Physiological Characteristics and Transcriptomic Responses of Pinus yunnanensis Lateral Branching to Different Shading Environments
Abstract
1. Introduction
2. Results
2.1. Effect of Shading Treatment on Lateral Branching Ability of P. yunnanensis Seedlings
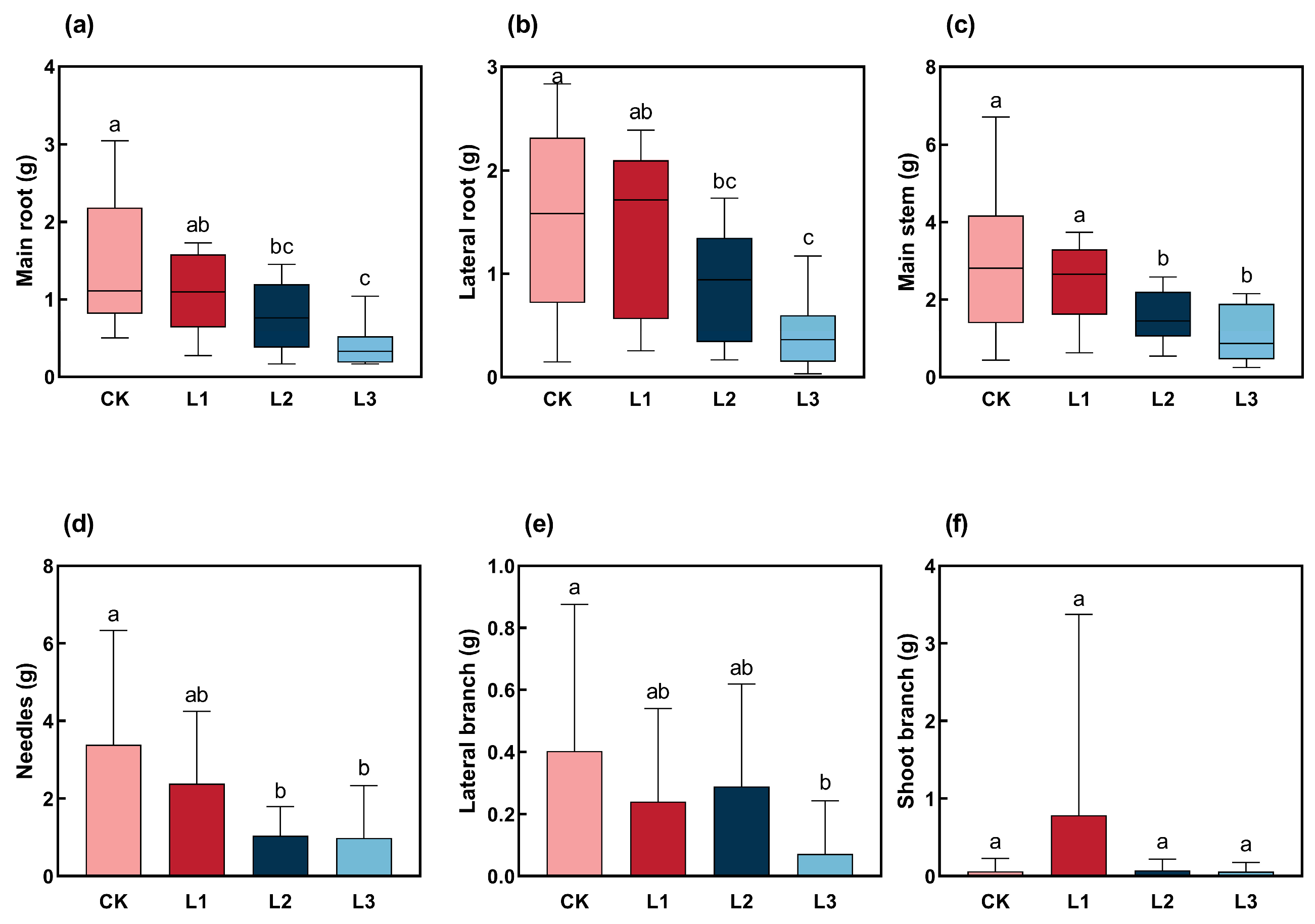
2.2. Response of Biomass Accumulation in Various Organs of P. yunnanensis to Different Shading Intensities
2.3. Effect of Shading Treatments on Photosynthesis of P. yunnanensis Seedlings
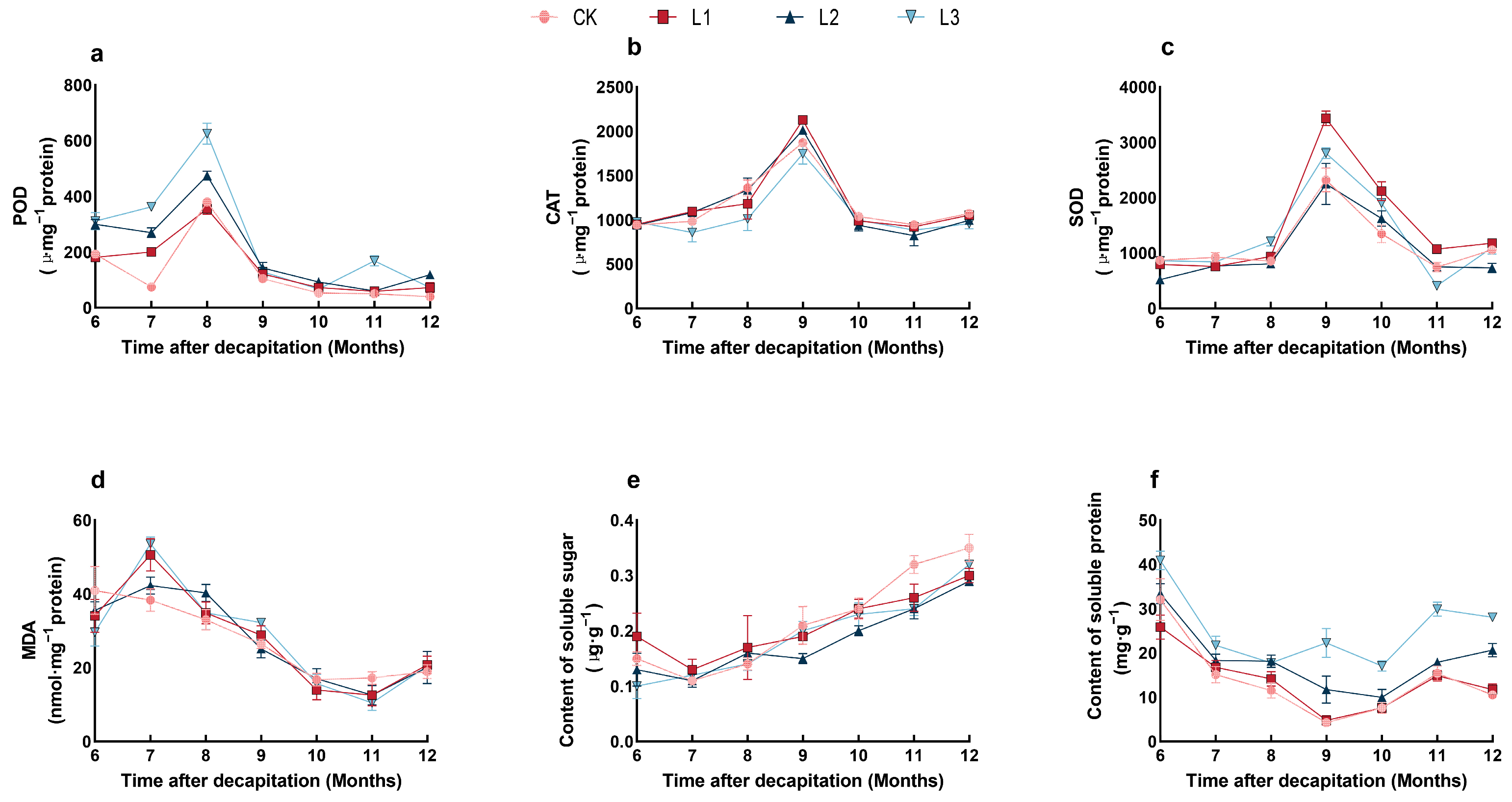
2.4. Effect of Shading Treatments on the Physiological Characteristics of P. yunnanensis Seedlings
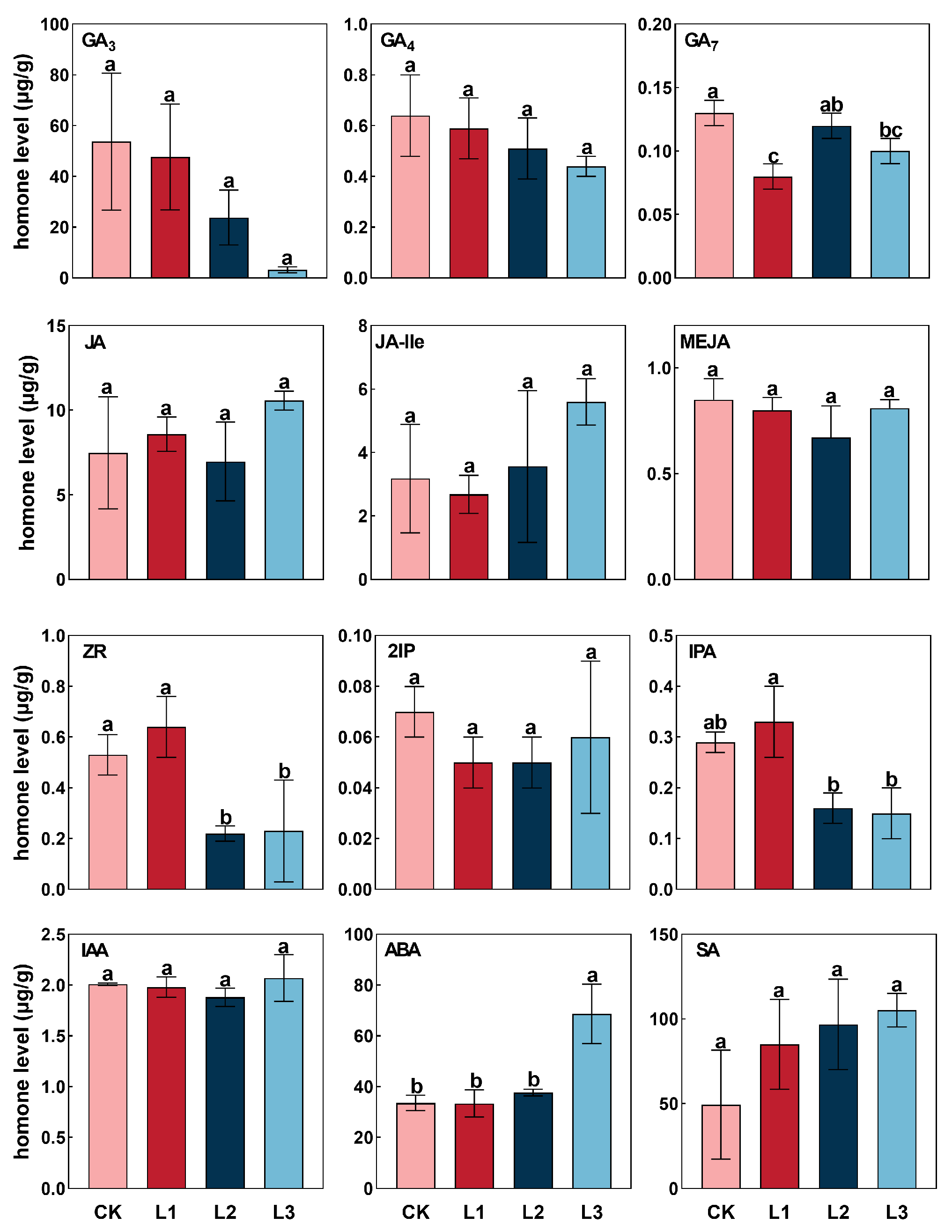
2.5. Response of Endogenous Hormone to Shading Treatments
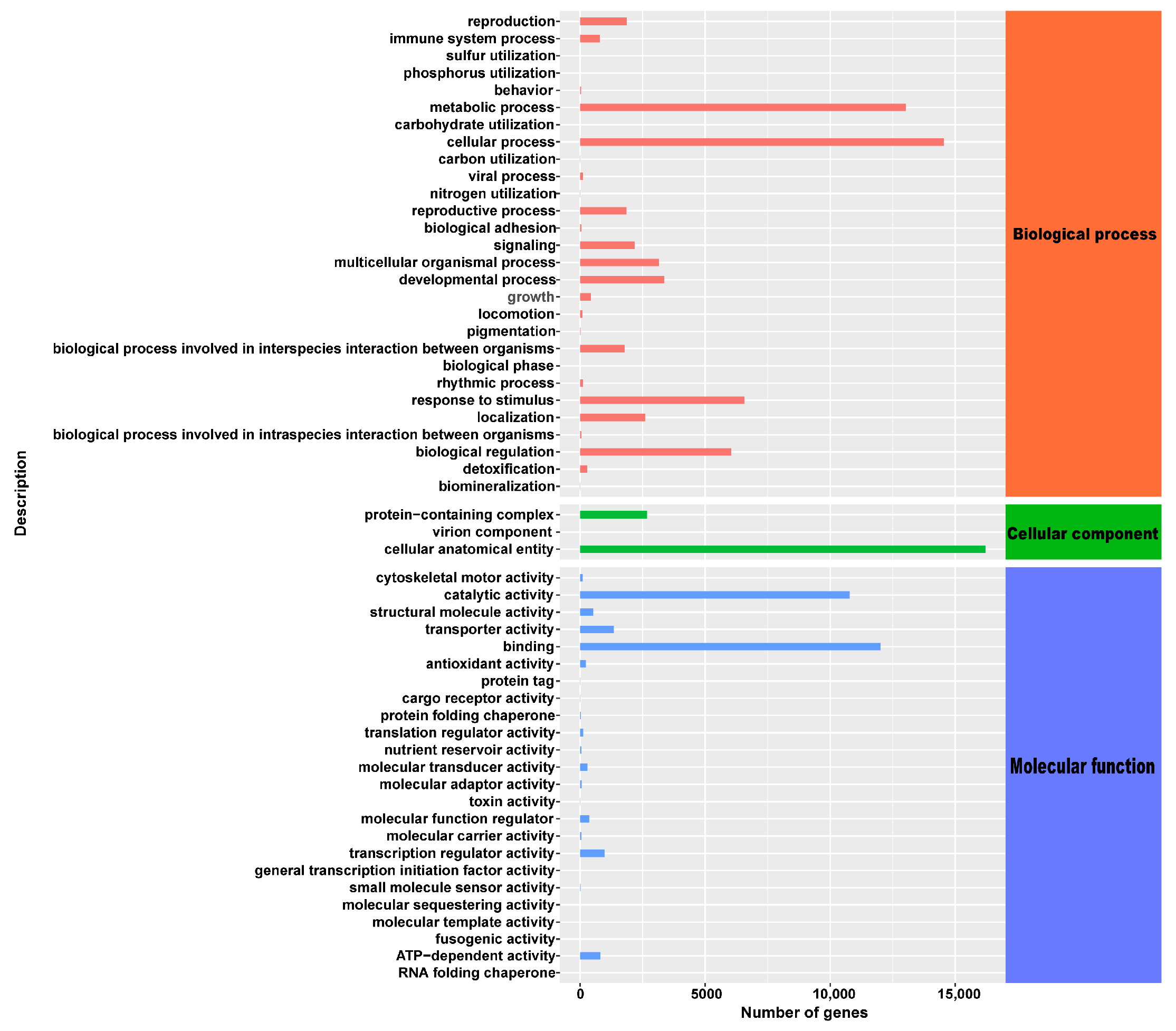
2.6. Transcriptomic Sequencing and De Novo Assembly and Annotation
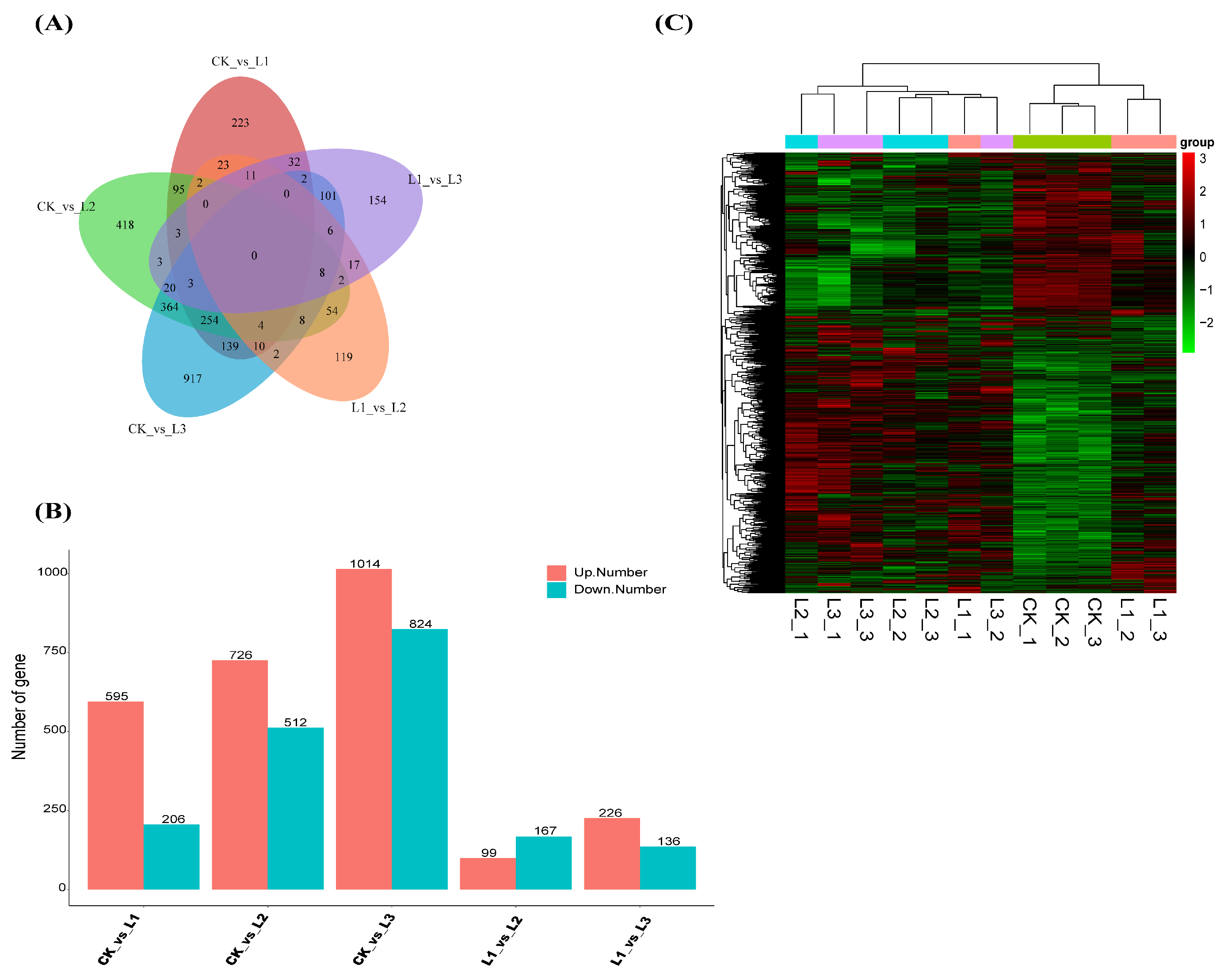
2.7. Analysis of Differentially Expressed Genes in P. yunnanensis under Different Shadings
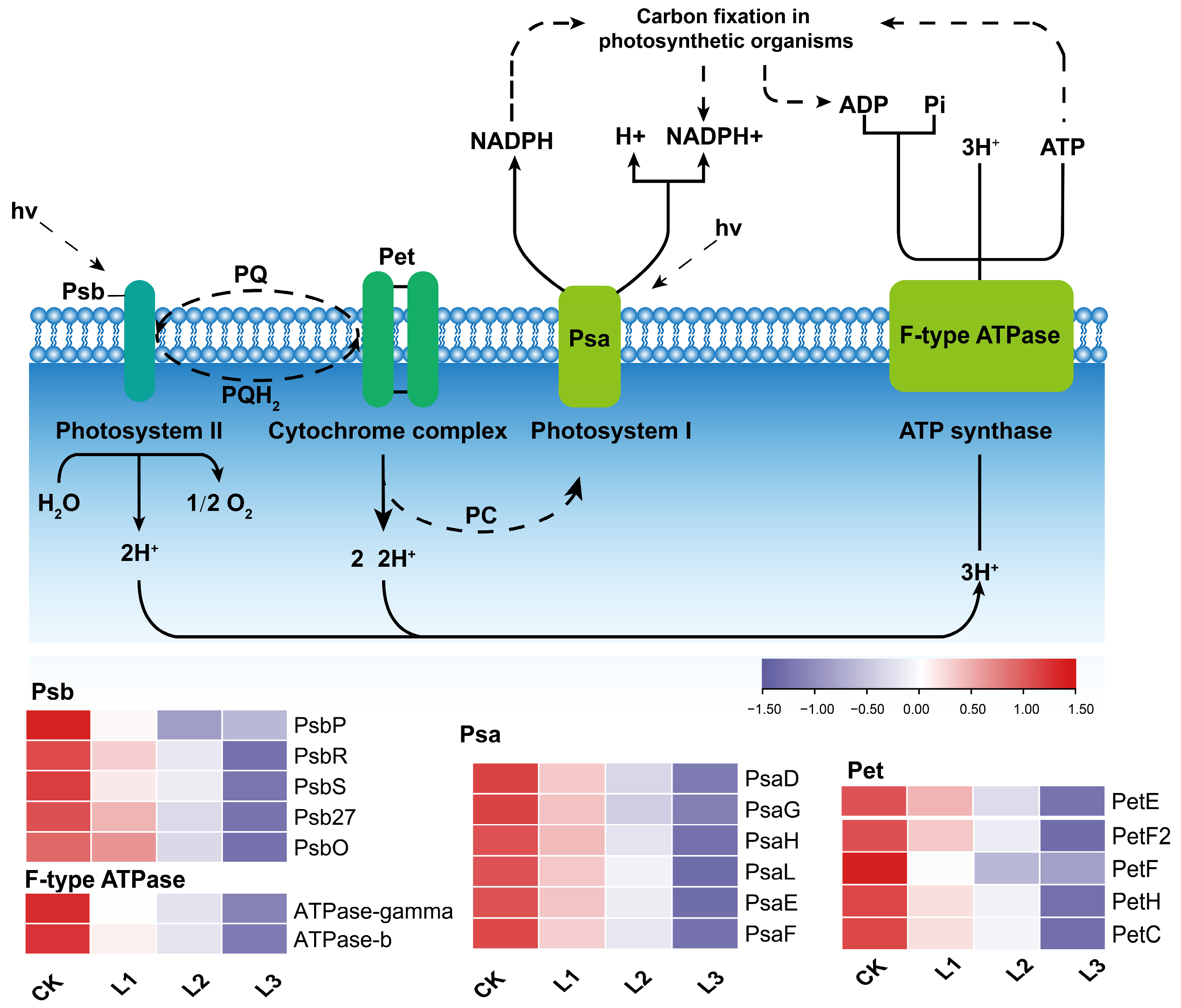
2.8. Response of Related Genes in the Photosynthesis Pathway to Different Shade Treatments
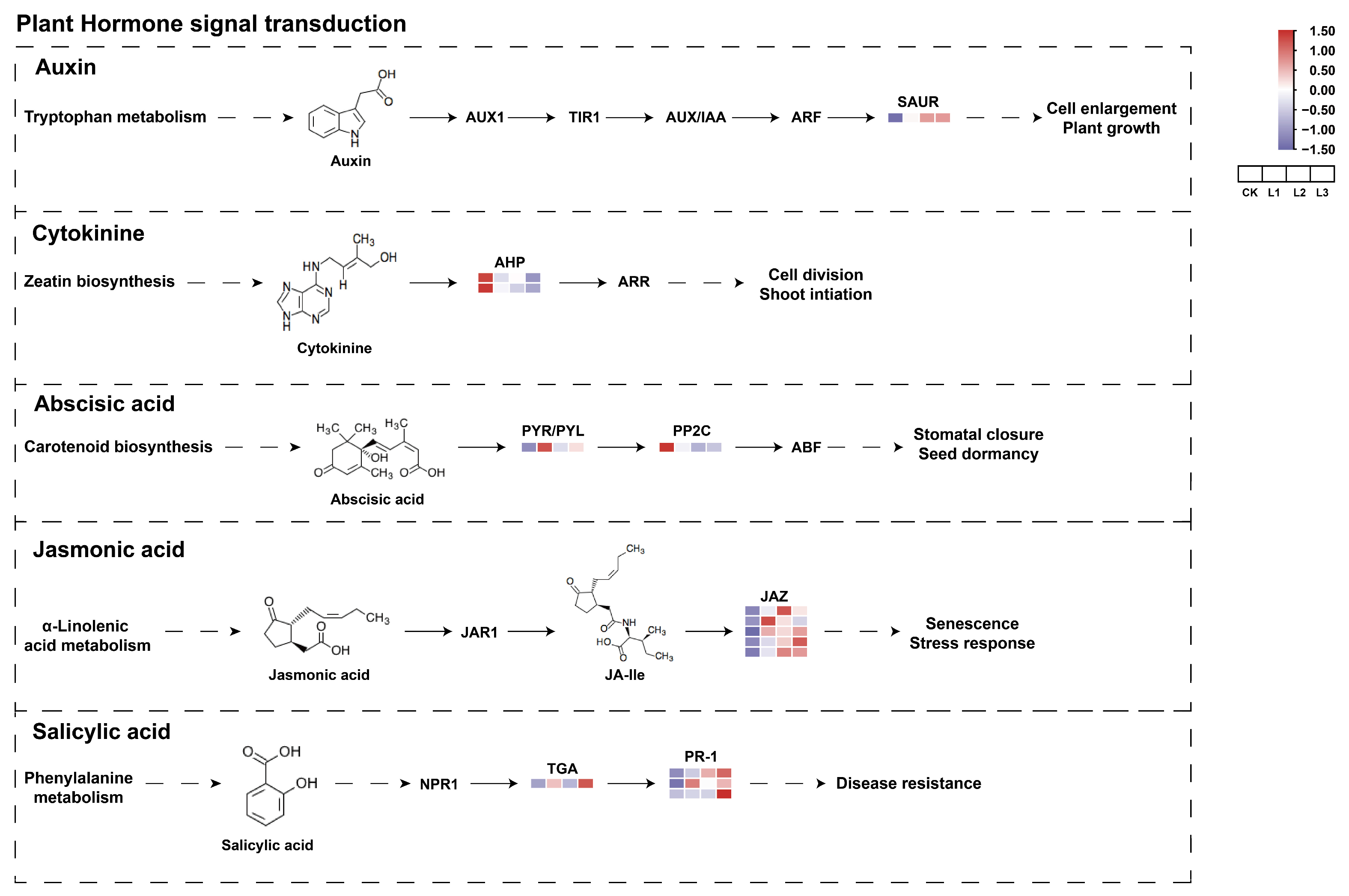
2.9. Response of Related Genes on the Hormone Signal Transduction Pathway to Different Shade Treatments
2.10. RT-qPCR Validation of Differential Genes in P. yunnanensis under Different Shading Levels
3. Discussion
3.1. Lateral Branching in Response to Different Shading Treatments
3.2. Comparison of Photosynthetic Pigment and Physiological Characteristics under Shading
3.3. Role of Phytohormones in the Growth and Development of Lateral Branching
3.4. The Response of Genes Related to Hormone and Photosynthesis Pathways in Pinus yunnanensis to Different Shading Levels
4. Material and Methods
4.1. Plant Material and Shading Treatment
4.2. Growth Indexes and Biomass Determination
4.3. Assay of Physiological Index Contents
4.4. Evaluation of Endogenous Hormone Content
4.5. Transcriptome Sequencing Analysis
4.6. RT-qPCR Validation of DEGs
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hammer, G.L.; McLean, G.; Kholova, J.; van Oosterom, E. Modelling the dynamics and phenotypic consequences of tiller outgrowth and cessation in sorghum. Silico Plants 2023, 5, diad019. [Google Scholar] [CrossRef]
- Ward, S.P.; Leyser, O. Shoot branching. Curr. Opin. Plant Biol. 2004, 7, 73–78. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, Y.; Tu, J.; Liang, D.; Li, M.; Wu, F. Comparative analysis of differential gene expression reveals novel insights into the heteroblastic foliage functional traits of Pinus massoniana seedlings. Int. J. Biol. Macromol. 2024, 264, 130762. [Google Scholar] [CrossRef]
- Leyser, O. The control of shoot branching: An example of plant information processing. Plant Cell Environ. 2009, 32, 694–703. [Google Scholar] [CrossRef]
- Rameau, C.; Bertheloot, J.; Leduc, N.; Andrieu, B.; Foucher, F.; Sakr, S. Multiple pathways regulate shoot branching. Front. Plant Sci. 2015, 5, 15. [Google Scholar] [CrossRef]
- Wang, B.; Smith, S.M.; Li, J.Y. Genetic Regulation of Shoot Architecture. In Annual Review of Plant Biology; Merchant, S.S., Ed.; Annual Review of Plant Biology; Annual Reviews: Palo Alto, CA, USA, 2018; Volume 69, pp. 437–468. [Google Scholar]
