Decoding Arabidopsis thaliana CPK/SnRK Superfamily Kinase Client Signaling Networks Using Peptide Library and Mass Spectrometry
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis and Development of the 2.1K Peptide Library
2.2. Preparation of Recombinant Protein Kinases
2.3. Purity and Activity Assessment of Recombinant Kinases
2.4. The Kinase Client Assay
2.5. LC-MS/MS Analysis
2.6. Database Search
2.7. Motif Analysis
2.8. Identification of Kinase Client Network
3. Results
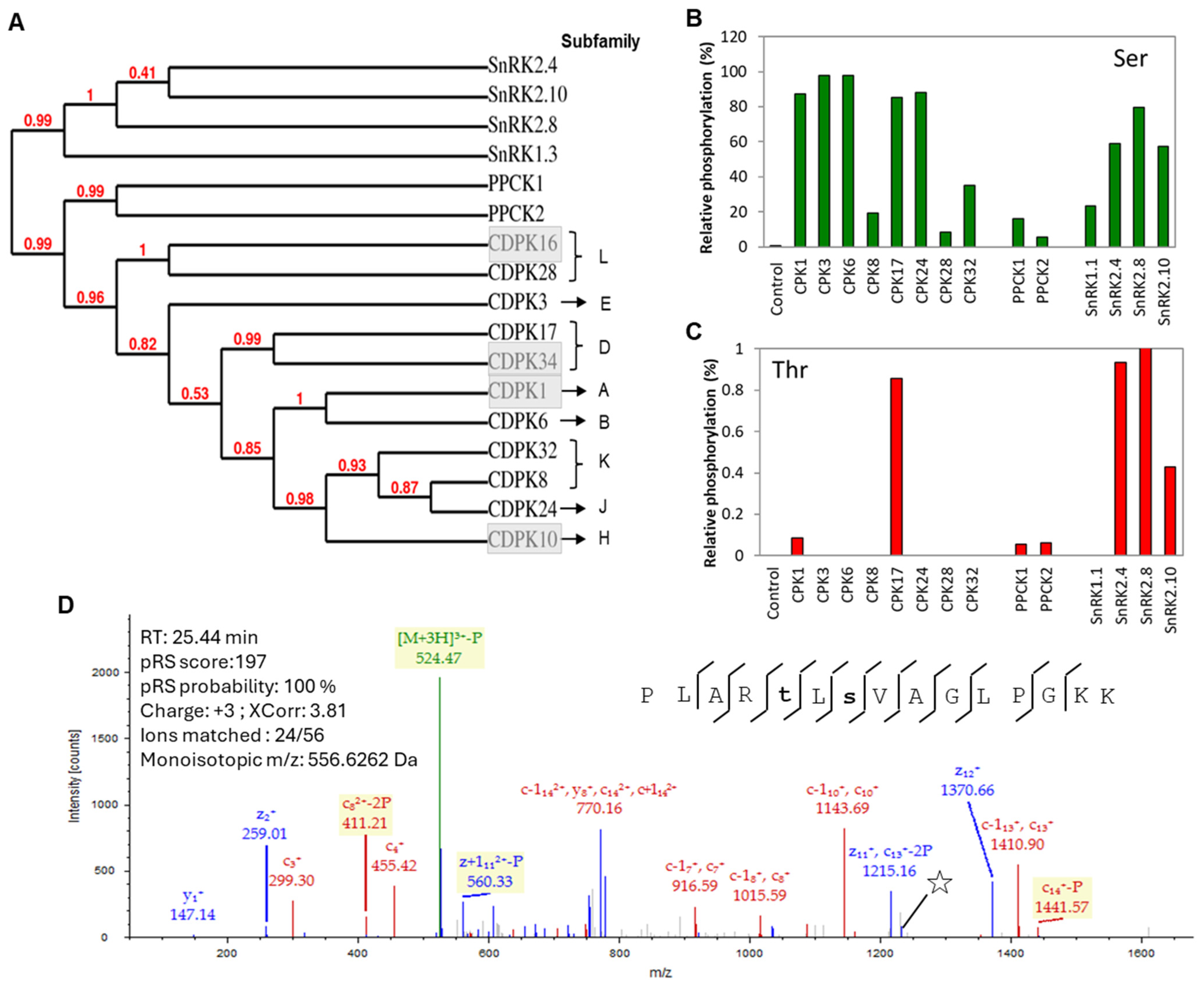
3.1. Mass Spectrometry-Based Kinase Activity Confirmation of the Recombinant Kinases
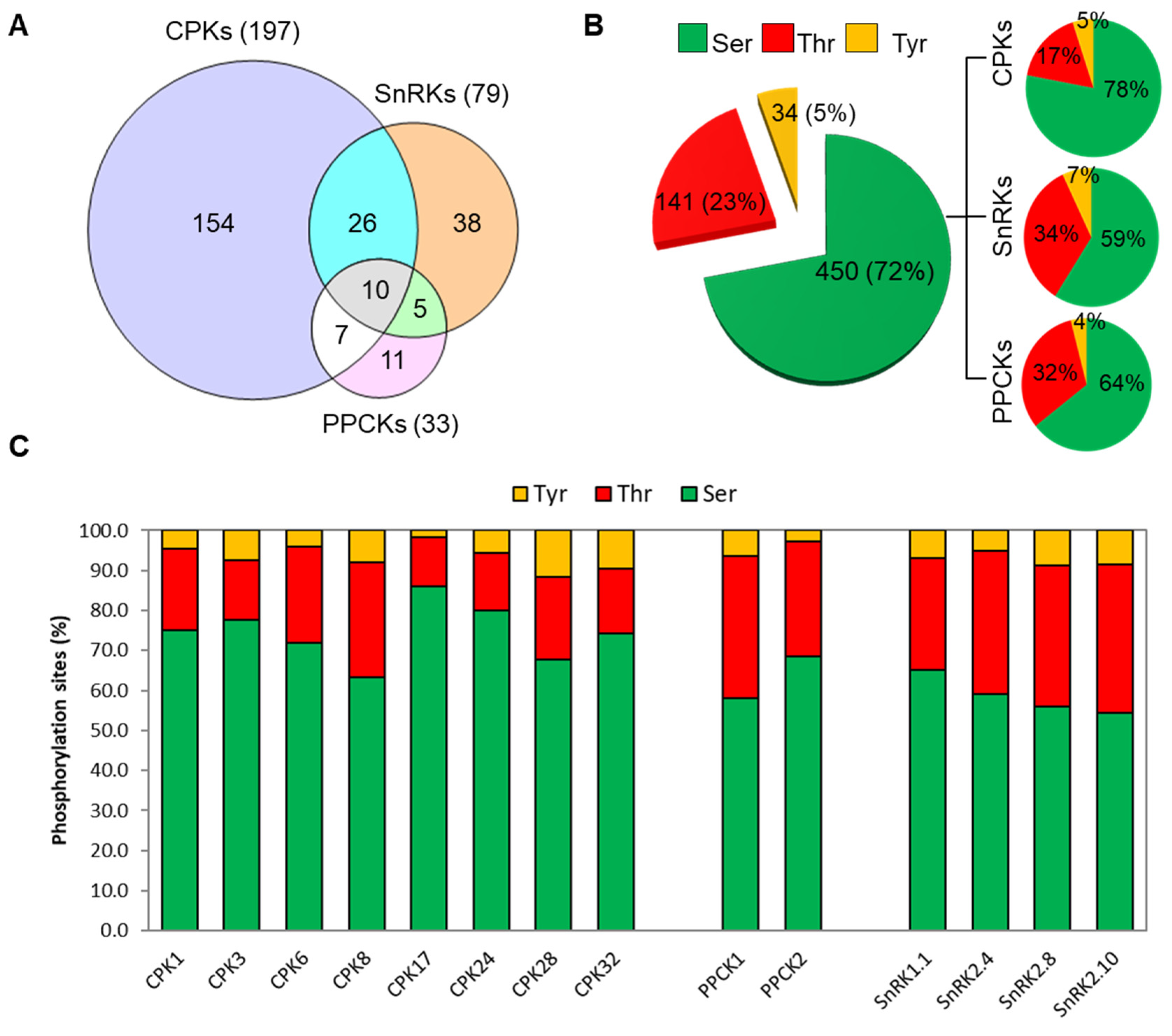
3.2. Identification of Putative Clients for CDPKs, SnRKs, and PPCKs
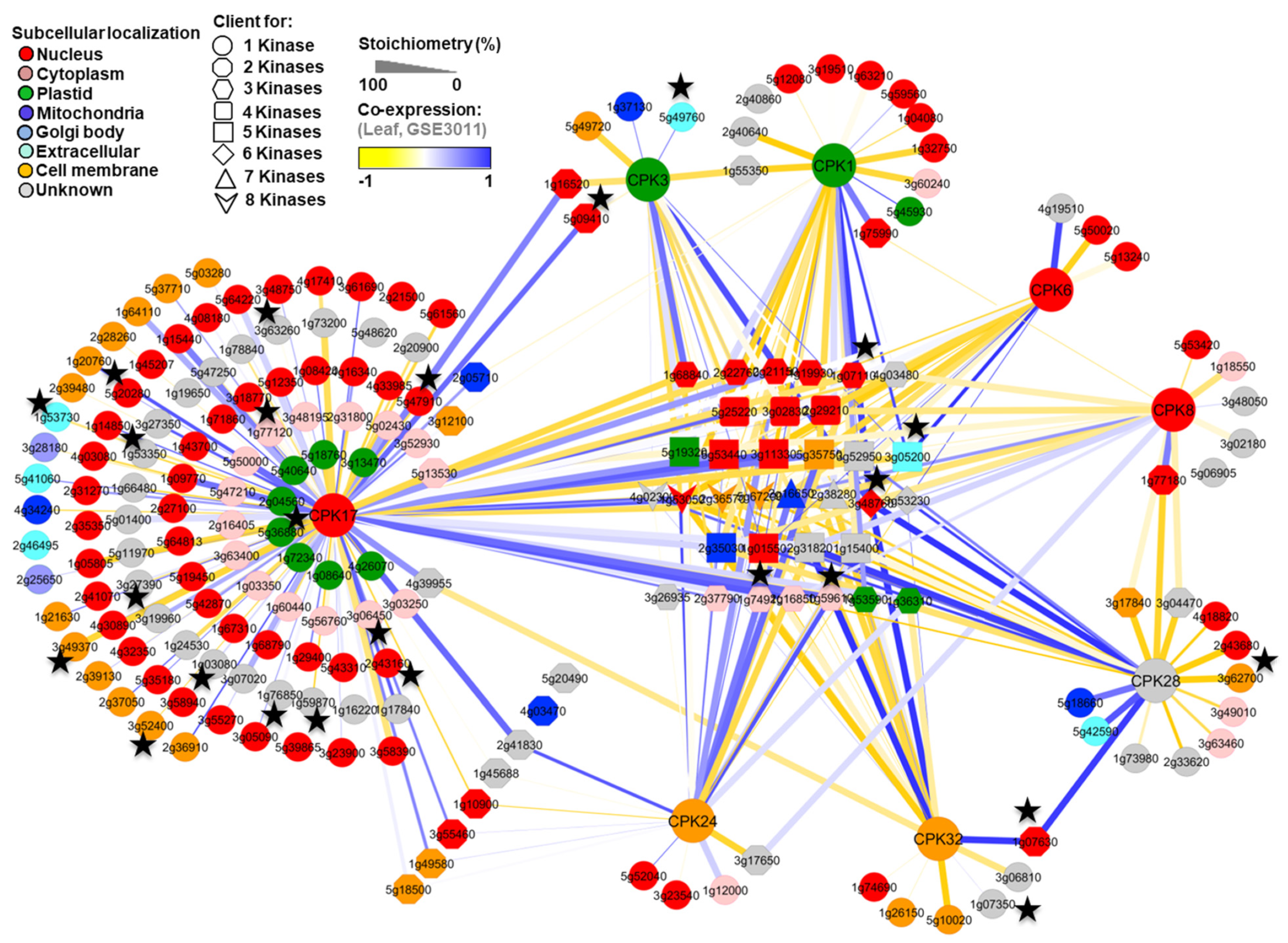
3.3. Reconstruction of In Silico Kinase Client Signaling Networks
3.4. Cross-Validation of the CPK Clients
3.5. Identification of Kinase-Specific Motifs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balderas-Hernández, V.E.; Alvarado-Rodríguez, M.; Fraire-Velázquez, S. Conserved versatile master regulators in signalling pathways in response to stress in plants. AoB Plants 2013, 5, plt033. [Google Scholar] [CrossRef] [PubMed]
- Sheen, J. Discover and connect cellular signaling. Plant Physiol. 2010, 154, 562–566. [Google Scholar] [CrossRef] [PubMed]
- Van Norman, J.M.; Breakfield, N.W.; Benfey, P.N. Intercellular communication during plant development. Plant Cell 2011, 23, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Collaudin, S.; Mirabet, V. Models to reconcile plant science and stochasticity. Front. Plant Sci. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.A.; Jones, A.; Godin, C.; Traas, J. Systems analysis of shoot apical meristem growth and development: Integrating hormonal and mechanical signaling. Plant Cell 2012, 24, 3907–3919. [Google Scholar] [CrossRef] [PubMed]
- Popik, O.V.; Saik, O.V.; Petrovskiy, E.D.; Sommer, B.; Hofestädt, R.; Lavrik, I.N.; Ivanisenko, V.A. Analysis of signaling networks distributed over intracellular compartments based on protein-protein interactions. BMC Genom. 2014, 15, S7. [Google Scholar] [CrossRef] [PubMed]
- Plattner, H.; Verkhratsky, A. The ancient roots of calcium signalling evolutionary tree. Cell Calcium 2015, 57, 123–132. [Google Scholar] [CrossRef]
- Spalding, E.P.; Harper, J.F. The ins and outs of cellular Ca2+ transport. Curr. Opin. Plant Biol. 2011, 14, 715–720. [Google Scholar] [CrossRef]
- Liese, A.; Romeis, T. Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim. Biophys. Acta 2013, 1833, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Hepler, P.K. Calcium: A central regulator of plant growth and development. Plant Cell 2005, 17, 2142–2155. [Google Scholar] [CrossRef]
