Genetic Diversity and Phylogeography of the Relict Tree Fern Culcita macrocarpa: Influence of Clonality and Breeding System on Genetic Variation
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction, Microsatellite Genotyping, and Plastid DNA Sequencing
2.3. Clonality and Genetic Diversity
2.3.1. Microsatellites
2.3.2. ptDNA
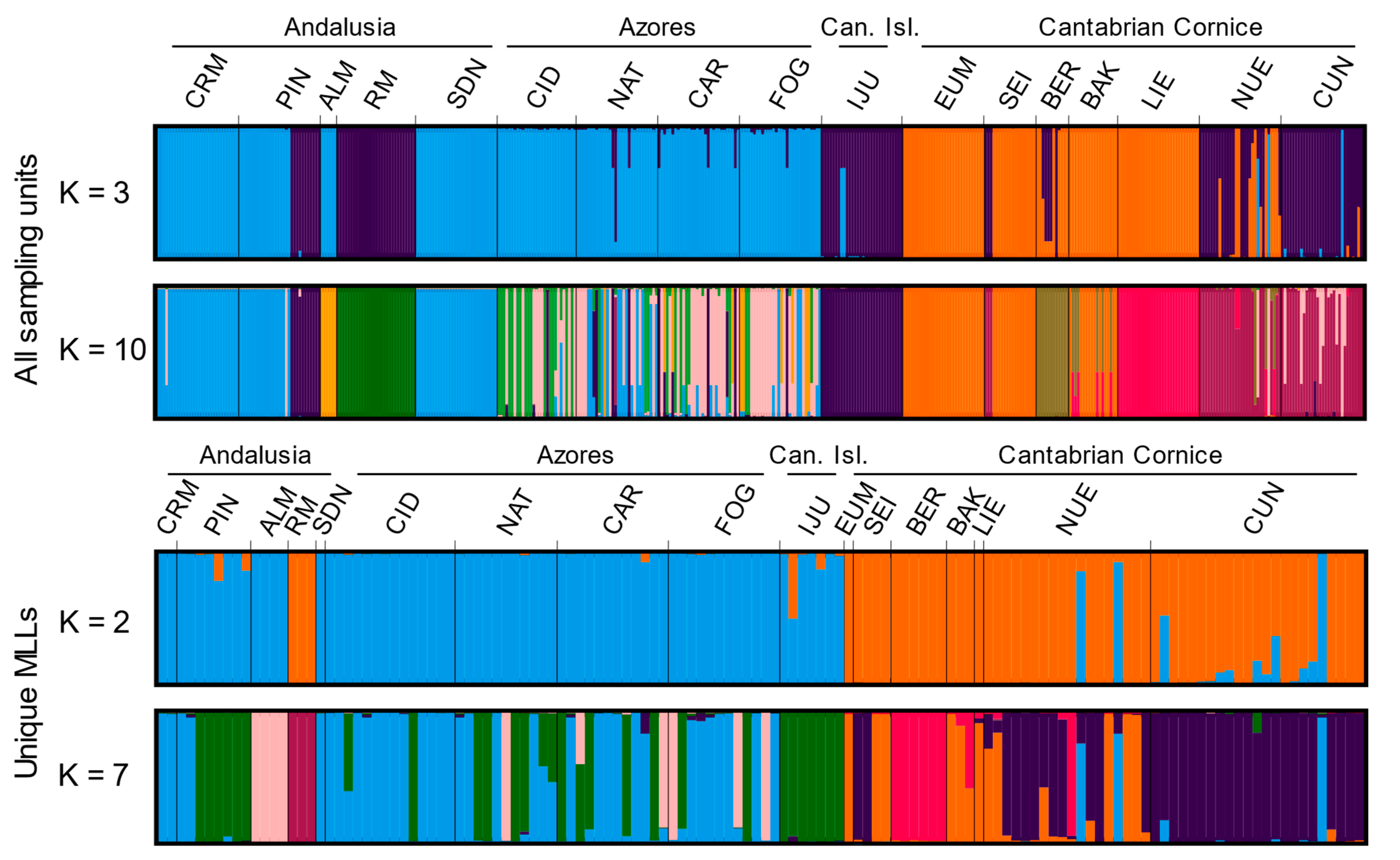
2.4. Genetic Structure and Phylogeography
2.5. Gene Flow Using Microsatellite Data
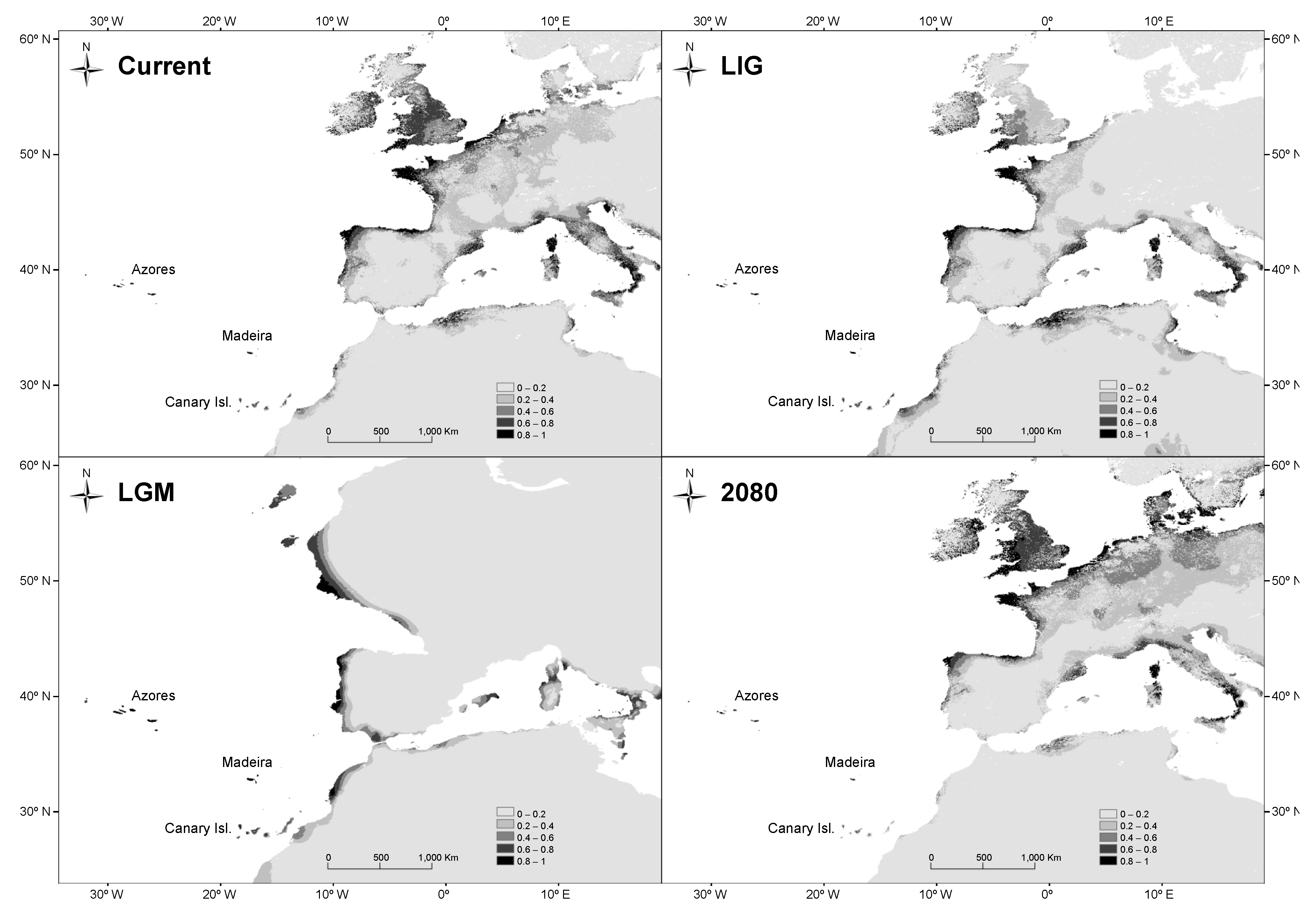
2.6. Species Distribution Modeling
3. Results
3.1. Clonality and Genetic Diversity
Microsatellites
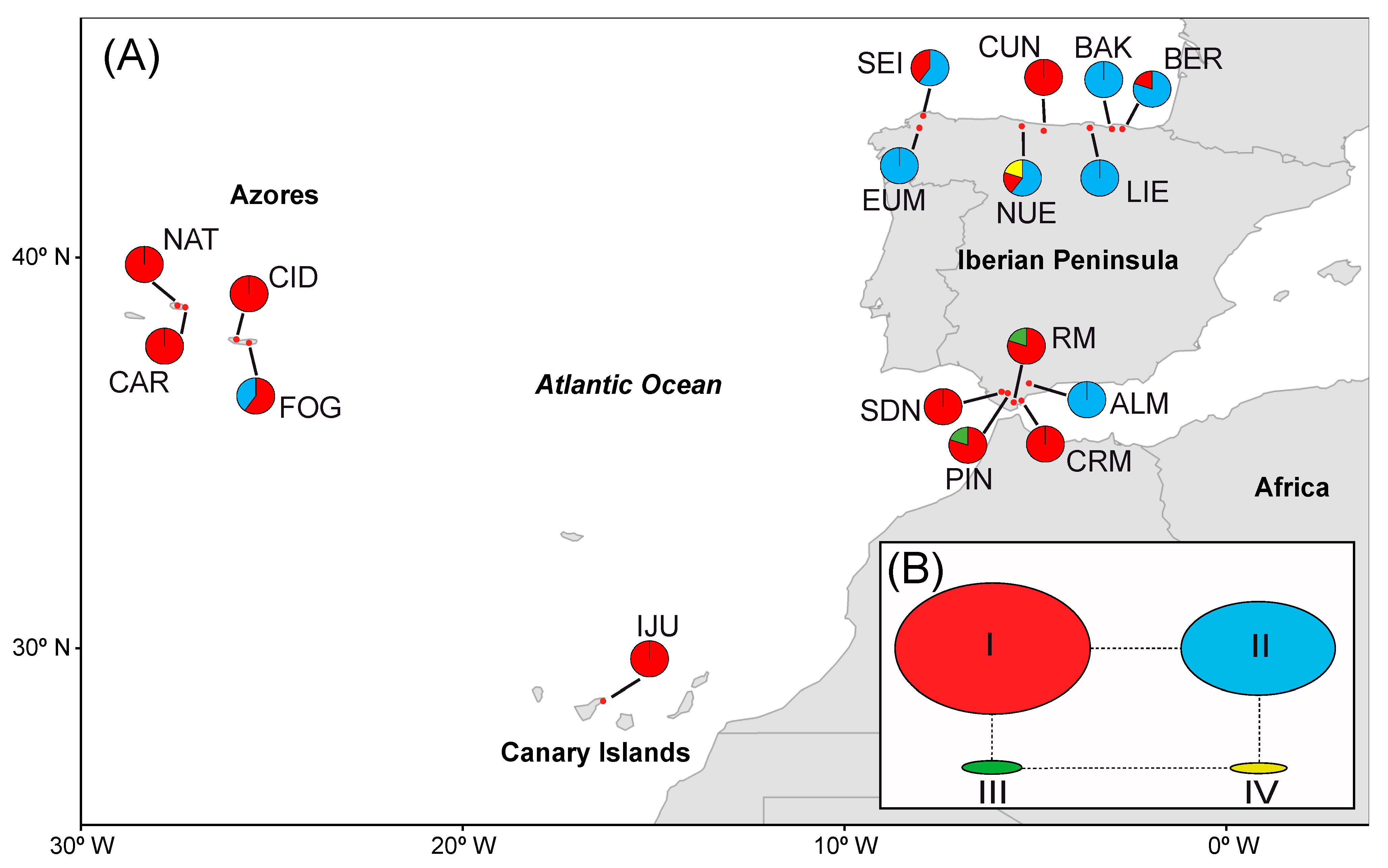
3.2. Genetic Structure and Phylogeography
3.3. Gene Flow
3.4. Species Distribution Modeling
4. Discussion
4.1. Clonality Effect on Genetic Diversity
4.2. Phylogeography and Population Dynamics
4.3. Conservation and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schuettpelz, E.; Schneider, H.; Smith, A.R.; Hovenkamp, P.; Prado, J.; Rouhan, G.; Salino, A.; Sundue, M.; Almeida, T.E.; Parris, B.; et al. A Community-Derived Classification for Extant Lycophytes and Ferns. J. Syst. Evol. 2016, 54, 563–603. [Google Scholar] [CrossRef]
- Pelosi, J.A.; Sessa, E.B. From Genomes to Populations: A Meta-Analysis and Review of Fern Population Genetics. Int. J. Plant Sci. 2021, 182, 325–343. [Google Scholar] [CrossRef]
- Pichi-Sermolli, R.E.G. A Survey of the Pteridological Flora of the Mediterranean Region: Rassegna Della Flora Pteridologica Della Regione Mediterranea. Webbia 1979, 34, 175–242. [Google Scholar] [CrossRef]
- Pichi-Sermolli, R.E.G. Considerazioni Sull’affinitá Ed Origine Della Flora Pteridologica Della Regione Mediterranea. Acta Botánica Malacit. 1991, 16, 235–280. [Google Scholar] [CrossRef]
- Benito Garzón, M. Potencialidad del Elemento Lauroide en la Península Ibérica. Predicción Mediante Redes Neuronales Artificiales en el Entorno de un Sistema de Información Geográfica. Ph.D. Thesis, Universidad Autónoma de Madrid, Madrid, Spain, 2002. [Google Scholar]
- Barrón, E. Evolución de Las Floras Terciarias En La Península Ibérica. Monogr. Jardín Botánico Córdoba 2003, 11, 63–74. [Google Scholar]
- Barrón, E.; Peyrot, D. La Vegetación Forestal En El Terciario. In Paleoambientes y Cambio Climático; Carrión, J.S., Fernández, S., Fuentes, N., Eds.; Fudación Séneca: Murcia, Spain, 2006; pp. 55–76. ISBN 84-932456-6-6. [Google Scholar]
- Vogel, J.C.; Rumsey, F.J.; Schneller, J.J.; Barrett, J.A.; Gibby, M. Where Are the Glacial Refugia in Europe? Evidence from Pteridophytes. Biol. J. Linn. Soc. 1999, 66, 23–37. [Google Scholar] [CrossRef]
- Wolf, P.G.; Schneider, H.; Ranker, T.A. Geographic Distributions of Homosporous Ferns: Does Dispersal Obscure Evidence of Vicariance? J. Biogeogr. 2001, 28, 263–270. [Google Scholar] [CrossRef]
- Soltis, D.E.; Soltis, P.S. Polyploidy, Breeding Systems, and Genetic Differentiation in Homosporous Pteridophytes. In Isozymes in Plant Biology; Soltis, D.E., Soltis, P.S., Eds.; Dioscorides Press: Portland, OR, USA, 1989; pp. 241–258. [Google Scholar]
- Trewick, S.A.; Morgan-Richards, M.; Russell, S.J.; Henderson, S.; Rumsey, F.J.; Pintér, I.; Barrett, J.A.; Gibby, M.; Vogel, J.C. Polyploidy, Phylogeography and Pleistocene Refugia of the Rockfern Asplenium ceterach: Evidence from Chloroplast DNA. Mol. Ecol. 2002, 11, 2003–2012. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, L.D.; Perrie, L.R.; Brownsey, P.J. Fire and Ice: Volcanic and Glacial Impacts on the Phylogeography of the New Zealand Forest Fern Asplenium hookerianum. Mol. Ecol. 2007, 16, 4536–4549. [Google Scholar] [CrossRef]