- Stirnberg, P.; van de Sande, K.; Leyser, H.M.O. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 2002, 129, 1131–1141. [Google Scholar] [CrossRef]
- Mason, M.G.; Ross, J.J.; Babst, B.A.; Wienclaw, B.N.; Beveridge, C.A. Sugar demand, not auxin, is the initial regulator of apical dominance. Proc. Natl. Acad. Sci. USA 2014, 111, 6092–6097. [Google Scholar] [CrossRef]
- Zhou, Z.; Feng, J. Light Response Characteristics of Ash in Ardisia giantifolia and Its Correlation with Growth Indexes. J. Trop. Subtrop. Bot. 2024, 32, 111–117. [Google Scholar] [CrossRef]
- Sims, D.A.; Pearcy, R.W. Response of leaf anatomy and photosynthetic capacity in alocasia-macrorrhiza (araceae) to a transfer from low to high light. Am. J. Bot. 1992, 79, 449–455. [Google Scholar] [CrossRef]
- Saura-Sánchez, M.; Gomez-Ocampo, G.; Pereyra, M.E.; Barraza, C.E.; Rossi, A.H.; Córdoba, J.P.; Botto, J.F. B-Box transcription factor BBX28 requires CONSTITUTIVE PHOTOMORPHOGENESIS1 to induce shade-avoidance response in Arabidopsis thaliana. Plant Physiol. 2024, 13, kiae216. [Google Scholar] [CrossRef]
- Fernández-Milmanda, G.L.; Ballaré, C.L. Shade Avoidance: Expanding the Color and Hormone Palette. Trends Plant Sci. 2021, 26, 509–523. [Google Scholar] [CrossRef]
- Ugarte, C.C.; Trupkin, S.A.; Ghiglione, H.; Slafer, G.; Casal, J.J. Low red/far-red ratios delay spike and stem growth in wheat. J. Exp. Bot. 2010, 61, 3151–3162. [Google Scholar] [CrossRef]
- Wang, X.C.; Wu, X.L.; Ding, G.H.; Yang, F.; Yong, T.W.; Wang, X.C.; Yang, W.Y. Analysis of Grain Yield Differences Among Soybean Cultivars under Maize-Soybean Intercropping. Agronomy 2020, 10, 110. [Google Scholar] [CrossRef]
- Yao, X.D.; Zhou, H.L.; Zhu, Q.; Li, C.H.; Zhang, H.J.; Wu, J.J.; Xie, F.T. Photosynthetic Response of Soybean Leaf to Wide Light-Fluctuation in Maize-Soybean Intercropping System. Front. Plant Sci. 2017, 8, 10. [Google Scholar] [CrossRef]
- Yao, X.D.; Li, C.H.; Li, S.Y.; Zhu, Q.; Zhang, H.J.; Wang, H.Y.; Yu, C.M.; St Martin, S.K.; Xie, F.T. Effect of shade on leaf photosynthetic capacity, light-intercepting, electron transfer and energy distribution of soybeans. Plant Growth Regul. 2017, 83, 409–416. [Google Scholar] [CrossRef]
- Deng, B.; Shang, X.L.; Fang, S.Z.; Li, Q.Q.; Fu, X.X.; Su, J. Integrated Effects of Light Intensity and Fertilization on Growth and Flavonoid Accumulation in Cyclocarya paliurus. J. Agric. Food Chem. 2012, 60, 6286–6292. [Google Scholar] [CrossRef]
- Li, Y.M.; Zheng, Y.J.; Liu, H.C.; Zhang, Y.T.; Hao, Y.W.; Song, S.W.; Lei, B.F. Effect of supplemental blue light intensity on the growth and quality of Chinese kale. Hortic. Environ. Biotechnol. 2019, 60, 49–57. [Google Scholar] [CrossRef]
- Roeber, V.M.; Bajaj, I.; Rohde, M.; Schmülling, T.; Cortleven, A. Light acts as a stressor and influences abiotic and biotic stress responses in plants. Plant Cell Environ. 2021, 44, 645–664. [Google Scholar] [CrossRef]
- Wu, Y.S.; Gong, W.Z.; Yang, W.Y. Shade Inhibits Leaf Size by Controlling Cell Proliferation and Enlargement in Soybean. Sci. Rep. 2017, 7, 10. [Google Scholar] [CrossRef]