- Galon, Y.; Finkler, A.; Fromm, H. Calcium-regulated transcription in plants. Mol. Plant 2010, 3, 653–669. [Google Scholar] [CrossRef] [PubMed]
- Saidi, Y.; Finka, A.; Muriset, M.; Bromberg, Z.; Weiss, Y.G.; Maathuis, F.J.; Goloubinoff, P. The heat shock response in moss plants is regulated by specific calcium-permeable channels in the plasma membrane. Plant Cell 2009, 21, 2829–2843. [Google Scholar] [CrossRef] [PubMed]
- Harada, A.; Shimazaki, K.-I. Measurement of changes in cytosolic Ca2+ in Arabidopsis guard cells and mesophyll cells in response to blue light. Plant Cell Physiol. 2009, 50, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Toyota, M.; Furuichi, T.; Tatsumi, H.; Sokabe, M. Cytoplasmic calcium increases in response to changes in the gravity vector in hypocotyls and petioles of Arabidopsis seedlings. Plant Physiol. 2008, 146, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Romeis, T.; Herde, M. From local to global: CDPKs in systemic defense signaling upon microbial and herbivore attack. Curr. Opin. Plant Biol. 2014, 20, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kiep, V.; Vadassery, J.; Lattke, J.; Maaß, J.; Boland, W.; Peiter, E.; Mithöfer, A. Systemic cytosolic Ca2+ elevation is activated upon wounding and herbivory in Arabidopsis. New Phytol. 2015, 207, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Hamel, L.-P.; Sheen, J.; Séguin, A. Ancient signals: Comparative genomics of green plant CDPKs. Trends Plant Sci. 2014, 19, 79–89. [Google Scholar] [CrossRef]
- Harper, J.F.; Breton, G.; Harmon, A. Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 2004, 55, 263–288. [Google Scholar] [CrossRef]
- Lillo, C.; Kataya, A.R.A.; Heidari, B.; Creighton, M.T.; Nemie-Feyissa, D.; Ginbot, Z.; Jonassen, E.M. Protein phosphatases PP2A, PP4 and PP6: Mediators and regulators in development and responses to environmental cues. Plant Cell Environ. 2014, 37, 2631–2648. [Google Scholar] [CrossRef]
- Uhrig, R.G.; Labandera, A.-M.; Moorhead, G.B. Arabidopsis PPP family of serine/threonine protein phosphatases: Many targets but few engines. Trends Plant Sci. 2013, 18, 505–513. [Google Scholar] [CrossRef]
- Zulawski, M.; Schulze, G.; Braginets, R.; Hartmann, S.; Schulze, W.X. The Arabidopsis Kinome: Phylogeny and evolutionary insights into functional diversification. BMC Genom. 2014, 15, 548. [Google Scholar] [CrossRef] [PubMed]
- Nagata, T.; Iizumi, S.; Satoh, K.; Ooka, H.; Kawai, J.; Carninci, P.; Hayashizaki, Y.; Otomo, Y.; Murakami, K.; Matsubara, K.; et al. Comparative analysis of plant and animal calcium signal transduction element using plant full-length cDNA data. Mol. Biol. Evol. 2004, 21, 1855–1870. [Google Scholar] [CrossRef] [PubMed]
