Bot. 2006, 98, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P. Roots of the second green revolution. Aust. J. Bot. 2007, 55, 493–512. [Google Scholar] [CrossRef]
- Bansal, P.; Bansal, R.; Arora, M. Traditional Nutritional Approaches and Nutriomics Evidence: Nutrition from Inception to Evidence. In Nutriomics; CRC Press: Boca Raton, FL, USA, 2022; pp. 23–47. [Google Scholar]
- Kabir, G. Genetic approaches of increasing nutrient use efficiency especially nitrogen in cereal crops a review. J. Bio-Sci. 2014, 22, 111–125. [Google Scholar] [CrossRef]
- Parker, D.; Beckmann, M.; Zubair, H.; Enot, D.P.; Caracuel-Rios, Z.; Overy, D.P.; Snowdon, S.; Talbot, N.J.; Draper, J. Metabolomic analysis reveals a common pattern of metabolic re-programming during invasion of three host plant species by Magnaporthe grisea. Plant J. 2009, 59, 723–737. [Google Scholar] [CrossRef]
- López-Arredondo, D.L.; Leyva-González, M.A.; Alatorre-Cobos, F.; Herrera-Estrella, L. Biotechnology of nutrient uptake and assimilation in plants. Int. J. Dev. Biol. 2013, 57, 595–610. [Google Scholar] [CrossRef]
- Khan, I.U.; Qi, S.-S.; Gul, F.; Manan, S.; Rono, J.K.; Naz, M.; Shi, X.-N.; Zhang, H.; Dai, Z.-C.; Du, D.-L. A Green Approach Used for Heavy Metals ‘Phytoremediation’Via Invasive Plant Species to Mitigate Environmental Pollution: A Review. Plants 2023, 12, 725. [Google Scholar] [CrossRef]
- Andrzejczak, O.; Krasuska, U.; Olechowicz, J.; Staszek, P.; Ciacka, K.; Bogatek, R.; Hebelstrup, K.; Gniazdowska, A. Destabilization of ROS metabolism in tomato roots as a phytotoxic effect of meta-tyrosine. Plant Physiol. Biochem. 2018, 123, 369–377. [Google Scholar] [CrossRef]
- Ghayal, N.; Dhumal, K. Morphophysiological investigations in some dominant alien invasive weeds. In Plants and Environment; InTech Open: Rijeka, Croatia, 2011; pp. 15–48. [Google Scholar]
- Borišev, M.; Pajević, S.; Nikolić, N.; Pilipović, A.; Arsenov, D.; Župunski, M. Mine site restoration using silvicultural approach. In Bio-Geotechnologies for Mine Site Rehabilitation; Elsevier: Amsterdam, The Netherlands, 2018; pp. 115–130. [Google Scholar]
- van Wijk, K.J.; Kessler, F. Plastoglobuli: Plastid microcompartments with integrated functions in metabolism, plastid developmental transitions, and environmental adaptation. Annu. Rev. Plant Biol. 2017, 68, 253–289. [Google Scholar] [CrossRef]
- Li, J.; Yu, Z.; Choo, S.; Zhao, J.; Wang, Z.; Xie, R. Chemico-proteomics reveal the enhancement of salt tolerance in an invasive plant species via H2S signaling. ACS Omega 2020, 5, 14575–14585. [Google Scholar] [CrossRef]
- Ma, N.; Dong, L.; Lü, W.; Lü, J.; Meng, Q.; Liu, P. Transcriptome analysis of maize seedling roots in response to nitrogen-, phosphorus-, and potassium deficiency. Plant and Soil. 2020, 447, 637–658. [Google Scholar] [CrossRef]
- Delaunois, B.; Jeandet, P.; Clément, C.; Baillieul, F.; Dorey, S.; Cordelier, S. Uncovering plant-pathogen crosstalk through apoplastic proteomic studies. Frontiers in Plant Science. 2014, 5, 249. [Google Scholar] [CrossRef]
- Wang, B.; He, T.; Zheng, X.; Song, B.; Chen, H. Proteomic analysis of potato responding to the invasion of Ralstonia solanacearum UW551 and its type III secretion system mutant. Mol. Plant-Microbe Interact. 2021, 34, 337–350. [Google Scholar] [CrossRef]