- Pawlowska, B.; Zupnik, M.; Szewczyk-Taranek, B.; Cioc, M. Impact of LED light sources on morphogenesis and levels of photosynthetic pigments in Gerbera jamesonii grown in vitro. Hortic. Environ. Biotechnol. 2018, 59, 115–123. [Google Scholar] [CrossRef]
- Shin, K.S.; Murthy, H.N.; Heo, J.W.; Hahn, E.J.; Paek, K.Y. The effect of light quality on the growth and development of in vitro cultured Doritaenopsis plants. Acta Physiol. Plant. 2008, 30, 339–343. [Google Scholar] [CrossRef]
- Siringam, K.; Juntawong, N.; Cha-um, S.; Kirdmanee, C. Salt stress induced ion accumulation, ion homeostasis, membrane injury and sugar contents in salt-sensitive rice (Oryza sativa L. spp. indica) roots under iso-osmotic conditions. Afr. J. Biotechnol. 2011, 10, 1340–1346. [Google Scholar]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef]
- Huang, Y.J.; Zhai, Y.N.; Huang, Y.; Huang, Y.F.; Liu, K.; Zhang, J.; Zhou, J.H. Effects of Light Intensity on Physiological Characteristics and Expression of Genes in Coumarin Biosynthetic Pathway of Angelica dahurica. Int. J. Mol. Sci. 2022, 23, 15912. [Google Scholar] [CrossRef]
- Liu, W.G.; Hussain, S.; Ting, L.I.; Zou, J.L.; Ren, M.L.; Tao, Z.H.; Jiang, L.I.; Feng, Y.A.; Yang, W.Y. Shade stress decreases stem strength of soybean through restraining lignin biosynthesis. J. Integr. Agric. 2019, 18, 43. [Google Scholar] [CrossRef]
- Thévenin, J.; Pollet, B.; Letarnec, B.; Saulnier, L.; Gissot, L.; Maia-Grondard, A.; Lapierre, C.; Jouanin, L. The Simultaneous Repression of CCR and CAD, Two Enzymes of the Lignin Biosynthetic Pathway, Results in Sterility and Dwarfism in Arabidopsis thaliana. Mol. Plant. 2011, 4, 70–82. [Google Scholar] [CrossRef]
- Sharma, P.; Dubey, R.S. Involvement of oxidative stress and role of antioxidative defense system in growing rice seedlings exposed to toxic concentrations of aluminum. Plant Cell Rep. 2007, 26, 2027–2038. [Google Scholar] [CrossRef]
- Deng, Y.M.; Li, C.C.; Shao, Q.S.; Ye, X.Q.; She, J.M. Differential responses of double petal and multi petal jasmine to shading: I. Photosynthetic characteristics and chloroplast ultrastructure. Plant Physiol. Biochem. 2012, 55, 93–102. [Google Scholar] [CrossRef]
- Ghorbel, M.; Brini, F.; Brestic, M.; Landi, M. Interplay between low light and hormone-mediated signaling pathways in shade avoidance regulation in plants. Plant Stress 2023, 9, 100178. [Google Scholar] [CrossRef]
- Gao, J.; Liu, Z.; Zhao, B.; Liu, P.; Zhang, J.W. Physiological and comparative proteomic analysis provides new insights into the effects of shade stress in maize (Zea mays L.). BMC Plant Biol. 2020, 20, 13. [Google Scholar] [CrossRef]
- Keuskamp, D.H.; Pollmann, S.; Voesenek, L.; Peeters, A.J.M.; Pierik, R. Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc. Natl. Acad. Sci. USA 2010, 107, 22740–22744. [Google Scholar] [CrossRef]
- Tao, Y.; Ferrer, J.L.; Ljung, K.; Pojer, F.; Hong, F.X.; Long, J.A.; Li, L.; Moreno, J.E.; Bowman, M.E.; Ivans, L.J.; et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 2008, 133, 164–176. [Google Scholar] [CrossRef]
- Kurepin, L.V.; Emery, R.J.N.; Pharis, R.P.; Reid, D.M. The interaction of light quality and irradiance with gibberellins, cytokinins and auxin in regulating growth of Helianthus annuus hypocotyls. Plant Cell Environ. 2007, 30, 147–155. [Google Scholar] [CrossRef]