- Norris, V.; Grant, S.; Freestone, P.; Canvin, J.; Sheikh, F.N.; Toth, I.; Trinei, M.; Modha, K.; Norman, R.I. Calcium signalling in bacteria. J. Bacteriol. 1996, 178, 3677–3682. [Google Scholar] [CrossRef]
- Hrabak, E.M.; Chan, C.W.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.-J.; Wei, F.-J.; Wang, C.; Wu, J.-J.; Ratnasekera, D.; Liu, W.-X.; Wu, W.-H. Arabidopsis calcium-dependent protein kinase CPK10 functions in abscisic acid- and Ca2+-mediated stomatal regulation in response to drought stress. Plant Physiol. 2010, 154, 1232–1243. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.-H.; Willmann, M.R.; Chen, H.-C.; Sheen, J. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002, 129, 469–485. [Google Scholar] [CrossRef] [PubMed]
- Bender, K.W.; Blackburn, R.K.; Monaghan, J.; Derbyshire, P.; Menke, F.L.; Zipfel, C.; Goshe, M.B.; Zielinski, R.E.; Huber, S.C. Autophosphorylation-based Calcium (Ca2+) Sensitivity Priming and Ca2+/Calmodulin Inhibition of Arabidopsis thaliana Ca2+-dependent Protein Kinase 28 (CPK28). J. Biol. Chem. 2017, 2292, 3988–4002. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Xiong, L.; Song, C.-P.; Gong, D.; Halfter, U.; Zhu, J.-K. A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev. Cell 2002, 3, 233–244. [Google Scholar] [CrossRef]
- Curran, A.; Chang, I.-F.; Chang, C.-L.; Garg, S.; Miguel, R.M.; Barron, Y.D.; Li, Y.; Romanowsky, S.; Cushman, J.C.; Gribskov, M.; et al. Calcium-dependent protein kinases from Arabidopsis show substrate specificity differences in an analysis of 103 substrates. Front. Plant Sci. 2011, 2, 36. [Google Scholar] [CrossRef]
- Matsoukas, I.G. Interplay between sugar and hormone signaling pathways modulate floral signal transduction. Front. Genet. 2014, 5, 218. [Google Scholar] [CrossRef]
- Umezawa, T.; Sugiyama, N.; Takahashi, F.; Anderson, J.C.; Ishihama, Y.; Peck, S.C.; Shinozaki, K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 2013, 6, rs8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Chen, S.; Harmon, A.C. Protein phosphorylation in stomatal movement. Plant Signal. Behav. 2014, 9, e972845. [Google Scholar] [CrossRef] [PubMed]
- Arsova, B.; Schulze, W.X. Current status of the plant phosphorylation site database PhosPhAt and its use as a resource for molecular plant physiology. Front. Plant Sci. 2012, 3, 132. [Google Scholar] [CrossRef]
- Wang, P.; Xue, L.; Batelli, G.; Lee, S.; Hou, Y.-J.; Van Oosten, M.J.; Zhang, H.; Tao, W.A.; Zhu, J.-K. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 2013, 110, 11205–11210. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Ge, H.; Wu, S.; Zhang, N.; Chen, W.; Xu, C.; Gao, J.; Thelen, J.J.; Xu, D. P3DB 3.0: From plant phosphorylation sites to protein networks. Nucleic Acids Res. 2014, 42, D1206–D1213. [Google Scholar] [CrossRef] [PubMed]