- Hunt, H.V.; Ansell, S.W.; Russell, S.J.; Schneider, H.; Vogel, J.C. Genetic Diversity and Phylogeography in Two Diploid Ferns, Asplenium fontanum Subsp. fontanum and A. petrarchae subsp. bivalens, in the Western Mediterranean. Mol. Ecol. 2009, 18, 4940–4954. [Google Scholar] [CrossRef]
- Jiménez, A.; Holderegger, R.; Csencsics, D.; Quintanilla, L.G. Microsatellites Reveal Substantial Among-Population Genetic Differentiation and Strong Inbreeding in the Relict Fern Dryopteris aemula. Ann. Bot. 2010, 106, 149–155. [Google Scholar] [CrossRef]
- Wang, Z.J.; Guan, K.Y. Genetic Structure and Phylogeography of a Relict Tree Fern, Sphaeropteris brunoniana (Cyatheaceae) from China and Laos Inferred from cpDNA Sequence Variations: Implications for Conservation. J. Syst. Evol. 2011, 49, 72–79. [Google Scholar] [CrossRef]
- Bystriakova, N.; Ansell, S.W.; Russell, S.J.; Grundmann, M.; Vogel, J.C.; Schneider, H. Present, Past and Future of the European Rock Fern Asplenium fontanum: Combining Distribution Modelling and Population Genetics to Study the Effect of Climate Change on Geographic Range and Genetic Diversity. Ann. Bot. 2014, 113, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Maccagni, A.; Parisod, C.; Grant, J.R. Phylogeography of the Moonwort Fern Botrychium lunaria (Ophioglossaceae) Based on Chloroplast DNA in the Central-European Mountain System. Alp. Bot. 2017, 127, 185–196. [Google Scholar] [CrossRef]
- Ben-Menni Schuler, S.; García-López, M.d.C.; López-Flores, I.; Nieto-Lugilde, M.; Suárez-Santiago, V.N. Genetic Diversity and Population History of the Killarney Fern, Vandenboschia speciosa (Hymenophyllaceae), at Its Southern Distribution Limit in Continental Europe. Bot. J. Linn. Soc. 2017, 183, 94–105. [Google Scholar] [CrossRef][Green Version]
- Ben-Menni Schuler, S.; Hamza, H.; Blanca, G.; Romero-García, A.T.; Suárez-Santiago, V.N. Phylogeographical Analyses of a Relict Fern of Palaeotropical Flora (Vandenboschia speciosa): Distribution and Diversity Model in Relation to the Geological and Climate Events of the Late Miocene and Early Pliocene. Plants 2022, 11, 839. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Zhang, X.; Wei, R. Ecological Adaptation Shaped the Genetic Structure of Homoploid Ferns against Strong Dispersal Capacity. Mol. Ecol. 2022, 31, 2679–2697. [Google Scholar] [CrossRef]
- Suter, M.; Schneller, J.J.; Vogel, J.C. Investigations into the Genetic Variation, Population Structure, and Breeding Systems of the Fern Asplenium trichomanes subsp. quadrivalens. Int. J. Plant Sci. 2000, 161, 233–244. [Google Scholar] [CrossRef] [PubMed]
- De Groot, G.A.; Verduyn, B.; Wubs, E.J.; Erkens, R.H.J.; During, H.J. Inter-and Intraspecific Variation in Fern Mating Systems after Long-Distance Colonization: The Importance of Selfing. BMC Plant Biol. 2012, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Silvertown, J. The Evolutionary Maintenance of Sexual Reproduction: Evidence from the Ecological Distribution of Asexual Reproduction in Clonal Plants. Int. J. Plant Sci. 2008, 169, 157–168. [Google Scholar] [CrossRef]
- Balloux, F.; Lehmann, L.; De Meeûs, T. The Population Genetics of Clonal and Partially Clonal Diploids. Genetics 2003, 164, 1635–1644. [Google Scholar] [CrossRef]
- De Meeûs, T.; Prugnolle, F.; Agnew, P. Asexual Reproduction: Genetics and Evolutionary Aspects. Cell. Mol. Life Sci. 2007, 64, 1355–1372. [Google Scholar] [CrossRef] [PubMed]
- Laínz, M. Culcita C. Presl. In Flora Iberica; Castroviejo, S., Laínz, M., López González, G., Montserrat, P., Muñoz Garmendia, F., Paiva, J., Villar, L., Eds.; Real Jardín Botánico, C.S.I.C.: Madrid, Spain, 1986; Volume 1, pp. 75–76. [Google Scholar]