- Lorrain, S.; Allen, T.; Duek, P.D.; Whitelam, G.C.; Fankhauser, C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008, 53, 312–323. [Google Scholar] [CrossRef]
- Kohnen, M.V.; Schmid-Siegert, E.; Trevisan, M.; Petrolati, L.A.; Sénéchal, F.; Müller-Moulé, P.; Maloof, J.; Xenarios, I.; Fankhauser, C. Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 2016, 28, 2889–2904. [Google Scholar] [CrossRef]
- Ortiz-Alcaide, M.; Llamas, E.; Gomez-Cadenas, A.; Nagatani, A.; Martínez-García, J.F.; Rodríguez-Concepción, M. Chloroplasts modulate elongation responses to canopy shade by retrograde pathways involving HY5 and abscisic acid. Plant Cell 2019, 31, 384–398. [Google Scholar] [CrossRef]
- Yi, R.; Yan, J.B.; Xie, D.X. Light promotes jasmonate biosynthesis to regulate photomorphogenesis in Arabidopsis. Sci. China Life Sci. 2020, 63, 943–952. [Google Scholar] [CrossRef]
- Liu, Y.X.; Jing, H.Q.; Wu, J.W. Non-structural carbohydrate (NSC) content and C:N:P stoichiometry of Pinus yunnanensis seedling needles in response to shade treatment. Ind. Crop. Prod. 2024, 210, 8. [Google Scholar] [CrossRef]
- Zhao, Z.J.; Wang, L.A.; Liu, Y.X.; Sun, J.L.; Xiao, J.D.; Dong, Q.; Li, L.F.; Zhang, W.J.; Wang, C.; Wu, J.W. Effects of drought on non-structural carbohydrates and C, N, and P stoichiometric characteristics of Pinus yunnanensis seedlings. J. For. Res. 2024, 35, 13. [Google Scholar] [CrossRef]
- Xiao, F.; Zhao, Y.; Wang, X.R.; Jian, X.Y.; Yang, Y. Physiological responses to drought stress of three pine species and comparative transcriptome analysis of Pinus yunnanensis var. pygmaea. BMC Genom. 2024, 25, 16. [Google Scholar] [CrossRef]
- Xu, Y.L.; Cai, N.A.H.; Woeste, K.; Kang, X.Y.; He, C.Z.; Li, G.Q.; Chen, S.; Duan, A.A. Genetic Diversity and Population Structure of Pinus yunnanensis by Simple Sequence Repeat Markers. For. Sci. 2016, 62, 38–47. [Google Scholar] [CrossRef]
- Xu, Y.L.; Zhang, Y.M.; Li, Y.F.; Li, G.Q.; Liu, D.Y.; Zhao, M.C.; Cai, N.H. Growth promotion of yunnan pine early seedlings in response to foliar application of iaa and iba. Int. J. Mol. Sci. 2012, 13, 6507–6520. [Google Scholar] [CrossRef]
- Dong, L.; Wu, Y.; Zhang, J.; Deng, X.; Wang, T. Transcriptome Analysis Revealed Hormone Pathways and bZIP Genes Responsive to Decapitation in Sunflower. Genes 2022, 13, 1737. [Google Scholar] [CrossRef]
- Uenaka, H.; Wada, M.; Kadota, A. Four distinct photoreceptors contribute to light-induced side branch formation in the moss Physcomitrella patens. Planta 2005, 222, 623–631. [Google Scholar] [CrossRef]
- de Wit, M.; Galvão, V.C.; Fankhauser, C. Light-Mediated Hormonal Regulation of Plant Growth and Development. Annu. Rev. Plant Biol. 2016, 67, 513–537. [Google Scholar] [CrossRef]
- Paciullo, D.S.C.; Fernandes, P.B.; Gomide, C.A.D.; de Castro, C.R.T.; Sobrinho, F.D.; de Carvalho, C.A.B. The growth dynamics in Brachiaria species according to nitrogen dose and shade. Rev. Bras. Zootecn. 2011, 40, 270–276. [Google Scholar] [CrossRef]