- Ghosh, S.; Narula, K.; Sinha, A.; Ghosh, R.; Jawa, P.; Chakraborty, N.; Chakraborty, S. Proteometabolomic study of compatible interaction in tomato fruit challenged with Sclerotinia rolfsii illustrates novel protein network during disease progression. Front. Plant Sci. 2016, 7, 1034. [Google Scholar] [CrossRef]
- Louis, B.; Waikhom, S.D.; Roy, P.; Bhardwaj, P.K.; Singh, M.W.; Chandradev, S.K.; Talukdar, N.C. Invasion of Solanum tuberosum L. by Aspergillus terreus: A microscopic and proteomics insight on pathogenicity. BMC Res. Notes 2014, 7, 350. [Google Scholar] [CrossRef]
- Meijer, H.J.G.; Mancuso, F.M.; Espadas, G.; Seidl, M.F.; Chiva, C.; Govers, F.; Sabidó, E. Profiling the Secretome and Extracellular Proteome of the Potato Late Blight Pathogen Phytophthora infestans. Mol. Cell. Proteom. 2014, 13, 2101–2113. [Google Scholar] [CrossRef]
- Anderson, J.P.; Hane, J.K.; Stoll, T.; Pain, N.; Hastie, M.L.; Kaur, P.; Hoogland, C.; Gorman, J.J.; Singh, K.B. Proteomic Analysis of Rhizoctonia solani Identifies Infection-specific, Redox Associated Proteins and Insight into Adaptation to Different Plant Hosts*. Mol. Cell. Proteom. 2016, 15, 1188–1203. [Google Scholar] [CrossRef]
- Mehta, A.; Brasileiro, A.C.M.; Souza, D.S.L.; Romano, E.; Campos, M.A.; Grossi-de-Sá, M.F.; Silva, M.S.; Franco, O.L.; Fragoso, R.R.; Bevitori, R.; et al. Plant–pathogen interactions: What is proteomics telling us? FEBS J. 2008, 275, 3731–3746. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, W.; Stanley, B.A.; Assmann, S.M. Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 2008, 20, 3210–3226. [Google Scholar] [CrossRef]
- Shrivastava, N.; Jiang, L.; Li, P.; Sharma, A.K.; Luo, X.; Wu, S.; Pandey, R.; Gao, Q.; Lou, B. Proteomic approach to understand the molecular physiology of symbiotic interaction between Piriformospora indica and Brassica napus. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Wang, L.; Zhao, M.; Gu, S.; Wang, C.; Zhao, J.; Tang, Z.; Gao, H.; Zhang, L.; Fu, L. iTRAQ proteomics reveals the regulatory response to Magnaporthe oryzae in durable resistant vs. susceptible rice genotypes. PLoS ONE 2020, 15, e0227470. [Google Scholar] [CrossRef] [PubMed]
- Butt, Y.K.-C.; Lo, S.C.-L. Proteomic studies on plant-pathogen interaction in compatible and incompatible systems. Curr. Proteom. 2007, 4, 141–156. [Google Scholar] [CrossRef]
- Barel, G.; Ginzberg, I. Potato skin proteome is enriched with plant defence components. J. Exp. Bot. 2008, 59, 3347–3357. [Google Scholar] [CrossRef]
- Afroz, A.; Zahur, M.; Zeeshan, N.; Komatsu, S. Plant-bacterium interactions analyzed by proteomics. Front. Plant Sci. 2013, 4, 21. [Google Scholar] [CrossRef]
- Fan, K.-T.; Hsu, Y.; Yeh, C.-F.; Chang, C.-H.; Chang, W.-H.; Chen, Y.-R. Quantitative Proteomics Reveals the Dynamic Regulation of the Tomato Proteome in Response to Phytophthora infestans. Int. J. Mol. Sci. 2021, 22, 4174. [Google Scholar] [CrossRef]
- Lebecka, R.; Kistowski, M.; Dębski, J.; Szajko, K.; Murawska, Z.; Marczewski, W. Quantitative proteomic analysis of differentially expressed proteins in tubers of potato plants differing in resistance to Dickeya solani. Plant Soil 2019, 441, 317–329. [Google Scholar] [CrossRef]