- Testo, W.; Sundue, M. A 4000-Species Dataset Provides New Insight into the Evolution of Ferns. Mol. Phylogenet. Evol. 2016, 105, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Barahona, S. Incorporating Fossils into the Joint Inference of Phylogeny and Biogeography of the Tree Fern Order Cyatheales. Evolution 2024, 78, 919–933. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. Convention of the Conservation of European Wildlife and Natural Habitats. Eur. Treaty Ser. 1979, 104, 1–27. [Google Scholar]
- Anonymous. Council Directive 92/43/EEC of 21 May 1992 on the Conservation of Habitats and Wild Fauna and Flora. Off. J. Eur. Communities L 1992, 206, 7–50. [Google Scholar]
- Bañares, Á.; Blanca, G.; Güemes, J.; Moreno, J.C.; Ortíz, S. Lista Roja de La Flora Vascular Española. Actualización Con Los Datos de La Adenda 2010 al Atlas y Libro Rojo de La Flora Vascular Amenazada; Dirección General de Medio Natural y Política Forestal (Ministerio de Medio Ambiente, y Medio Rural y Marino); Sociedad Española de Biología de la Conservación de Plantas: Madrid, Spain, 2011. [Google Scholar]
- Bueno Sánchez, A.; Valderrábano Luque, J.; Fernández Prieto, J.A. Culcita macrocarpa C. Presl. In Atlas y Libro Rojo de la Flora Vascular amenazada de España. Adenda 2017; Moreno Saiz, J.C., Iriondo Alegría, J.M., Martínez García, F., Martínez Rodríguez, J., Salazar Mendías, C., Eds.; Ministerio para la Transición Ecológica-Sociedad Española de Biología de la Conservación de Plantas: Madrid, Spain, 2019; pp. 50–51. [Google Scholar]
- Carapeto, A.; Francisco, A.; Pereira, P.; Porto, M. Lista Vermelha Da Flora Vascular de Portugal Continental; Imprensa Nacional: Lisboa, Portugal, 2020; ISBN 978-972-27-2876-8. [Google Scholar]
- Stokey, A.G. Prothallia of the Cyatheaceae. Bot. Gaz. 1930, 90, 1–45. [Google Scholar] [CrossRef]
- Quintanilla, L.G.; Pangua, E.; Amigo, J.; Pajarón, S. Comparative Study of the Sympatric Ferns Culcita macrocarpa and Woodwardia radicans: Sexual Phenotype. Flora 2005, 200, 187–194. [Google Scholar] [CrossRef]
- Klekowski, E.J., Jr.; Lloyd, R.M. Reproductive Biology of the Pteridophyta: 1. General Considerations and a Study of Onoclea sensibilis L. Bot. J. Linn. Soc. 1968, 60, 315–324. [Google Scholar] [CrossRef]
- Hornych, O.; Testo, W.L.; Sessa, E.B.; Watkins, J.E.; Campany, C.E.; Pittermann, J.; Ekrt, L. Insights into the Evolutionary History and Widespread Occurrence of Antheridiogen Systems in Ferns. New Phytol. 2021, 229, 607–619. [Google Scholar] [CrossRef]
- Quintanilla, L.G.; Pajarón, S.; Pangua, E.; Amigo, J. Allozyme Variation in the Sympatric Ferns Culcita macrocarpa and Woodwardia radicans at the Northern Extreme of Their Ranges. Plant Syst. Evol. 2007, 263, 135–144. [Google Scholar] [CrossRef]
- Quintanilla, L.G.; Pajarón, S.; Pangua, E.; Amigo, J. Effect of Temperature on Germination in Northernmost Populations of Culcita macrocarpa and Woodwardia radicans. Plant Biol. 2000, 2, 612–617. [Google Scholar] [CrossRef]
- Arseneau, J.-R.E.; Steeves, R.; Laflamme, M. Modified Low-Salt CTAB Extraction of High-Quality DNA from Contaminant-Rich Tissues. Mol. Ecol. Resour. 2017, 17, 686–693. [Google Scholar] [CrossRef] [PubMed]
- Hall, T. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R Package for Genetic Analysis of Populations with Clonal, Partially Clonal, and/or Sexual Reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [PubMed]
- Stenberg, P.; Lundmark, M.; Saura, A. MLGsim: A Program for Detecting Clones Using a Simulation Approach. Mol. Ecol. Notes 2003, 3, 329–331. [Google Scholar] [CrossRef]