- Gao, Z.; Khalid, M.; Jan, F.; Jiang, X.; Yu, X. Effects of light-regulation and intensity on the growth, physiological and biochemical properties of Aralia elata (miq.) seedlings. S. Afr. J. Bot. 2019, 121, 456–462. [Google Scholar] [CrossRef]
- Poorter, H.; Niklas, K.J.; Reich, P.B.; Oleksyn, J.; Poot, P.; Mommer, L. Biomass allocation to leaves, stems and roots: Meta-analyses of interspecific variation and environmental control. New Phytol. 2012, 193, 30–50. [Google Scholar] [CrossRef]
- Puglielli, G.; Laanisto, L.; Poorter, H.; Niinemets, Ü. Global patterns of biomass allocation in woody species with different tolerances of shade and drought: Evidence for multiple strategies. New Phytol. 2021, 229, 308–322. [Google Scholar] [CrossRef]
- Liu, B.; Liu, Q.Q.; Daryanto, S.; Guo, S.; Huang, Z.J.; Wang, Z.N.; Wang, L.X.; Ma, X.Q. Responses of Chinese fir and Schima superba seedlings to light gradients: Implications for the restoration of mixed broadleaf-conifer forests from Chinese fir monocultures. For. Ecol. Manag. 2018, 419, 51–57. [Google Scholar] [CrossRef]
- Chmura, D.J.; Modrzynski, J.; Chmielarz, P.; Tjoelker, M.G. Plasticity in seedling morphology, biomass allocation and physiology among ten temperate tree species in response to shade is related to shade tolerance and not leaf habit. Plant Biol. 2017, 19, 172–182. [Google Scholar] [CrossRef]
- Rosado, D.; Ackermann, A.; Spassibojko, O.; Rossi, M.; Pedmale, U.V. WRKY transcription factors and ethylene signaling modify root growth during the shade-avoidance response. Plant Physiol. 2022, 188, 1294–1311. [Google Scholar] [CrossRef]
- Youn, W.B.; Hernandez, J.O.; Park, B.B. Effects of Shade and Planting Methods on the Growth of Heracleum moellendorffii and Adenophora divaricata in Different Soil Moisture and Nutrient Conditions. Plants 2021, 10, 2203. [Google Scholar] [CrossRef]
- Chory, J. Light signal transduction: An infinite spectrum of possibilities. Plant J. 2010, 61, 982–991. [Google Scholar] [CrossRef]
- Moustakas, M.; Sperdouli, I.; Adamakis, I.D.S.; Sas, B.; Isgören, S.; Moustaka, J.; Morales, F. Mechanistic approach on melatonin-induced hormesis of photosystem II function in the medicinal plant Mentha spicata. Plants 2023, 12, 4025. [Google Scholar] [CrossRef]
- Lazár, D. Parameters of photosynthetic energy partitioning. J. Plant Physiol. 2015, 175, 131–147. [Google Scholar] [CrossRef]
- Fan, Y.F.; Chen, J.X.; Wang, Z.L.; Tan, T.T.; Li, S.L.; Li, J.F.; Wang, B.B.; Zhang, J.W.; Cheng, Y.J.; Wu, X.L.; et al. Soybean (Glycine max L. Merr.) seedlings response to shading: Leaf structure, photosynthesis and proteomic analysis. BMC Plant Biol. 2019, 19, 12. [Google Scholar] [CrossRef]
- Tanaka, R.; Tanaka, A. Chlorophyll cycle regulates the construction and destruction of the light-harvesting complexes. Biochim. Biophys. Acta Bioenerg. 2011, 1807, 968–976. [Google Scholar] [CrossRef]
- Othman, R.; Zaifuddin, F.A.M.; Hassan, N.M. Carotenoid Biosynthesis Regulatory Mechanisms in Plants. J. Oleo Sci. 2014, 63, 753–760. [Google Scholar] [CrossRef]
- Na, Z.; Xu, Q.G.; Xu, W. Ecological and physiological characteristics of Pinellia ternata under different shade conditions. Pak. J. Agric. Sci. 2022, 59, 925–933. [Google Scholar] [CrossRef]