- Miernyk, J.A.; Randall, D.D. A synthetic peptide-directed antibody as a probe of the phosphorylation site of pyruvate dehydrogenase. J. Biol. Chem. 1989, 264, 9141–9144. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, N.; Swatek, K.N.; Zhang, J.; Miernyk, J.A.; Xu, D.; Thelen, J.J. “Scanning mutagenesis” of the amino acid sequences flanking phosphorylation site 1 of the mitochondrial pyruvate dehydrogenase complex. Front. Plant Sci. 2012, 3, 153. [Google Scholar] [CrossRef]
- Gribskov, M.; Fana, F.; Harper, J.; Hope, D.A.; Harmon, A.C.; Smith, D.W.; Tax, F.E.; Zhang, G. PlantsP: A functional genomics database for plant phosphorylation. Nucleic Acids Res. 2001, 29, 111–113. [Google Scholar] [CrossRef] [PubMed]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef]
- Durek, P.; Schmidt, R.; Heazlewood, J.L.; Jones, A.; MacLean, D.; Nagel, A.; Kersten, B.; Schulze, W.X. PhosPhAt: The Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 2010, 38, D828–D834. [Google Scholar] [CrossRef]
- Kemp, B.E.; Graves, D.J.; Benjamini, E.; Krebs, E.G. Role of multiple basic residues in determining the substrate specificity of cyclic AMP-dependent protein kinase. J. Biol. Chem. 1977, 252, 4888–4894. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Soderling, T.R. Calcium·calmodulin-dependent protein kinase II and calcium·phospholipid-dependent protein kinase activities in rat tissues assayed with a synthetic peptide. Arch. Biochem. Biophys. 1987, 252, 418–425. [Google Scholar] [CrossRef]
- Wang, Z. The peptide microarray-based assay for kinase functionality and inhibition study. Methods Mol. Biol. 2009, 570, 329–337. [Google Scholar]
- Xu, J.; Sun, L.; Ghosh, I.; Xu, M.-Q.; Kochinyan, S.; Barshevsky, T. Western blot analysis of Src kinase assays using peptide substrates ligated to a carrier protein. Biotechniques 2004, 36, 976–981. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.A.; Biggs, W.H., 3rd; Treiber, D.K.; Atteridge, C.E.; Azimioara, M.D.; Benedetti, M.G.; Carter, T.A.; Ciceri, P.; Edeen, P.T.; Floyd, M.; et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 2005, 23, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Kunz, R.C.; McAllister, F.E.; Rush, J.; Gygi, S.P. A High-Throughput, Multiplexed Kinase Assay Using a Benchtop Orbitrap Mass Spectrometer To Investigate the Effect of Kinase Inhibitors on Kinase Signaling Pathways. Anal. Chem. 2012, 84, 6233–6239. [Google Scholar] [CrossRef]