- Arnaud-Haond, S.; Duarte, C.M.; Alberto, F.; Serrão, E.A. Standardizing Methods to Address Clonality in Population Studies. Mol. Ecol. 2007, 16, 5115–5139. [Google Scholar] [CrossRef]
- Agapow, P.; Burt, A. Indices of Multilocus Linkage Disequilibrium. Mol. Ecol. Notes 2001, 1, 101–102. [Google Scholar] [CrossRef]
- De Meeûs, T.; Lehmann, L.; Balloux, F. Molecular Epidemiology of Clonal Diploids: A Quick Overview and a Short DIY (Do It Yourself) Notice. Infect. Genet. Evol. 2006, 6, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. HIERFSTAT, a Package for R to Compute and Test Hierarchical F-Statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef]
- Meirmans, P.G.; Van Tienderen, P.H. GENOTYPE and GENODIVE: Two Programs for the Analysis of Genetic Diversity of Asexual Organisms. Mol. Ecol. Notes 2004, 4, 792–794. [Google Scholar] [CrossRef]
- Douhovnikoff, V.; Leventhal, M. The Use of Hardy-Weinberg Equilibrium in Clonal Plant Systems. Ecol. Evol. 2016, 6, 1173–1180. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of Molecular Variance Inferred from Metric Distances among DNA Haplotypes: Application to Human Mitochondrial DNA Restriction Data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of Population Structure Using Multilocus Genotype Data: Linked Loci and Correlated Allele Frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Liu, J.X. StructureSelector: A Web-Based Software to Select and Visualize the Optimal Number of Clusters Using Multiple Methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Puechmaille, S.J. The Program Structure Does Not Reliably Recover the Correct Population Structure When Sampling Is Uneven: Subsampling and New Estimators Alleviate the Problem. Mol. Ecol. Resour. 2016, 16, 608–627. [Google Scholar] [CrossRef] [PubMed]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A Program for Identifying Clustering Modes and Packaging Population Structure Inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant Analysis of Principal Components: A New Method for the Analysis of Genetically Structured Populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef]
- Templeton, A.R.; Crandall, K.A.; Sing, C.F. A Cladistic Analysis of Phenotypic Associations with Haplotypes Inferred from Restriction Endonuclease Mapping and DNA Sequence Data. III. Cladogram Estimation. Genetics 1992, 132, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A Computer Program to Estimate Gene Genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.A.; Rannala, B. Bayesian Inference of Recent Migration Rates Using Multilocus Genotypes. Genetics 2003, 163, 1177–1191. [Google Scholar] [CrossRef] [PubMed]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very High Resolution Interpolated Climate Surfaces for Global Land Areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- van Vuuren, D.P.; Edmonds, J.; Kainuma, M.; Riahi, K.; Thomson, A.; Hibbard, K.; Hurtt, G.C.; Kram, T.; Krey, V.; Lamarque, J.-F.; et al. The Representative Concentration Pathways: An Overview. Clim. Change 2011, 109, 5. [Google Scholar] [CrossRef]
- Hengl, T.; de Jesus, J.M.; MacMillan, R.A.; Batjes, N.H.; Heuvelink, G.B.M.; Ribeiro, E.; Samuel-Rosa, A.; Kempen, B.; Leenaars, J.G.B.; Walsh, M.G.; et al. SoilGrids1km—Global Soil Information Based on Automated Mapping. PLoS ONE 2014, 9, e105992. [Google Scholar] [CrossRef] [PubMed]
- Soñora, F.X. Notas Pteridológicas de Galicia. IV. Acta Botánica Malacit. 1992, 17, 282–283. [Google Scholar] [CrossRef]