- Dong, C.J.; Wang, X.L.; Shang, Q.M. Salicylic acid regulates sugar metabolism that confers tolerance to salinity stress in cucumber seedlings. Sci. Hortic. 2011, 129, 629–636. [Google Scholar] [CrossRef]
- Asghar, M.A.; Jiang, H.K.; Shui, Z.W.; Cao, X.Y.; Huang, X.Y.; Imran, S.; Ahmad, B.; Zhang, H.; Yang, Y.N.; Shang, J.; et al. Interactive effect of shade and PEG-induced osmotic stress on physiological responses of soybean seedlings. J. Integr. Agric. 2021, 20, 2382–2394. [Google Scholar] [CrossRef]
- Liu, B.; Li, Y.S.; Liu, X.B.; Wang, C.; Jin, J.; Herbert, S.J. Lower total soluble sugars in vegetative parts of soybean plants are responsible for reduced pod number under shading conditions. Aust. J. Crop Sci. 2011, 5, 1852–1857. [Google Scholar]
- Silva, E.N.; Silveira, J.A.G.; Aragao, R.M.; Vieira, C.F.; Carvalho, F.E.L. Photosynthesis impairment and oxidative stress in Jatropha curcas exposed to drought are partially dependent on decreased catalase activity. Acta Physiol. Plant 2019, 41, 12. [Google Scholar] [CrossRef]
- Wang, Y.B.; Huang, R.D.; Zhou, Y.F. Effects of shading stress during the reproductive stages on photosynthetic physiology and yield characteristics of peanut (Arachis hypogaea Linn.). J. Integr. Agric. 2021, 20, 1250–1265. [Google Scholar] [CrossRef]
- Zhu, H.F.; Li, X.F.; Zhai, W.; Liu, Y.; Gao, Q.Q.; Liu, J.P.; Ren, L.; Chen, H.Y.; Zhu, Y.Y. Effects of low light on photosynthetic properties, antioxidant enzyme activity, and anthocyanin accumulation in purple pak-choi (Brassica campestris ssp Chinensis Makino). PLoS ONE 2017, 12, 17. [Google Scholar] [CrossRef]
- Zhang, J.F.; Ge, J.R.; Dayananda, B.; Li, J.Q. Effect of light intensities on the photosynthesis, growth and physiological performances of two maple species. Front. Plant Sci. 2022, 13, 999026. [Google Scholar] [CrossRef]
- Dong, L.L.; Xiong, L.B.; Sun, X.Y.; Shah, S.; Guo, Z.X.; Zhao, X.Y.; Liu, L.Z.; Cheng, L.T.; Tian, Z.; Xie, F.C.; et al. Morphophysiological Responses of Two Cool-Season Turfgrasses with Different Shade Tolerances. Agronomy 2022, 12, 959. [Google Scholar] [CrossRef]
- El-Beltagi, H.S.; Mohamed, H.I.; Sofy, M.R. Role of Ascorbic acid, Glutathione and Proline Applied as Singly or in Sequence Combination in Improving Chickpea Plant through Physiological Change and Antioxidant Defense under Different Levels of Irrigation Intervals. Molecules 2020, 25, 1702. [Google Scholar] [CrossRef]
- Yu, L.; Zhou, J.K.; Lin, J.L.; Chen, M.W.; Liu, F.; Zheng, X.L.; Zhou, L.P.; Wang, R.Z.; Xiao, L.T.; Liu, Y.H. Perception of strigolactones and the coordinated phytohormonal regulation on rice (Oryza sativa) tillering is affected by endogenous ascorbic acid. Funct. Plant Biol. 2024, 51, FP23148. [Google Scholar] [CrossRef]
- Selwal, N.; Wani, A.K.; Akhtar, N.; Kaur, M.; Jassal, P.S. Molecular insights of Strigolactone biosynthesis, signalling pathways, regulatory roles, and hormonal crosstalks in plant systems. S. Afr. J. Bot. 2023, 160, 9–22. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, T.; Liu, J.; Li, H.Y.; Liu, B. Shade-Induced Leaf Senescence in Plants. Plants 2023, 12, 1550. [Google Scholar] [CrossRef]