- Huang, Y.; Houston, N.L.; Tovar-Mendez, A.; Stevenson, S.E.; Miernyk, J.A.; Randall, D.D.; Thelen, J.J. A quantitative mass spectrometry-based approach for identifying protein kinase clients and quantifying kinase activity. Anal. Biochem. 2010, 402, 69–76. [Google Scholar] [CrossRef]
- Feilner, T.; Hultschig, C.; Lee, J.; Meyer, S.; Immink, R.G.H.; Koenig, A.; Possling, A.; Seitz, H.; Beveridge, A.; Scheel, D.; et al. High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol. Cell. Proteom. 2005, 4, 1558–1568. [Google Scholar] [CrossRef] [PubMed]
- Panse, S.; Dong, L.; Burian, A.; Carus, R.; Schutkowski, M.; Reimer, U.; Schneider-Mergener, J. Profiling of generic anti-phosphopeptide antibodies and kinases with peptide microarrays using radioactive and fluorescence-based assays. Mol. Divers. 2004, 8, 291–299. [Google Scholar] [CrossRef]
- Ptacek, J.; Devgan, G.; Michaud, G.; Zhu, H.; Zhu, X.; Fasolo, J.; Guo, H.; Jona, G.; Breitkreutz, A.; Sopko, R.; et al. Global analysis of protein phosphorylation in yeast. Nature 2005, 438, 679–684. [Google Scholar] [CrossRef]
- Huang, Y.; Thelen, J.J. KiC assay: A quantitative mass spectrometry-based approach for kinase client screening and activity analysis. Methods Mol. Biol. 2012, 893, 359–370. [Google Scholar] [PubMed]
- Ahsan, N.; Huang, Y.; Tovar-Mendez, A.; Swatek, K.N.; Zhang, J.; Miernyk, J.A.; Xu, D.; Thelen, J.J. A versatile mass spectrometry-based method to both identify kinase client-relationships and characterize signaling network topology. J. Proteome Res. 2013, 12, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.V.; Blagoev, B.; Gnad, F.; Macek, B.; Kumar, C.; Mortensen, P.; Mann, M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006, 127, 635–648. [Google Scholar] [CrossRef]
- Zhu, L.; Li, N. Quantitation, networking, and function of protein phosphorylation in plant cell. Front. Plant Sci. 2013, 3, 302. [Google Scholar] [CrossRef] [PubMed]
- Brauer, E.K.; Ahsan, N.; Dale, R.; Kato, N.; Coluccio, A.E.; Piñeros, M.A.; Kochian, L.V.; Thelen, J.J.; Popescu, S.C. The Raf-like Kinase ILK1 and the High Affinity K+ Transporter HAK5 Are Required for Innate Immunity and Abiotic Stress Response. Plant Physiol. 2016, 171, 1470–1484. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Cao, Y.; Li, H.; Kim, D.; Ahsan, N.; Thelen, J.; Stacey, G. Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat. Commun. 2017, 8, 2265. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, Y.; Huang, J.; Ahsan, N.; Biener, G.; Paprocki, J.; Thelen, J.J.; Raicu, V.; Zhao, D. Two SERK Receptor-Like Kinases Interact with EMS1 to Control Anther Cell Fate Determination. Plant Physiol. 2017, 173, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Hao, F.; Mu, H.; Ahsan, N.; Thelen, J.J.; Stacey, G. S-acylation of P2K1 mediates extracellular ATP-induced immune signaling in Arabidopsis. Nat. Commun. 2021, 12, 2750. [Google Scholar] [CrossRef]