- Quintanilla, L.; Amigo, J. Culcita macrocarpa C. Presl (Dicksoniaceae, Pteridophyta) En La Cuenca Del Xubia (NO de La Peninsula Iberica). Nov. Acta Científica Compostel. (Biol.) 2001, 11, 267–268. [Google Scholar]
- Romero, M.; Rodríguez Guitián, M.; Rubinos, M. Adiciones al Catálogo Pteridológico Gallego. Bot. Complut. 2004, 28, 51–55. [Google Scholar]
- Rodríguez Guitián, M.A.; Ferreiro da Costa, J.; Ramil Rego, P.; Lijó Pose, G. Caracterización Ambiental, Demografía y Amenazas Para Su Conservacion de La Población Lucense de Culcita macrocarpa C. Presl. (NW Ibérico). Recur. Rurais 2011, 7, 15–25. [Google Scholar]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum Entropy Modeling of Species Geographic Distributions. Ecol. Modell. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Martínez, B.; Viejo, R.M.; Carreño, F.; Aranda, S.C. Habitat Distribution Models for Intertidal Seaweeds: Responses to Climatic and Non-Climatic Drivers. J. Biogeogr. 2012, 39, 1877–1890. [Google Scholar] [CrossRef]
- Hurlbert, S.H. The Nonconcept of Species Diversity: A Critique and Alternative Parameters. Ecology 1971, 52, 577–586. [Google Scholar] [CrossRef]
- Dorken, M.E.; Eckert, C.G. Severely Reduced Sexual Reproduction in Northern Populations of a Clonal Plant, Decodon verticillatus (Lythraceae). J. Ecol. 2001, 89, 339–350. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Pielou, E.C. Ecological Diversity; A Wiley-Interscience publication; John Wiley & Sons: New York, NY, USA, 1975; Volume viii, ISBN 9780471689256. [Google Scholar]
- Ludwig, J.A.; Reynolds, J.F. Statistical Ecology: A Primer in Methods and Computing; Wiley: New York, NY, USA, 1988; ISBN 9780471832355. [Google Scholar]
- Grünwald, N.J.; Goodwin, S.B.; Milgroom, M.G.; Fry, W.E. Analysis of Genotypic Diversity Data for Populations of Microorganisms. Phytopathology 2003, 93, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987; ISBN 9780231886710. [Google Scholar]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Carrillo-Angeles, I.G.; Golubov, J.; Milligan, B.G.; Mandujano, M.C. Spatial Distribution Pattern of a Clonal Species: Effects of Differential Production of Clonal and Sexual Offspring. Evol. Ecol. 2011, 25, 1357–1383. [Google Scholar] [CrossRef]
- Petit, R.J.; Mousadik, A.E.L.; Pons, O. Identifying Populations for Conservation on the Basis of Genetic Markers. Conserv. Biol. 1998, 12, 844–855. [Google Scholar] [CrossRef]
- Prugnolle, F.; De Meeûs, T. The Impact of Clonality on Parasite Population Genetic Structure. Parasite 2008, 15, 455–457. [Google Scholar] [CrossRef]
- Provan, J.; Bennett, K.D. Phylogeographic Insights into Cryptic Glacial Refugia. Trends Ecol. Evol. 2008, 23, 564–571. [Google Scholar] [CrossRef]
- Schneller, J.J.; Holderegger, R. Genetic Variation in Small, Isolated Fern Populations. J. Veg. Sci. 1996, 7, 113–120. [Google Scholar] [CrossRef]
- Vogel, J.C.; Rumsey, F.J.; Russell, S.J.; Cox, C.J.; Holmes, J.S.; Bujnoch, W.; Stark, C.; Barrett, J.A.; Gibby, M. Genetic Structure, Reproductive Biology and Ecology of Isolated Populations of Asplenium csikii (Aspleniaceae, Pteridophyta). Heredity 1999, 83, 604–612. [Google Scholar] [CrossRef] [PubMed]
- García-Verdugo, C.; Sajeva, M.; La Mantia, T.; Harrouni, C.; Msanda, F.; Caujapé-Castells, J. Do Island Plant Populations Really Have Lower Genetic Variation than Mainland Populations? Effects of Selection and Distribution Range on Genetic Diversity Estimates. Mol. Ecol. 2015, 24, 726–741. [Google Scholar] [CrossRef] [PubMed]
- Hutsemékers, V.; Szövényi, P.; Shaw, A.J.; González-Mancebo, J.-M.; Muñoz, J.; Vanderpoorten, A. Oceanic Islands Are Not Sinks of Biodiversity in Spore-Producing Plants. Proc. Natl. Acad. Sci. USA 2011, 108, 18989–18994. [Google Scholar] [CrossRef] [PubMed]