- Ni, J.; Gao, C.C.; Chen, M.S.; Pan, B.Z.; Ye, K.Q.; Xu, Z.F. Gibberellin Promotes Shoot Branching in the Perennial Woody Plant Jatropha curcas. Plant Cell Physiol. 2015, 56, 1655–1666. [Google Scholar] [CrossRef]
- Sirhindi, G.; Mir, M.A.; Abd-Allah, E.F.; Ahmad, P.; Gucel, S. Jasmonic Acid Modulates the Physio-Biochemical Attributes, Antioxidant Enzyme Activity, and Gene Expression in Glycine max under Nickel Toxicity. Front. Plant Sci. 2016, 7, 190643. [Google Scholar] [CrossRef]
- Dun, E.A.; de Saint Germain, A.; Rameau, C.; Beveridge, C.A. Antagonistic Action of Strigolactone and Cytokinin in Bud Outgrowth Control. Plant Physiol. 2012, 158, 487–498. [Google Scholar] [CrossRef]
- Barbier, F.F.; Dun, E.A.; Beveridge, C.A. Apical dominance. Curr. Biol. 2017, 27, R864–R865. [Google Scholar] [CrossRef]
- Carabelli, M.; Possenti, M.; Sessa, G.; Ciolfi, A.; Sassi, M.; Morelli, G.; Ruberti, I. Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev. 2007, 21, 1863–1868. [Google Scholar] [CrossRef]
- Holalu, S.V.; Reddy, S.K.; Blackman, B.K.; Finlayson, S.A. Phytochrome interacting factors 4 and 5 regulate axillary branching via bud abscisic acid and stem auxin signalling. Plant Cell Environ. 2020, 43, 2224–2238. [Google Scholar] [CrossRef]
- Lu, Z.; Wang, Q.; Yang, Z.; Chen, L.; Cai, N.; Xu, Y. Endogenous Phytohormone and Transcriptome Analysis Provided Insights into Seedling Height Growth of Pinus yunnanensis. Forests 2024, 15, 489. [Google Scholar] [CrossRef]
- Liu, X.L.; Wu, Y.F.; Wang, L.Y.; Zhang, Y.T.; Tang, X.L.; Yan, C. Comparative transcriptome analyses revealed photosynthesis- and sucrose metabolism-related responses of endangered evergreen tree species phoebe bournei (hemsl.) Yang-lauraceae to shade. Pak. J. Bot. 2023, 55, 2283–2293. [Google Scholar] [CrossRef]
- Schreiber, U. Redox changes of ferredoxin, P700, and plastocyanin measured simultaneously in intact leaves. Photosynth. Res. 2017, 134, 343–360. [Google Scholar] [CrossRef]
- Silveira, J.A.G.; Carvalho, F.E.L. Proteomics, photosynthesis and salt resistance in crops: An integrative view. J. Proteom. 2016, 143, 24–35. [Google Scholar] [CrossRef]
- An, J.; Wei, X.L.; Huo, H.H. Transcriptome analysis reveals the accelerated expression of genes related to photosynthesis and chlorophyll biosynthesis contribution to shade-tolerant in Phoebe bournei. BMC Plant Biol. 2022, 22, 14. [Google Scholar] [CrossRef]
- Mi, W.B.; Luo, F.; Liu, W.H.; Liu, K.Q. A Transcriptome Reveals the Mechanism of Nitrogen Regulation in Tillering. Genes 2024, 15, 223. [Google Scholar] [CrossRef]
- Tian, H.Y.; Tang, B.X.; Fan, W.W.; Pan, Z.Y.; Peng, J.T.; Wang, Y.X.; Liu, F.; Liu, G.Q. The role of strigolactone analog (GR24) in endogenous hormone metabolism and hormone-related gene expression in tobacco axillary buds. Plant Cell Rep. 2024, 43, 24. [Google Scholar] [CrossRef]
- Wei, X.; Yang, J.; Lei, D.; Feng, H.; Yang, Z.; Wen, G.; He, Z.; Zeng, W.; Zou, J. The SlTCP26 promoting lateral branches development in tomato. Plant Cell Rep. 2021, 40, 1115–1126. [Google Scholar] [CrossRef]
- Kant, S.; Bi, Y.M.; Zhu, T.; Rothstein, S.J. SAUR39, a Small Auxin-Up RNA Gene, Acts as a Negative Regulator of Auxin Synthesis and Transport in Rice. Plant Physiol. 2009, 151, 691–701. [Google Scholar] [CrossRef]