- Brauer, E.K.; Ahsan, N.; Popescu, G.V.; Thelen, J.J.; Popescu, S.C. Back From the Dead: The Atypical Kinase Activity of a Pseudokinase Regulator of Cation Fluxes During Inducible Immunity. Front. Plant Sci. 2022, 13, 931324. [Google Scholar] [CrossRef]
- Kim, D.; Chen, D.; Ahsan, N.; Jorge, G.L.; Thelen, J.J.; Stacey, G. The Raf-like MAPKKK INTEGRIN-LINKED KINASE 5 regulates purinergic receptor-mediated innate immunity in Arabidopsis. Plant Cell 2023, 35, 1572–1592. [Google Scholar] [CrossRef]
- Bahk, S.; Ahsan, N.; An, J.; Kim, S.H.; Ramadany, Z.; Hong, J.C.; Thelen, J.J.; Chung, W.S. Identification of mitogen-activated protein kinases substrates in Arabidopsis using kinase client assay. Plant Signal. Behav. 2024, 19, 2326238. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, Y.; Tanford, C. The solubility of amino acids and two glycine peptides in aqueous ethanol and dioxane solutions. Establishment of a hydrophobicity scale. J. Biol. Chem. 1971, 246, 2211–2217. [Google Scholar] [CrossRef] [PubMed]
- Bucholc, M.; Ciesielski, A.; Goch, G.; Anielska-Mazur, A.; Kulik, A.; Krzywińska, E.; Dobrowolska, G. SNF1-related protein kinases 2 are negatively regulated by a plant-specific calcium sensor. J. Biol. Chem. 2011, 286, 3429–3441. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, F.; Galvan-Ampudia, C.S.; Julkowska, M.M.; Caarls, L.; van der Does, D.; Laurière, C.; Munnik, T.; Haring, M.A.; Testerink, C. The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J. 2012, 72, 436–449. [Google Scholar] [CrossRef] [PubMed]
- Swaney, D.L.; McAlister, G.C.; Coon, J.J. Decision tree–driven tandem mass spectrometry for shotgun proteomics. Nat. Methods 2008, 5, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, D.; Gygi, S.P. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 2005, 23, 1391–1398. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.S.P.; Møller, I.M. Large-scale analysis of phosphorylation site occupancy in eukaryotic proteins. Biochim. Biophys. Acta 2012, 1824, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Lotia, S.; Pico, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods 2012, 9, 1069–1076. [Google Scholar] [CrossRef]
- Furlotte, N.A.; Kang, H.M.; Ye, C.; Eskin, E. Mixed-model coexpression: Calculating gene coexpression while accounting for expression heterogeneity. Bioinformatics 2011, 27, i288–i294. [Google Scholar] [CrossRef]
- Uno, Y.; Rodriguez Milla, M.A.; Maher, E.; Cushman, J.C. Identification of proteins that interact with catalytically active calcium-dependent protein kinases from Arabidopsis. Mol. Genet. Genomics 2009, 281, 375–390. [Google Scholar] [CrossRef]