- Hu, J.; Rampitsch, C.; Bykova, N.V. Advances in plant proteomics toward improvement of crop productivity and stress resistancex. Front. Plant Sci. 2015, 6, 209. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Fernie, A.R. Regulation of metabolic networks: Understanding metabolic complexity in the systems biology era. New Phytol. 2005, 168, 9–24. [Google Scholar] [CrossRef]
- van Wijk, K.J. Challenges and prospects of plant proteomics. Plant Physiol. 2001, 126, 501–508. [Google Scholar] [CrossRef]
- Li, L.-Q.; Lyu, C.-C.; Li, J.-H.; Tong, Z.; Lu, Y.-F.; Wang, X.-Y.; Ni, S.; Yang, S.-M.; Zeng, F.-C.; Lu, L.-M. Physiological analysis and proteome quantification of alligator weed stems in response to potassium deficiency stress. Int. J. Mol. Sci. 2019, 20, 221. [Google Scholar] [CrossRef]
- Bai, H.; Si, H.; Zang, J.; Pang, X.; Yu, L.; Cao, H.; Xing, J.; Zhang, K.; Dong, J. Comparative proteomic analysis of the defense response to Gibberella stalk rot in maize and reveals that ZmWRKY83 is involved in plant disease resistance. Front. Plant Sci. 2021, 12, 694973. [Google Scholar] [CrossRef]
- Vo, K.T.X.; Rahman, M.M.; Rahman, M.M.; Trinh, K.T.T.; Kim, S.T.; Jeon, J.-S. Proteomics and metabolomics studies on the biotic stress responses of rice: An update. Rice 2021, 14, 30. [Google Scholar] [CrossRef]
- Castro-Díez, P.; Godoy, O.; Saldaña, A.; Richardson, D.M. Predicting invasiveness of Australian acacias on the basis of their native climatic affinities, life history traits and human use. Divers. Distrib. 2011, 17, 934–945. [Google Scholar] [CrossRef]
- Li, Z.; Xu, C.; Wang, J. Integrated physiological, transcriptomic and proteomic analyses revealed molecular mechanism for salt resistance in Solidago canadensis L. Environ. Exp. Bot. 2020, 179, 104211. [Google Scholar] [CrossRef]
- Lodge, D.M.; Simonin, P.W.; Burgiel, S.W.; Keller, R.P.; Bossenbroek, J.M.; Jerde, C.L.; Kramer, A.M.; Rutherford, E.S.; Barnes, M.A.; Wittmann, M.E. Risk analysis and bioeconomics of invasive species to inform policy and management. Annu. Rev. Environ. Resour. 2016, 41, 453–488. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, S.; Wang, J.; Zhang, Y.; Naz, M.; Afzal, M.R.; Du, D.; Dai, Z. Omics Approaches in Invasion Biology: Understanding Mechanisms and Impacts on Ecological Health. Plants 2023, 12, 1860. https://doi.org/10.3390/plants12091860
Qi S, Wang J, Zhang Y, Naz M, Afzal MR, Du D, Dai Z. Omics Approaches in Invasion Biology: Understanding Mechanisms and Impacts on Ecological Health. Plants. 2023; 12(9):1860. https://doi.org/10.3390/plants12091860
Chicago/Turabian StyleQi, Shanshan, Jiahao Wang, Yi Zhang, Misbah Naz, Muhammad Rahil Afzal, Daolin Du, and Zhicong Dai. 2023. "Omics Approaches in Invasion Biology: Understanding Mechanisms and Impacts on Ecological Health" Plants 12, no. 9: 1860. https://doi.org/10.3390/plants12091860
APA StyleQi, S., Wang, J., Zhang, Y., Naz, M., Afzal, M. R., Du, D., & Dai, Z. (2023). Omics Approaches in Invasion Biology: Understanding Mechanisms and Impacts on Ecological Health. Plants, 12(9), 1860. https://doi.org/10.3390/plants12091860