- Nishiyama, R.; Watanabe, Y.; Leyva-Gonzalez, M.A.; Ha, C.V.; Fujita, Y.; Tanaka, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Herrera-Estrella, L.; et al. Arabidopsis AHP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought stress response. Proc. Natl. Acad. Sci. USA 2013, 110, 4840–4845. [Google Scholar] [CrossRef]
- Ha, C.V.; Mostofa, M.G.; Nguyen, K.H.; Tran, C.D.; Watanabe, Y.; Li, W.; Osakabe, Y.; Sato, M.; Toyooka, K.; Tanaka, M.; et al. The histidine phosphotransfer AHP4 plays a negative role in Arabidopsis plant response to drought. Plant J. 2022, 111, 1732–1752. [Google Scholar] [CrossRef]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 2013, 147, 15–27. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Pierik, R.; Van Wees, S.C. Different shades of JAZ during plant growth and defense. New Phytol. 2014, 204, 261–264. [Google Scholar] [CrossRef]
- Arshad, M.N.; Naegele, J.R. Correction notice: Measurements of Proline and Malondialdehyde Contents and Antioxidant Enzyme Activities in Leaves of Drought Stressed Cotton. Bio Protoc. 2023, 5, e4752. [Google Scholar] [CrossRef]
- Su, S.Q.; Zhou, Y.M.; Qin, J.G.; Yao, W.Z.; Ma, Z.H. Optimization of the Method for Chlorophyll Extraction in Aquatic Plants. J. Freshw. Ecol. 2010, 25, 531–538. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, C.X.; Yan, X.J.; Zhang, J.R.; Xu, J.L. Simultaneous analysis of ten phytohormones in Sargassum horneri by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry. J. Sep. Sci. 2016, 39, 1804–1813. [Google Scholar] [CrossRef]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




| Treatments | Shade Cloth Specification | Shade Level (%) |
|---|---|---|
| CK | None | 0 |
| L1 | Black polyethylene net curtains (1 layer 2 stitches) | 25 |
| L2 | Black polyethylene net curtains (1 layer 3 stitches) | 50 |
| L3 | Black polyethylene net curtains (1 layer 4 stitches) | 75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, C.; Gu, X.; Li, J.; Su, X.; Chen, S.; Tang, J.; Chen, L.; Cai, N.; Xu, Y. Physiological Characteristics and Transcriptomic Responses of Pinus yunnanensis Lateral Branching to Different Shading Environments. Plants 2024, 13, 1588. https://doi.org/10.3390/plants13121588
Zhou C, Gu X, Li J, Su X, Chen S, Tang J, Chen L, Cai N, Xu Y. Physiological Characteristics and Transcriptomic Responses of Pinus yunnanensis Lateral Branching to Different Shading Environments. Plants. 2024; 13(12):1588. https://doi.org/10.3390/plants13121588
Chicago/Turabian StyleZhou, Chiyu, Xuesha Gu, Jiangfei Li, Xin Su, Shi Chen, Junrong Tang, Lin Chen, Nianhui Cai, and Yulan Xu. 2024. "Physiological Characteristics and Transcriptomic Responses of Pinus yunnanensis Lateral Branching to Different Shading Environments" Plants 13, no. 12: 1588. https://doi.org/10.3390/plants13121588
APA StyleZhou, C., Gu, X., Li, J., Su, X., Chen, S., Tang, J., Chen, L., Cai, N., & Xu, Y. (2024). Physiological Characteristics and Transcriptomic Responses of Pinus yunnanensis Lateral Branching to Different Shading Environments. Plants, 13(12), 1588. https://doi.org/10.3390/plants13121588