- Ben-Menni Schuler, S.; Picazo-Aragonés, J.; Rumsey, F.J.; Romero-García, A.T.; Suárez-Santiago, V.N. Macaronesia Acts as a Museum of Genetic Diversity of Relict Ferns: The Case of Diplazium caudatum (Athyriaceae). Plants 2021, 10, 2425. [Google Scholar] [CrossRef]
- Bauert, M.R.; Kälin, M.; Baltisberger, M.; Edwards, P.J. No Genetic Variation Detected within Isolated Relict Populations of Saxifraga cernua in the Alps Using RAPD Markers. Mol. Ecol. 1998, 7, 1519–1527. [Google Scholar] [CrossRef]
- Paun, O.; Stuessy, T.F.; Hörandl, E. The Role of Hybridization, Polyploidization and Glaciation in the Origin and Evolution of the Apomictic Ranunculus cassubicus Complex. New Phytol. 2006, 171, 223–236. [Google Scholar] [CrossRef]
- Peck, J.H.; Peck, C.J.; Farrar, D.R. Influences of Life History Attributes on Formation of Local and Distant Fern Populations. Am. Fern. J. 1990, 80, 126. [Google Scholar] [CrossRef]
- Sessa, E.B.; Testo, W.L.; Watkins, J.E. On the Widespread Capacity for, and Functional Significance of, Extreme Inbreeding in Ferns. New Phytol. 2016, 211, 1108–1119. [Google Scholar] [CrossRef] [PubMed]
- De Groot, G.A.; During, H.J.; Ansell, S.W.; Schneider, H.; Bremer, P.; Wubs, E.R.J.; Maas, J.W.; Korpelainen, H.; Erkens, R.H.J. Diverse Spore Rains and Limited Local Exchange Shape Fern Genetic Diversity in a Recently Created Habitat Colonized by Long-Distance Dispersal. Ann. Bot. 2012, 109, 965–978. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.B.; Beck, J.T.; Farmer, S.B.; Liu, W.; Miller, J.; Siripun, K.C.; Winder, C.T.; Schilling, E.E.; Small, R.L. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005, 92, 142–166. [Google Scholar] [CrossRef]



| Sample Size | |||||
|---|---|---|---|---|---|
| Code | Location | Voucher | Geographical Coordinates | Microsatellites | ptDNA |
| Andalusia | |||||
| ALM | Cádiz: Almoraima | GDA 65361 | N 36.304°/W 5.520° | 6 | 3 |
| CRM | Cádiz: Cabecera del río de la Miel | GDA 65363 | N 36.105°/W 5.528° | 30 | 5 |
| PIN | Cádiz: Laja del Pinarejo | GDA 65360 | N 36.188°/W 5.589° | 30 | 5 |
| RM | Cádiz: Río de la Miel | GDA 65359 | N 36.112°/W 5.507° | 29 | 5 |
| SDN | Cádiz: Sierra del Niño | GDA 65362 | N 36.186°/W 5.610° | 30 | 5 |
| Azores | |||||
| CAR | Terceira: Algar do Carvão | GDA 63533 | N 38.727°/W 27.215° | 30 | 5 |
| CID | São Miguel: Sete Cidades | GDA 63534 | N 37.835°/W 25.788° | 29 | 5 |
| FOG | São Miguel: Lagoa do Fogo | GDA 63532 | 30 | 5 | |
| NAT | Terceira: Gruta do Natal | GDA 63531 | N 38.738°/W 27.264° | 30 | 5 |
| Canary Isl. | |||||
| IJU | Tenerife: Ijuana | GDA 63536 | N 28.560°/W 16.172° | 30 | 4 |
| Cantabrian Cornice | |||||
| BER | Bizkaia: Bermeo | GDA 63539 | N 43.392°/W 2.734° | 12 | 5 |
| BAK | Bizkaia: Bakio | GDA 65364 | N 43.425°/W 2.845° | 18 | 5 |
| CUN | Asturias: San Esteban de Cuñaba | GDA 63537 | N 43.277°/W 4.676° | 30 | 5 |
| EUM | A Coruña: Eume | GDA 63535 | N 43.404°/W 8.087° | 30 | 5 |
| LIE | Cantabria: Liendo | GDA 65365 | N 43.375°/W 3.383° | 30 | 5 |
| NUE | Asturias: Nueva de Llanes | GDA 63538 | N 43.421°/W 4.954° | 30 | 5 |
| SEI | A Coruña: Seixo | GDA 63530 | N 43.706°/W 7.946° | 19 | 5 |
| Clonal Richness | Genotype Diversity | Linkage Disequilibrium | ||||||
|---|---|---|---|---|---|---|---|---|
| Population | N | MLL (Private) | eMLL | R | Lambda | E.5 | rd | grd |
| Andalusia | 125 | 16 (12) | 6.56 | 0.121 | 0.579 | 0.464 | 0.402 * | 0.186 * |
| ALM | 6 | 4 (4) | 4 | 0.6 | 0.8 | 0.812 | NA | NA |