- Vlad, F.; Turk, B.E.; Peynot, P.; Leung, J.; Merlot, S. A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J. 2008, 55, 104–117. [Google Scholar] [CrossRef]
- Mehlmer, N.; Wurzinger, B.; Stael, S.; Hofmann-Rodrigues, D.; Csaszar, E.; Pfister, B.; Bayer, R.; Teige, M. The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 2010, 63, 484–498. [Google Scholar] [CrossRef]
- Xie, K.; Chen, J.; Wang, Q.; Yang, Y. Direct phosphorylation and activation of a mitogen-activated protein kinase by a calcium-dependent protein kinase in rice. Plant Cell 2014, 26, 3077–3089. [Google Scholar] [CrossRef]
- Sebastià, C.H.; Hardin, S.C.; Clouse, S.D.; Kieber, J.J.; Huber, S.C. Identification of a new motif for CDPK phosphorylation in vitro that suggests ACC synthase may be a CDPK substrate. Arch. Biochem. Biophys. 2004, 428, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, N.; Nakagami, H.; Mochida, K.; Daudi, A.; Tomita, M.; Shirasu, K.; Ishihama, Y. Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol. Syst. Biol. 2008, 4, 193. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, H.; Sugiyama, N.; Mochida, K.; Daudi, A.; Yoshida, Y.; Toyoda, T.; Tomita, M.; Ishihama, Y.; Shirasu, K. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 2010, 153, 1161–1174. [Google Scholar] [CrossRef] [PubMed]
- Van Wijk, K.J.; Friso, G.; Walther, D.; Schulze, W.X. Meta-Analysis of Arabidopsis thaliana Phospho-Proteomics Data Reveals Compartmentalization of Phosphorylation Motifs. Plant Cell 2014, 26, 2367–2389. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, N.; Wilson, R.S.; Rao, R.S.P.; Salvato, F.; Sabila, M.; Ullah, H.; Miernyk, J.A. Mass Spectrometry-Based Identification of Phospho-Tyr in Plant Proteomics. J. Proteome Res. 2020, 19, 561–571. [Google Scholar] [CrossRef]
- Hunter, T. The genesis of tyrosine phosphorylation. Cold Spring Harb. Perspect. Biol. 2014, 6, a020644. [Google Scholar] [CrossRef]
- Coca, M.; Segundo, B.S. AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J. 2010, 63, 526–540. [Google Scholar] [CrossRef] [PubMed]
- Kataya, A.R.; Muench, D.G.; Moorhead, G.B. A Framework to Investigate Peroxisomal Protein Phosphorylation in Arabidopsis. Trends Plant Sci. 2019, 24, 366–381. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Peng, L.; Liu, Y.; Yao, R.; Liu, Z.; Li, X.; Yang, Y.; Wang, J. Arabidopsis calcium-dependent protein kinase AtCPK1 plays a positive role in salt/drought-stress response. Biochem. Biophys. Res. Commun. 2018, 498, 92–98. [Google Scholar] [CrossRef]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef]