| CRM | 30 | 2 | 1.33 | 0.034 | 0.067 | 0.438 | NA | NA |
| PIN | 30 | 8 (5) | 4.12 | 0.241 | 0.623 | 0.528 | 0.728 * | 0.193 * |
| RM | 29 | 3 (3) | 1.92 | 0.071 | 0.197 | 0.48 | 0.141 * | 0.5 |
| SDN | 30 | 1 | 1 | 0 | 0 | NA | NA | NA |
| Azores | 119 | 35 (25) | 16.01 | 0.288 | 0.904 | 0.5 | 0.041 * | −0.05 |
| CAR | 30 | 12 (4) | 6.53 | 0.379 | 0.881 | 0.727 | 0.062 * | −0.047 |
| CID | 29 | 14 (12) | 6.98 | 0.464 | 0.901 | 0.719 | 0.140 * | 0.003 |
| FOG | 30 | 12 (5) | 6.68 | 0.379 | 0.885 | 0.725 | 0.029 | −0.063 |
| NAT | 30 | 11 (4) | 6.03 | 0.345 | 0.855 | 0.713 | 0.005 | −0.067 |
| Canary Isl. | 30 | 7 (5) | 7 | 0.207 | 0.811 | 0.836 | −0.018 | −0.248 |
| IJU | 30 | 7 (5) | 4.86 | 0.207 | 0.811 | 0.836 | −0.018 | −0.248 |
| Cantabrian Cornice | 169 | 53 (44) | 14.93 | 0.31 | 0.853 | 0.347 | 0.185 * | 0.085 * |
| BER | 12 | 6 (6) | 5.33 | 0.454 | 0.803 | 0.762 | 0.3 * | −0.042 |
| BAK | 18 | 3 (2) | 2.55 | 0.118 | 0.503 | 0.757 | 0.536 * | 0 |
| CUN | 30 | 23 (16) | 9.2 | 0.759 | 0.982 | 0.913 | 0.04 * | 0.011 |
| EUM | 30 | 1 | 1 | 0 | 0 | NA | NA | NA |
| LIE | 30 | 1 (1) | 1 | 0 | 0 | NA | NA | NA |
| NUE | 30 | 18 (16) | 8.01 | 0.586 | 0.942 | 0.76 | 0.07 * | 0.024 |
| SEI | 19 | 4 (3) | 2.84 | 0.166 | 0.38 | 0.52 | 0.725 * | 0.12 |
| Total | 443 (130) | 104 (91) | 7.69 | 0.292 | 0.926 | 0.384 | 0.288 | |
| Microsatellites | ptDNA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | N | A (Private) | Ar | gAr | HO | gHO | HE | gHE | FIS | gFIS | ha | Hd | π |
| Andalusia | 125 | 18 (3) | 2.228 | 2.187 | 0.164 | 0.215 | 0.331 | 0.426 | 0.505 * | 0.494 * | 3 (1) | 0.38 | 0.17 |
| ALM | 6 | 11 (1) | 1.375 | 1.165 | 0.146 | 0.156 | 0.146 | 0.167 | 0.000 | 0.063 | 1 | 0.00 | 0.00 |
| CRM | 30 | 10 | 1.15 | 1.146 | 0.129 | 0.188 | 0.067 | 0.125 | −0.938 * | −0.500 | 1 | 0.00 | 0.00 |
| PIN | 30 | 12 | 1.498 | 1.255 | 0.163 | 0.250 | 0.232 | 0.267 | 0.301 * | 0.063 | 2 | 0.40 | 0.17 |
| RM | 29 | 11 (1) | 1.338 | 1.217 | 0.246 | 0.250 | 0.145 | 0.208 | −0.692 * | −0.200 | 2 | 0.40 | 0.17 |
| SDN | 30 | 9 | 1.125 | 1.125 | 0.125 | NA | 0.063 | NA | −1.000 * | NA | 1 | 0.00 | 0.00 |
| Azores | 119 | 21 (6) | 2.414 | 2.159 | 0.169 | 0.181 | 0.267 | 0.330 | 0.366 * | 0.452 * | 2 | 0.19 | 0.08 |
| CAR | 30 | 15 (1) | 1.708 | 1.307 | 0.175 | 0.177 | 0.235 | 0.308 | 0.257 * | 0.425 * | 1 | 0.00 | 0.00 |
| CID | 29 | 17 (2) | 1.932 | 1.365 | 0.198 | 0.205 | 0.327 | 0.371 | 0.394 * | 0.447 * | 1 | 0.00 | 0.00 |
| FOG | 30 | 16 (1) | 1.727 | 1.280 | 0.163 | 0.188 | 0.232 | 0.284 | 0.299 * | 0.340 * | 2 | 0.60 | 0.25 |
| NAT | 30 | 15 | 1.668 | 1.273 | 0.142 | 0.148 | 0.209 | 0.280 | 0.323 * | 0.472 * | 1 | 0.00 | 0.00 |
| Canary Isl. | 30 | 13 | 1.625 | 1.625 | 0.267 | 0.268 | 0.193 | 0.211 | −0.385 * | −0.268 | 1 | 0.00 | 0.00 |
| IJU | 30 | 13 | 1.498 | 1.216 | 0.267 | 0.268 | 0.193 | 0.211 | −0.385 * | −0.268 | 1 | 0.00 | 0.00 |
| Cantabrian Cornice | 169 | 26 (8) | 2.745 | 2.461 | 0.133 | 0.248 | 0.369 | 0.473 | 0.639 * | 0.476 * | 3 (1) | 0.42 | 0.18 |
| BER | 12 | 16 (1) | 1.815 | 1.333 | 0.125 | 0.208 | 0.288 | 0.346 | 0.566 * | 0.398 * | 2 | 0.40 | 0.17 |
| BAK | 18 | 11 | 1.320 | 1.2 | 0.000 | 0.000 | 0.132 | 0.250 | 1.000 * | 1.000 * | 1 | 0.00 | 0.00 |
| CUN | 30 | 16 (1) | 1.879 | 1.373 | 0.425 | 0.408 | 0.364 | 0.372 | −0.169 * | −0.096 | 1 | 0.00 | 0.00 |
| EUM | 30 | 8 | 1 | 1 | 0.000 | NA | 0.000 | NA | NA | NA | 1 | 0.00 | 0.00 |
| LIE | 30 | 9 | 1.125 | 1.125 | 0.125 | NA | 0.063 | NA | −1.000 * | NA | 1 | 0.00 | 0.00 |
| NUE | 30 | 19 (1) | 2.084 | 1.341 | 0.117 | 0.132 | 0.347 | 0.356 | 0.663 * | 0.630 * | 3 (1) | 0.70 | 0.34 |