- Lid, S.E.; Gruis, D.; Jung, R.; Lorentzen, J.A.; Ananiev, E.; Chamberlin, M.; Niu, X.; Meeley, R.; Nichols, S.; Olsen, O.-A. The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc. Natl. Acad. Sci. USA 2002, 99, 5460–5465. [Google Scholar] [CrossRef]
- Whalley, H.J.; Sargeant, A.W.; Steele, J.F.C.; Lacoere, T.; Lamb, R.; Saunders, N.J.; Knight, H.; Knight, M.R. Transcriptomic analysis reveals calcium regulation of specific promoter motifs in Arabidopsis. Plant Cell 2011, 23, 4079–4095. [Google Scholar] [CrossRef]
- Kaplan, B.; Davydov, O.; Knight, H.; Galon, Y.; Knight, M.R.; Fluhr, R.; Fromm, H. Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 2006, 18, 2733–2748. [Google Scholar] [CrossRef]
- Wang, Y.-M.; Yang, Q.; Liu, Y.-J.; Yang, H.-L. Molecular Evolution and Expression Divergence of the Aconitase (ACO) Gene Family in Land Plants. Front. Plant Sci. 2016, 7, 1879. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Sagar, S.; Biswas, D.K. Calcium Dependent Protein Kinase, a Versatile Player in Plant Stress Management and Development. Crit. Rev. Plant Sci. 2017, 36, 336–352. [Google Scholar] [CrossRef]
- Kline, K.G.; Barrett-Wilt, G.A.; Sussman, M.R. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. USA 2010, 107, 15986–15991. [Google Scholar] [CrossRef]
- Underwood, W.; Somerville, S.C. Phosphorylation is required for the pathogen defense function of the Arabidopsis PEN3 ABC transporter. Plant Signal. Behav. 2017, 12, e1379644. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zhang, H.; Yu, B.; Xiong, L.; Xia, Y. Proteomic identification of early salicylate- and flg22-responsive redox-sensitive proteins in Arabidopsis. Sci. Rep. 2015, 5, 8625. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahsan, N.; Kataya, A.R.A.; Rao, R.S.P.; Swatek, K.N.; Wilson, R.S.; Meyer, L.J.; Tovar-Mendez, A.; Stevenson, S.; Maszkowska, J.; Dobrowolska, G.; et al. Decoding Arabidopsis thaliana CPK/SnRK Superfamily Kinase Client Signaling Networks Using Peptide Library and Mass Spectrometry. Plants 2024, 13, 1481. https://doi.org/10.3390/plants13111481
Ahsan N, Kataya ARA, Rao RSP, Swatek KN, Wilson RS, Meyer LJ, Tovar-Mendez A, Stevenson S, Maszkowska J, Dobrowolska G, et al. Decoding Arabidopsis thaliana CPK/SnRK Superfamily Kinase Client Signaling Networks Using Peptide Library and Mass Spectrometry. Plants. 2024; 13(11):1481. https://doi.org/10.3390/plants13111481
Chicago/Turabian StyleAhsan, Nagib, Amr R. A. Kataya, R. Shyama Prasad Rao, Kirby N. Swatek, Rashaun S. Wilson, Louis J. Meyer, Alejandro Tovar-Mendez, Severin Stevenson, Justyna Maszkowska, Grazyna Dobrowolska, and et al. 2024. "Decoding Arabidopsis thaliana CPK/SnRK Superfamily Kinase Client Signaling Networks Using Peptide Library and Mass Spectrometry" Plants 13, no. 11: 1481. https://doi.org/10.3390/plants13111481