| SEI | 19 | 16 (1) | 1.605 | 1.339 | 0.053 | 0.188 | 0.149 | 0.365 | 0.646 * | 0.486 * | 2 | 0.60 | 0.25 |
| Total | 443 | 37 | 1.520 | 1.239 | 0.153 | 0.177 | 0.528 | 0.545 | 0.186 * | 0.357 * | 4 | 0.52 | 0.0023 |
| Source of Variation | d.f. | Sum of Squares | Percentage of Variation | Phi | p-Value |
|---|---|---|---|---|---|
| Microsatellites | |||||
| All sampling units (ramet-based analyses) | |||||
| Standard | |||||
| Within samples | 443 | 547 | 29,23 | 0.708 | <0.001 |
| Between samples within populations | 426 | 742.883 | 6.03 | 0.171 | <0.001 |
| Between populations | 16 | 2296.697 | 64.74 | 0.647 | <0.001 |
| Total | 885 | 3586.58 | 100 | ||
| Hierarchical (4 geographical regions) | |||||
| Within samples | 443 | 547 | 26.41 | 0.736 | <0.001 |
| Between samples within populations | 426 | 742.883 | 5.44 | 0.171 | <0.001 |
| Between population within regions | 13 | 949.498 | 30.18 | 0.486 | <0.001 |
| Between regions | 3 | 1347.199 | 37.97 | 0.379 | <0.001 |
| Total | 885 | 3586.58 | 100 | ||
| MLLs (genet-based analyses) | |||||
| Standard | |||||
| Within samples | 130 | 224 | 42.6 | 0.574 | <0.001 |
| Between samples within populations | 113 | 371.188 | 19.31 | 0.312 | <0.001 |
| Between populations | 16 | 413.658 | 38.09 | 0.381 | <0.001 |
| Total | 259 | 1008.846 | 100 | ||
| Hierarchical (4 geographical regions) | |||||
| Within samples | 130 | 224 | 40.44 | 0.596 | <0.001 |
| Between samples within populations | 113 | 371.188 | 18.33 | 0.312 | <0.001 |
| Between population within regions | 13 | 217.449 | 23.02 | 0.281 | <0.001 |
| Between regions | 3 | 196.208 | 18.21 | 0.182 | <0.001 |
| Total | 259 | 1008.846 | 100 | ||
| ptDNA | |||||
| Standard | |||||
| Between populations | 17 | 16.529 | 65.96 | <0.001 | |
| Within populations | 66 | 6.4 | 34.04 | <0.001 | |
| Total | 83 | 22.929 | 100 | ||
| Hierarchical (4 geographical regions) | |||||
| Between regions | 3 | 8.069 | 38.37 | 0.0123 | |
| Between populations within regions | 14 | 7.314 | 30.44 | <0.001 | |
| Within populations | 66 | 6.200 | 31.19 | <0.001 | |
| Total | 83 | 21.583 | 100 | ||
| Hierarchical (2 supra-regional groups) | |||||
| Between groups | 1 | 7.873 | 50.53 | 0.0017 | |
| Between populations within groups | 16 | 7.511 | 22.87 | <0.001 | |
| Within populations | 66 | 6.200 | 26.60 | <0.001 | |
| Total | 83 | 21.583 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suárez-Santiago, V.N.; Provan, J.; Romero-García, A.T.; Ben-Menni Schuler, S. Genetic Diversity and Phylogeography of the Relict Tree Fern Culcita macrocarpa: Influence of Clonality and Breeding System on Genetic Variation. Plants 2024, 13, 1587. https://doi.org/10.3390/plants13121587
Suárez-Santiago VN, Provan J, Romero-García AT, Ben-Menni Schuler S. Genetic Diversity and Phylogeography of the Relict Tree Fern Culcita macrocarpa: Influence of Clonality and Breeding System on Genetic Variation. Plants. 2024; 13(12):1587. https://doi.org/10.3390/plants13121587
Chicago/Turabian StyleSuárez-Santiago, Víctor N., Jim Provan, Ana Teresa Romero-García, and Samira Ben-Menni Schuler. 2024. "Genetic Diversity and Phylogeography of the Relict Tree Fern Culcita macrocarpa: Influence of Clonality and Breeding System on Genetic Variation" Plants 13, no. 12: 1587. https://doi.org/10.3390/plants13121587
APA StyleSuárez-Santiago, V. N., Provan, J., Romero-García, A. T., & Ben-Menni Schuler, S. (2024). Genetic Diversity and Phylogeography of the Relict Tree Fern Culcita macrocarpa: Influence of Clonality and Breeding System on Genetic Variation. Plants, 13(12), 1587. https://doi.org/10.3390/plants13121587







